Abstract
Background
Tsetse flies (Diptera: Glossinidae) and tabanids (Diptera: Tabanidae) are haematophagous insects of medical and veterinary importance due to their respective role in the biological and mechanical transmission of trypanosomes. Few studies on the distribution and relative abundance of both families have been conducted in Mozambique since the country’s independence. Despite Nicoadala, Mozambique, being a multiple trypanocidal drug resistance hotspot no information regarding the distribution, seasonality or infection rates of fly-vectors are available. This is, however, crucial to understanding the epidemiology of trypanosomosis and to refine vector management.
Methods
For 365 days, 55 traps (20 NGU traps, 20 horizontal traps and 15 Epsilon traps) were deployed in three grazing areas of Nicoadala District: Namitangurine (25 traps); Zalala (15 traps); and Botao (15 traps). Flies were collected weekly and preserved in 70% ethanol. Identification using morphological keys was followed by molecular confirmation using cytochrome c oxidase subunit 1 gene. Trap efficiency, species distribution and seasonal abundance were also assessed. To determine trypanosome infection rates, DNA was extracted from the captured flies, and submitted to 18S PCR-RFLP screening for the detection of Trypanosoma.
Results
In total, 4379 tabanids (of 10 species) and 24 tsetse flies (of 3 species), were caught. NGU traps were more effective in capturing both the Tabanidae and Glossinidae. Higher abundance and species diversity were observed in Namitangurine followed by Zalala and Botao. Tabanid abundance was approximately double during the rainy season compared to the dry season. Trypanosoma congolense and T. theileri were detected in the flies with overall infection rates of 75% for tsetse flies and 13% for tabanids. Atylotus agrestis had the highest infection rate of the tabanid species. The only pathogenic trypanosome detected was T. congolense.
Conclusions
Despite the low numbers of tsetse flies captured, it can be assumed that they are still the cyclical vectors of trypanosomosis in the area. However, the high numbers of tabanids captured, associated to their demonstrated capacity of transmitting trypanosomes mechanically, suggest an important role in the epidemiology of trypanosomosis in the Nicoadala district. These results on the composition of tsetse and tabanid populations as well as the observed infection rates, should be considered when defining strategies to control the disease.

Similar content being viewed by others
Background
Tsetse-transmitted animal African trypanosomosis is one the most important diseases in sub-Saharan Africa [1] where it causes the death of thousands of heads of cattle per year and reduces the availability of meat, milk and other cattle-derived products [2,3,4,5]. Additionally, fertile areas are often unsuitable for cattle keeping due to the presence of tsetse flies, the biological vector of the pathogen [6, 7]. The presence of tabanid flies, mechanical vectors of many human and veterinary disease-causing agents, including Trypanosoma species, the cause of animal African trypanosomosis, intensifies the problem [8, 9].
Tsetse flies (Diptera: Glossinidae), are thought to comprise 34 species and subspecies from a single genus, Glossina [10,11,12,13]. Glossina spp. males and females feed exclusively on blood, which is required for all metabolic processes, and are subsequently ideal pathogen vectors. In Mozambique, G. morsitans, G. pallidipes, G. brevipalpis and G. austeni have all been recorded [14,15,16]. Species of the Tabanidae, in contrast, need nectar for the metabolic processes, with only females of most species requiring blood for the production of eggs [8, 9]. Approximately 4400 species have been described, currently divided into the subfamilies Pangoniinae, Tabaninae and Chrysopsinae; however, the higher classification remains unsettled [9, 17, 18]. Tabanids have been incriminated as vectors of livestock pathogens, including trypanosomes, and are considered one of the most efficient mechanical vectors due to their high mobility, interrupted feeding and large mouthparts [19,20,21].
Scarce information regarding the distribution of tsetse flies and tabanids in Mozambique, particularly in the Nicoadala District, an area identified as trypanocidal resistance hotspot, through a block treatment study [22], is currently available. Furthermore, their role in the transmission of trypanosomes requires further elucidation, especially in the Afrotropics [23]. Moreover, little has been done to update the knowledge on the distribution and composition of hematophagous fly populations in Mozambique since the studies of Dias [24] and Oldroyd [25,26,27]. The lack of the aforementioned information, together with constant reports of high trypanosomosis prevalence in Nicoadala, urge for an accurate monitoring study targeting vectors of trypanosomosis and is the aim of this study. This study would allow an updated view on the respective vector species composition, with the ultimate goal of gaining a better understanding of the role said vectors play in maintaining and spreading trypanosomes in the trypanocidal drug resistance foci of Nicoadala District. Such results are of paramount importance for implementing regional vector control and/or management as well as subsequent policy making.
The use of trapping to determine the distribution of hematophagous insects have been extensively described in the scientific literature [28,29,30,31,32,33]. Currently, a plethora of different traps are available for collecting hematophagous brachyceran flies. Here, the horizontal or H trap, developed in South Africa for G. brevipalpis and G. austeni [34], the Epsilon trap, developed in Zimbabwe as an alternative trap for savanna species such as G. pallidipes and G. morsitans [35] and the NGU trap, developed in Kenya for catching savannah flies such as G. pallidipes and also effective for tabanids [36], were selected given the species present in Mozambique.
The 18S PCR-RFLP method is effective for detecting Trypanosoma, or mixed parasite infections, from fresh or ethanol-preserved flies. The method has been shown to be accurate for identification of parasites up to the subspecies level [37, 38].
This study aims to assess the species composition and abundance of the Tabanidae and Glossinidae in three foci within Nicoadala District, Mozambique. Additionally, the study aims to report on Trypanosoma infection rates of said vectors, trapping efficacy and cytochrome c oxidase subunit 1 (cox1) gene as a species delimitation tool.
Methods
Study area
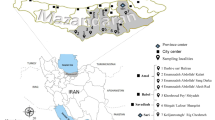
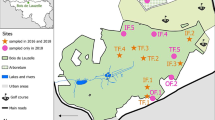
Three grazing areas, Botao, Namitangurine and Zalala, all located in the Nicoadala District (17.608°E, 36.820°N), were used for the survey. The region has a rainy tropical savannah climate according to the Köppen-Geiger system [39, 40], with two seasons: rainy and dry. The average temperature in Nicoadala is 25.6 °C with about 1428 mm annual rainfall. In Zalala, the vegetation is mostly composed of coconut trees, grasses and small shrubs covering approximately 420 ha. Botao (c.1300 ha) is an open forest with large grassland areas. Namitangurine (c.1600 ha) is a slightly closed canopy forest with a high diversity of trees and shrubs. The dominant tree species in the district are Pterocarpus angolensis, Swartzia madagascariensis, Afzelia quanzensis, Millettia stuhlmannii, Khaya nyasica, Pericopsis angolensis, Combretum imberbe, Brachystegia spiciformis and Pteleopsis myrtifolia. Both Botao and Namitangurine are areas with pasture and water availability with some wild animals present [41]. Recently, Nicoadala was identified as a multiple trypanocidal drug resistance hotspot, in a block treatment study where trypanosomes in cattle were tested to both isometamidium chloride and diminazene aceturate [22]. As there are no recent studies elucidating the epidemiology of trypanosomosis in the region, it is of crucial importance to develop a study to identify possible vectors of the disease. In addition, this study will help in the understanding of the role of mechanical transmission of the disease in the presence of resistant isolates.
Sampling
For 12 months, a total of 5 H traps, 5 Epsilon traps and 5 NGU traps were deployed in Botao, an equal number of traps were deployed in Zalala, and 10 H traps, 5 Epsilon traps and 10 NGU traps were deployed in Namitangurine. Trap numbers were defined by the size of the sampling areas and all traps were placed at least 200 m apart to prevent interference [33, 42]. Trapping sites were georeferenced using a Garmin-GPSMAP®76 (Garmin Ltd., Kansas, USA) (Fig. 1). The traps were left at the deployment sites for the duration of the study [43, 44]. Each trap was baited with 4 polyethylene sachets (150 µm thick, surface area of 30 cm2) containing 4 ml of 1-octen-3-ol and one 15 ml plastic bottle with a 5 mm hole on top, containing acetone. Flies were collected weekly and those captured were placed in 1.5 ml tubes containing 1 ml of 70% ethanol until further analysis [16, 45]. All the bottles containing trapped flies were labeled with location, trap number, date and time and then replaced with empty ones.
Identification of hematophagous insects
All tsetse flies captured were identified using the tsetse control personnel training manual key for Glossina species identification, through the analysis of the fly’s morphological traits [11, 46]. Tabanids were identified to the genus and species level using the morphological keys of Oldroyd [25,26,27] supplemented by Taioe et al. [23] and Dias [24]. To conduct the molecular species delimitation, DNA was extracted using the Chelex® protocol modified from Walsh et al. [47] and Ravel et al. [48]. Polymerase chain reaction (PCR) targeting the cytochrome c oxidase subunit 1 (cox1) gene was conducted using an Applied Biosystems® thermal cycler (Thermo Fisher Scientific, Göteborg, Sweden). Reactions were performed in a total volume of 20 µl containing, 10 µl of 2× Phusion Flash Master Mix, 0.5 µM of each primer and 6.5 µl of double distilled water (ddH2O). PCR conditions were as follows: 98 °C for 10 s, 30 cycles of 98 °C for 1 s, 50 °C for 5 s and 72 °C for 15 s, followed by a final elongation step of 72 °C for 1 min. The primers used were LCO1490 (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′) and HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) and the expected size of the fragment was approximately 653 bp [23, 49]. Genomic DNA from Glossina brevipalpis and G. austeni (from the Biotechnology Center, Eduardo Mondlane University reference collection) were used as a positive control and ddH2O as a negative control. For PCR product visualization, the samples were analysed on 2% agarose gels, where 2 µl of loading dye was mixed with 5 µl of PCR product and loaded onto the gel. A 1 Kb DNA ladder was also loaded (4 µl) for fragment size determination; gel electrophoresis was performed for 45 min at 100 volts. The gels were stained with GelRed (Biotium, Inc., Fremont, CA, USA) using 4 µl per 100 ml of agarose solution directly added before polymerization. Four replicates of the PCR amplicons from the cox1 PCR amplification were produced and pooled. After confirmation on agarose gel, 50 µl of the product was sent for sequencing at Inqaba Biotec™ (Pretoria, South Africa).
Phylogenetic analysis
With the aim of verifying monophyly of the species using molecular methods, and subsequently, infer species identification, all retrieved sequences were viewed and assembled using BioEdit version 7.0.9 [50]. Assembled sequences from species of both the Tabanidae and Glossinidae were separately aligned with various sequences from the GenBank database using the online version of MAFFT with default parameters [51]. The aligned matrices were viewed, edited and truncated in MEGA 7 [52] and used for all analysis. Data-display networks (DDN) were constructed in SplitsTree version 4 [53], from uncorrected p-distances using both parsimony-informative and -uninformative characters. Bootstrap (bs) support for the DDN was calculated from 1000 replicates. jModelTest version 2.1.10, via the Cipres Science gateway [54], was used for model estimation. A maximum likelihood (ML) analysis was performed in RAxML version 8 using the estimated models (GTR + G + I). The auto MRE function was invoked for the calculation of bootstrap support [55]. A Bayesian approach for phylogenetic inference (BI) was conducted in MrBayes version 3 [56]. Four simultaneous cold Monte-Carlo Markov Chains searched for 10 million generations, with each 1000th tree sampled. In both cases, TIM1 + G + I estimated using the jModelTest was employed as the substitution model. Before tree construction, the first 15% of the trees were discarded as ‘burn-in’ for the subsequent calculation of the posterior probabilities (pp). Effective sample sizes were calculated and viewed using Tracer version 1.6 [57] where values of > 200 were considered sufficient.
Glossinidae and Tabanidae diversity and seasonal abundance and trap efficiency
To access species richness and evenness from the grazing areas, alpha (α) diversity was calculated using the Shannon index [58, 59], respectively. Data was processed with Vegan 2.5-2 in R [60]. A comparison of trap efficiency was performed considering both the total number of individuals and the number of species captured by each type of trap. Seasonal abundance was assessed and compared.
Detection of trypanosome infection rates
A total of 480 (160 specimens from each site) tabanids and all tsetse captured (24 in total) were screened. To estimate the sample size and get a statistically representative number of 160 individuals to be sampled per area, the formula from Cannon & Roe [61] was used. The number was determined when the expected prevalence was set at 10% with a 5% confidence interval and a 5% desired absolute precision. These were selected equally into the four most captured species namely Tabanus par, T. taeniola, Atylotus agrestis and Ancala africana. The 24 tsetse flies comprised 17 Glossina specimens from Namitangurine (11 G. brevipalpis, 4 G. morsitans and 2 G. pallidipes), 6 G. brevipalpis from Botao and 1 G. brevipalpis from Zalala. DNA was extracted from individual flies using the ammonium acetate precipitation protocol modified from Bruford et al. [62]. For the molecular detection of trypanosomes in the flies, semi-nested rRNA PCRs were run targeting a fragment of the 18S ribosomal RNA gene. PCR conditions and gel visualization followed the description by Mulandane et al. [22] and Geysen et al. [38].
Statistical analysis
Mean infection rate per species, per species/area and per total tabanids/area were calculated. To determine whether observed differences in tsetse and tabanids infection rates, infection rates per area and per species, tabanids distribution, trap efficiency and seasonal abundance were statistically significant, ANOVA with Tukey’s HSD post-hoc test was used; Studentʼs t-test was used to determine if significant differences were observed between the seasons. All analyses were carried out with Statistica version 13.3 [63].
Results
Over a period of 12 months using 55 traps, a total of 4379 tabanids and 24 tsetse flies were captured in three areas of Nicoadala District of which 35.9% in Zalala, 33.4% in Namitangurine and 30.7% in Botao.
Based upon morphological characterization, the captured tabanids belonged to four genera: Tabanus, Ancala, Atylotus and Haematopota. The genus Tabanus accounted for 73.2% of the total capture (n = 3207), followed by the genera Atylotus (23.2%; n = 1017), Ancala (2.4%; n = 149) and Hematopota (0.14%; n = 6). Tabanus par and A. agrestis were the most frequently caught species (66.8% and 23.2% of the captured flies, respectively). Tabanus par, T. taeniola, A. agrestis and Ancala africana were captured in all the three habitats (Table 1). From the 4379 tabanids captured, 16 Tabanus specimens were damaged and/or discolored by alcohol that made the morphological identification to the species level unreliable.
Three of the four Glossina species present in Mozambique were captured. These species were G. brevipalpis (n = 18), G. pallidipes (n = 2) and G. morsitans morsitans (n = 4). In Botao (n = 6) and Zalala (n = 1), only G. brevipalpis was captured. Namitangurine contributed with more than 50% of the total captured Glossina (11 G. brevipalpis, 4 G. morsitans morsitans and 2 G. pallidipes), and thus, the only area where all three Glossina species were captured.
The dimensions of the final alignment matrices were 113 taxa and 658 characters and 18 taxa and 659 characters for the Tabanidae and Glossinidae, respectively. Identical substitution models, GTR + G + I (AIC criterion) and TIM1 + G + I (BIC criterion) were recovered for both datasets. Congruent topologies across all methods were recovered (Figs. 2, 3). Three specimens could be confidently identified as Ancala africana (nos. 7, 32, 37: DDN bs = 100; ML bs = 98; BI pp = 1) (Fig. 2, Additional file 1: Figure S1). Two Atylotus clades were recovered across all methods. All ten specimens sequenced for this study fell within a collapsed node representing A. agrestis, A. diurnus and A. nigromaculatus (DDN bs = 100; ML bs = 99; BI pp = 1) (Fig. 2, Additional file 1: Figure S1). Three specimens (nos. 21–23) could confidently be identified as T. par (DDN bs = 100; ML bs = 100; BI pp = 1) (Fig. 2, Additional file 1: Figure S1). An unverified sequence (GenBank: KY555744) submitted by Mugasa et al. [64], as T. taeniatus consistently fell within the T. par group. Seven specimens (nos. 8, 24, 25, 27–29 and 31) were confidently recovered as T. taeniola (DDN bs = 94; ML bs = 98; BI pp = 0.99), and four specimens were identified as T. fraternus (nos. 18, 53–55) (DDN bs = 100; ML bs = 100; BI pp = 1) (Fig. 2, Additional file 1: Figure S1). Together, T. taeniola and T. fraternus formed a well-supported monophyletic clade (DDN bs = 98; ML bs = 97; BI pp = 1). A single specimen (57), formed a well-supported clade with Haematopota fenestralis (DDN bs = 100; ML bs = 95; BI pp = 1) (Fig. 2, Additional file 1: Figure S1). The specimen morphologically identified as T. ustus (no. 14) could not be verified as belonging to any species-group due to a lack of reference sequences on GenBank.
A consensus phylogram constructed in a Bayesian inference analysis using the Tabanidae cox1 data. Nodal support presented on the branches are bootstraps (bs) calculated a RAxML analysis (autoMRE function) and posterior probabilities (pp) calculated from a MrBayes analysis. Only bs values > 75 and pp values > 0.95 are shown (bs/pp)
A consensus phylogram recovered from a MrBayes analysis using the Glossinidae cox1 data. Nodal support presented on the branches are bootstraps (bs) calculated from a RAxML analysis (autoMRE function) and posterior probabilities (pp) calculated from a MrBayes analysis. Only bs values > 75 and pp values > 0.95 are shown (bs/pp). Red arrows indicate the specimen sequences from this study
Congruent topologies, analyzing the Glossinidae cox1 data, from all three analyses were recovered (Fig. 3, Additional file 2: Figure S2). In all analyses, the Fusca and Palpalis groups were well-supported monophyletic clades (DDN bs = 99; ML bs = 85; BI pp = 0.98 and DDN bs = 100; ML bs = 100; BI pp = 1, respectively). Two well-supported monophyletic clades representing the Morsitans group were consistently found across all analyses (Clade 1: DDN bs = 94; ML bs = 87; BI pp = 1; and Clade 2: DDN bs = 100; ML bs = 86; BI pp = 0.99) (Fig. 3, Additional file 2: Figure S2). All four specimens could be confidently identified. Three specimens (40, 42 and 43) were molecularly identified as G. morsitans (DDN bs = 100; ML bs = 99; BI pp = 1) and one specimen was identified as G. brevipalpis (DDN bs = 100; ML bs = 96; BI pp = 1) (Fig. 3, Additional file 2: Figure S2). No sequence data could be generated from the G. pallidipes specimen sampled in this study; however, morphological identification was assigned with a high degree of confidence. Metadata for all specimens analysed are provided in Additional file 3: Table S1.
Alpha diversity analysis revealed a slightly higher tabanid species richness and evenness in Zalala (Shannonʼs index of 1.08) and Namitangurine (Shannonʼs index 0.98), with 9 species of tabanids captured in each area. Botao with a Shannonʼs index of 0.64, revealed a lower diversity and evenness in the area.
Analyzing the trap efficiency, NGU (1735 tabanids and 9 tsetse flies captured) demonstrated better performance in general. This trap also successfully captured all three species of Glossina. The H traps only captured G. brevipalpis while Epsilon traps managed to capture the two tsetse savannah species present in the country (Fig. 4). There was a significant difference (Studentʼs t-test, t = 7.44, df = 658, P < 0.001) between the captures in dry and rainy seasons, with the latter accounting for 70.5% of the total captures. Tabanids had a high activity peak in March (755 tabanids captured) as compared to July (106 tabanids), during which the lowest density was recorded (Fig. 5).
From the 480 tabanids screened, 13.0% (66/480) tested positive for the presence of Trypanosoma spp. The overall trypanosome infection rate for tsetse flies was 75.0% (18/24). Only T. congolense infections were detected in Glossina spp. and only in G. brevipalpis and G. morsitans (Fig. 6). Namitangurine with 18.8% (30/160) presented the highest tabanid infection rate followed by Botao with 13.1% (21/160) and Zalala with 9.4% (15/160). There were no significant differences when comparing the infection rates of the tabanids between the three sites (ANOVA: F(2, 9) = 0.44, P = 0.66).
Species comparisons revealed that A. agrestis had the highest trypanosome infection rate with 6.25% (30/480), followed by T. par and T. taeniola, both yielding 3.8% of infected flies (18/480). No A. africana specimen tested positive for trypanosome infection. There was significant difference in infection rate between the three species where Trypanosoma infections were detected (ANOVA, F(3, 8) = 7.84, P = 0.009, post-hoc test: Tukey’s HSD, P = 1.0 for T. par vs T. taeniola, P = 0.19 for T. par vs A. agrestis, P = 0.23 for A. agrestis vs T. taeniola and P = 0.01 for A. agrestis vs A. africana). However, the post-hoc test only showed significant difference between A. agrestis and A. africana, which represents the difference between the species with the highest infection rate and the species with no infection. Comparing the infection rate per species/area, A. agrestis remained the species with the highest infection rate in both Namitangurine and Botao. In Zalala, T. par had a higher infection rate (Fig. 7). The trypanosome DNA detected in tabanids belonged to T. congolense (the only pathogenic trypanosome) and T. theileri (Fig. 8). Tabanids from Namitangurine presented the highest T. congolense infection rate (15.6%), followed by Botao (12.5%) and Zalala (5.6%).
Percentage of infected tabanid flies per species/area and per species of Trypanosoma (n = 480) as detected by 18S PCR-RFLP. Abbreviations: T. par, Tabanus par; T. taeniola, Tabanus taeniola; A. africana, Ancala africana; A. agrestis, Atylotus agrestis; T. co, Trypanosoma congolense; T. th, Trypanosoma theileri; Zal, Zalala; Bot, Botao; Nam, Namitangurine
RFLP restriction enzyme analysis using MspI and Eco571 (a) and MboII (b) digestion of 18 SSU-rDNA from Trypanosoma congolense (Lanes 16, 18, 9) and T. theileri (Lanes 6, 7, 8, 9 and 10) in polyacrylamide gel. Positive control for T. congolense (Lane 17) and for T. theileri (Lanes C) and a 100-bp DNA ladder (Lanes M) were included on the gels. Abbreviations: M1, samples from the first DNA extraction group; M2, samples from the second DNA extraction group; M1/Lane 16, Glossina brevipalpis; M1/Lane 18, Atylotus agrestis; M2/Lane 19, Glossina brevipalpis; F11, fly 11 (Tabanus par); F16, fly 16 (Atylotus agrestis); F19, fly 19 (Tabanus par); F21, fly 21 (Tabanus taeniola); F22, fly 22 (Atylotus agrestis)
Discussion
Despite the important role of tabanids and tsetse flies in the transmission of diseases, little research attention has been devoted to these taxa in the Afrotropics, including Mozambique. Limited fundamental information on their classification and distribution patterns are available, with the last major work focused on elucidating these aspects having been conducted more than half a century ago, in Mozambique [24]. Few surveys aiming at the distribution [16], genetic characterization [15] or infection of tsetse flies [65, 66] were previously published.
The results regarding the diversity of tabanids from this study are similar to those of Ahmed et al. [67] in Nigeria, Lendzele et al. [33] in Cameroon, Koné et al. [68] in Burkina Faso and Taioe et al. [23] in South Africa and Zambia. These authors also recorded a greater abundance and diversity of Tabanus versus other tabanid genera. Tabanus par and Atylotus agrestis were the dominant tabanid species in West Africa, despite the large geographical separation, possibly indicative of these species preferring tropical climates. High numbers of T. par were also recorded by Taioe et al. [23] in KwaZulu Natal, South Africa, but not A. agrestis. KwaZulu Natal is part of the eastern tropical corridor, reinforcing the idea that T. par might be a dominant species in tropical Africa. In their studies, Ahmed et al. [67] and Taioe et al. [23] also recorded a low abundance and diversity of Haematopota. This is a clear contrast with the Catalogue of Afrotropical Tabanidae by Chainey & Oldroyd [69] that reports approximately 250 Haematopota species of a total of 700 tabanid species present in the Afrotropical region. The reason for this contrast is unclear, but possibly the trapping methodology might not be favorable for Haematopota species in the region. The complete lack of Chrysopsinae in the trapping is similarly curious and unexpected as it was reported by both Esterhuizen [70] and Taioe et al. [23], both using H traps. It might be possible that species of Chrysopsinae are not easily captured by blue traps, designed for Glossina spp., despite their well-known blood-feeding behavior.
Atylotus spp. could not be separated confidently and a collapsed group comprising A. agrestis, A. diurnus and A. nigromaculatus was recovered. All specimens in this study were morphologically identified as A. agrestis. This requires further study and might reflect incorrectly identified specimens uploaded to GenBank, or the inability of cox1 to distinguish between Atylotus species. Alternatively, the taxonomy may not correspond to the genetic isolation prescribed by the biological species concept. Cytochrome oxidase cox1 barcodes for Haematopota species from the Afrotropics are needed. The specimen sequenced in this study (specimen 57) fell sister to a monophyletic group of H. fenestralis sequences from a single study. This might reflect confirmation of H. fenestralis or a closely related sister species.
All specimens within the Fraternus group as described by Oldroyd [27] were here identified as T. taeniola, despite four specimens grouping separately (specimens 18, 53–55), with one specimen identified as T. fraternus. Due to a lack of T. fraternus reference sequences available and the subtle morphological difference, especially in specimens stored in 70% ethanol (T. fraternus has a slight additional pigment on the wings, lacking in T. taeniola), confident identification was impossible. This grouping remains an interesting one and the species boundaries are worth investigating. The species boundaries between T. par and T. thoracinus remains unclear and was not treated in the most recent work by Mugasa et al. [64], who recovered two “genetic variants” from Uganda in their study. Unfortunately, Mugasa et al. [64] did not include T. par specimens and could not find morphological differences between the genetic variants. With the sheer abundance of T. par in southern and eastern Africa, this species status also deserves investigation.
The performance of cox1 as a Tabanidae species delimitating barcode should be reassessed as it posed several problems of which an inadequate number of sequences from accurately identified specimens. Other molecular markers, such as the internal spacers in the rRNA gene (ITS1 and ITS2) are worth investigating as an alternative for species delimitation. It should be noted, however, that cox1 worked well considering Glossina, but Glossina is considerably less diverse and all species have several representative sequences on GenBank. The fact that no sequences from G. pallidipes were obtained may be due to the poor quality of the PCR product sent for sequencing.
The underlying reasons for the observed differences in tsetse and tabanids abundance between the selected sites are unclear. Vegetation type, vegetation cover and non-domesticated host species (host-preference), have all been shown to be important variables in the distribution and composition of haematophagous fly populations [71,72,73]. Zalala is an area consisting of small and widely dispersed pockets of shrubs and trees that may provide insufficient habitat for a high diversity of flies. Moreover, cattle being the only apparent food source available in the area is an additional limiting factor to the expansion of both tabanid- and tsetse fly assemblages. Finally, the dense human settlements that are rapidly expanding and changing the surrounding environment may in turn also negatively impact biodiversity. Haematophagous flies that possibly require a wide range of hosts may then be especially affected [74,75,76,77,78].
Namitangurine on the other hand, is characterized by dense thickets and is seemingly a less disturbed area with wild animals like sable, kudu, reedbuck, duiker and bush pig present [79]. Interestingly, the highest infection rates were also detected from this area, possibly reflecting the importance of wild animals as reservoirs of Trypanosoma species. In fact, salivarian trypanosomes can be found in a wide variety of hosts such as ruminants, carnivores, rodents and reptiles [80,81,82,83]. This might then indicate the important role of these animals for the circulation of trypanosomes in a certain habitat. Control measures directed to trypanosomes reservoirs have shown to be successful in the reduction of trypanosomosis prevalence, though they have proven not to be environmentally sustainable [84,85,86].
The efficacy of blue traps for determining Tabanidae diversity should perhaps be revisited, with all aforementioned studies being unable to reflect the “true” diversity of Tabanidae species as reported from museum data [25,26,27, 87]. Furthermore, the higher diversity recorded in regions from museum data might possibly reflect “false” diversity due to erroneous taxonomy and highlight the need for renewed taxonomic approaches as stipulated by Morital et al. [18].
It is important to note that the H trap had a higher efficacy in capturing G. brevipalpis, compared to both the Epsilon- and NGU traps. However, Malele et al. [88] reported that, in Tanzania, H traps did not capture any G. brevipalpis. Several other studies, in turn, have demonstrated good performance of H traps in the capture of G. brevipalpis in South Africa [34, 89, 90] and in Mozambique [16]. These contradicting results are important in control and surveillance studies and require clarification.
The low levels of Shannonʼs diversity index obtained in the present study were mainly due to the high frequency of T. par (66.8%) and Atylotus agrestis (23.2%), showing the existence of clear dominant species and a very low evenness of species. This is often found in areas where human disturbances are evident, and cattle grazing might form part of such a disturbance [91]. The rainy season accounted for two folds the total capture number of the dry season as recorded by several studies [28, 33, 67, 92]. On a finer scale, seasonal tabanid activity peaks is extremely important to understand the pathogen transmission dynamics. In this region of Mozambique, an early wet-season peak was observed in November, followed by a second more pronounced peak in March, at the end of the rainy season. This might be bivoltine behavior, with the emergence of the dry-season generation that probably included diapause (first peak), and a subsequent wet-season generation that lacks diapause (second peak). Investigating the drivers of these peaks might prove valuable for predicting, mitigating and controlling diseases spread by horse flies [93].
In this study, G. brevipalpis appears to be the most important vector since the species was most often collected and showed a high infection rate (72.2%, n = 18). Motloang et al. [94] reported contrasting results from an experimental study in South Africa; the authors concluded that, despite their higher abundance, the role of G. brevipalpis in the transmission of T. congolense was negligible. The results here, in turn, warrants additional investigation and to reassess the role of G. brevipalpis in the transmission of trypanosomosis. Over the course of a year’s uninterrupted collection, G. brevipalpis was the only species present in all three study areas with individuals from all three areas infected with Trypanosoma.
Although the number of captured G. morsitans flies was low (four individuals), their high infection rate (100%) could nevertheless indicate their important role as a vector of trypanosomosis in this area. Glossina morsitans was described by Vreysen et al. [7] as one of the most important species in the Morsitans group and the major vector of AAT in eastern and southern Africa, and it was experimentally proven by Reinfeberg et al. [95] that Morsitans flies were more susceptible to T. congolense infections compared to other groups, as Palpalis for example. Thus, the high infection rate found in G. morsitans from Namitangurine is somewhat expected. Even though infection rates differ from those found in this study, this is congruent with the findings of Salekwa et al. [96] in Tanzania and Shereni et al. [97] in Zimbabwe. These studies postulated that G. morsitans from conservation or less disturbed areas (like Namitangurine) are more likely to be infected with Trypanosoma species.
Since only one G. pallidipes specimen was captured, no accurate discussion can be presented on the infection rate of this species. However, in other studies, Trypanosoma infection were detected in G. pallidipes [31, 98], which shows that they can actively harbor and transmit trypanosomes. The presence/absence of infection in tsetse flies can be due to feeding preferences [99], genetic differences, the availability of reservoirs, the parasitaemia of the vertebrate host [100, 101] and the nutritional status of the flies [102, 103]. As discussed previously, tsetse flies respond demographically to habitat destruction or fragmentation. However, it is known that the flies also respond physiologically to this process and to other ecological pressures. In fact, it was experimentally proven that environmental stress, including starvation, causes increase in the susceptibility of tsetse flies to trypanosome infections [102,103,104,105]. This may explain the high infection rates detected in the present study despite the small sample size.
The only pathogenic trypanosome found in the present study was T. congolense. This is well in accordance with the study of Mulandane et al. [22] reporting T. congolense as the only circulating pathogenic Trypanosoma species in cattle in Nicoadala District. Additional to T. congolense, T. theileri was also found in tabanids in all the species screened except for A. africana, which is not an indication that this species cannot harbor trypanosomes as Firmino et al. [106] detected trypanosome sequences in A. africana flies captured in Ethiopia. Trypanosoma theileri is a stercorarian, non-pathogenic trypanosome transmitted to cattle by tabanids, where it undergoes a developmental cycle [107, 108], and highly prevalent around the globe [109,110,111,112].
Our results strongly suggest a role for tabanids in the transmission of T. congolense in the study area. In general, the participation of tabanids in the dynamics of African animal trypanosomosis (linked to T. vivax and T. evansi) is rightly becoming more important with the ecological transformations that are progressively affecting the tsetse densities and distribution in Africa [113,114,115]. Moreover, mechanical transmission of trypanosomosis, depending on the circumstances and conditions, may be as efficient as biological transmission, although its potential impact has never been estimated [8, 9, 19]. These inferences can be supported by the findings from Desquesnes & Dia [113, 116], where mechanical transmission of T. congolense and T. vivax by A. agrestis and A. fuscipes were experimentally demonstrated. They are also supported by Abebe & Jobre [117], in which high T. vivax and T. congolense infection where detected in tsetse free zones. Our study indicates that A. agrestis could have an important impact on the dynamics and/or epidemiology of trypanosomosis.
Conclusions
To the best of our knowledge, this is the first study conducted in Nicoadala District, an area identified as a drug resistance hotspot, involving the characterization of the Tabanidae and Glossinidae and the detection of the trypanosomes they harbor. As a pioneer study, it constitutes a starting point for future work on the subject. Considering the trapping effort, it can be concluded that a very small population of tsetse flies responsible for the biological transmission of trypanosomosis is present in the district. However, the strong presence of tabanids, including species that have previously been identified as trypanosomosis vectors, suggests their participation as mechanical vectors as a relay and amplification factor to tsetse flies resulting in a high prevalence of trypanosomosis in the district as reported by Mulandane et al. [22], Jamal et al. [118] and Specht [119].
Availability of data and materials
Captured flies and DNA extracted in this study are deposited at the Biotechnology Center Laboratory (Eduardo Mondlane University) in Maputo, Mozambique. The newly generated sequences were submitted to the GenBank database under the Accession numbers MT231157–MT231193.
Abbreviations
- PCR:
-
polymerase chain reaction
- H trap:
-
horizontal trap
- ddH2O:
-
double-distilled water
- rRNA:
-
ribosomal ribonucleic acid
- ANOVA:
-
analysis of variance
- SSU-rDNA:
-
small subunit of ribosomal deoxyribonucleic acid
- ITS:
-
internal transcribed spacer
References
Steverding D. The history of African trypanosomiasis. Parasites Vectors. 2008;1:3.
Elkarib AE. Animal trypanosomiasis in Sudan. Sud J Vet Sci Anim Husb. 1961;2:39–46.
Jordan AM. Tsetse eradication plans for southern Africa. Parasitol Today. 1985;1:121–3.
Cecchi G, Paone M, Feldmann U, Vreysen MJB, Diall O, Mattioli RC. Assembling a geospatial database of tsetse-transmitted animal trypanosomosis for Africa. Parasites Vectors. 2014;7:39.
Ahmed SK, Rahman AH, Hassan MA, Salih EM, Paone M, Cecchi G. An atlas of tsetse and bovine trypanosomosis in Sudan. Parasites Vectors. 2016;9:194.
Malele II. Fifty years of tsetse control in Tanzania: challenges and prospects for the future. Tanzan J Health Res. 2011;13:399–406.
Vreysen MJB, Talla M, Sall B, Bouyer J. Tsetse flies: their biology and control using area-wide integrated pest management approaches. J Invertebr Pathol. 2013;112:15–25.
Desquesnes M, Biteau-Coroller F, Bouyer J, Dia ML, Foil L. Development of a mathematical model for mechanical transmission of trypanosomes and other pathogens of cattle transmitted by tabanids. Int J Parasitol. 2009;39:333–46.
Baldacchino F, Desquesnes M, Mihok S, Foil LD, Duvallet G, Jittapalapong S. Tabanids: neglected subjects of research, but important vectors of disease agents! Infect Genet Evol. 2014;28:596–615.
Hamon J, Challier A, Mouchet J. Biology and control of tsetse flies. WHO inter-regional seminar on entomology methods in vector control. https://horizon.documentation.ird.fr/exl-doc/pleins_textes/pleins_textes_5/b_fdi_04-05/04683.pdf. Accessed 14 Jan 2019.
FAO. Training manual for tsetse control personnel: ecology and behavior of tsetse. Volume 2. In: Pollock JN, editor. Rome: FAO; 1991. http://www.fao.org/3/p5444e/P5444E.pdf. Accessed 14 Apr 2018.
Gooding RH, Krafsur ES. Tsetse genetics: contributions to biology, systematics, and control of tsetse flies. Annu Rev Entomol. 2005;50:101–23.
Krafsur ES. Tsetse flies: genetics, evolution, and role as vectors. Infect Genet Evol. 2010;9:124–41.
Lovemore DF. A regional approach to trypanosomiasis control: activities and progress of RTTCP. FAO: Rome. 1992. 1992. http://www.fao.org/3/T0599E/T0599E12.htm. Accessed 21 Mar 2020.
Wohlford DL, Krafsur ES, Griffiths NT, Griffiths NT, Marques JG, Baker MD. Genetic differentiation of some Glossina morsitans morsitans populations. Med Vet Entomol. 1999;13:377–85.
Sigauque I, Van den Bossche P, Moiana M, Jamal S, Neves L. The distribution of tsetse (Diptera: Glossinidae) and bovine trypanosomosis in the Matutuine District, Maputo Province, Mozambique. Onderstepoort J Vet Res. 2000;67:167–72.
Müller GC, Revay EE, Hogsette JA, Zeegers T, Kline D, Kravchenko VD, Schlein Y. An annotated checklist of the horse flies (Diptera: Tabanidae) of the Sinai Peninsula Egypt with remarks on ecology and zoogeography. Acta Trop. 2012;122:205–11.
Morita SI, Bayless KM, Yeates DK, Wiegmann BM. Molecular phylogeny of the horse flies: a framework for renewing tabanid taxonomy. Syst Entomol. 2016;41:56–72.
Foil LD. Tabanids as vectors of disease agents. Parasitol Today. 1989;5:88–96.
Dávila AMR, Silva RAMS. Animal trypanosomiasis in South America Current status, partnership, and information technology. Ann N Y Acad Sci. 2000;916:199–212.
Hall M. Vectors of Trypanosoma vivax in the new outbreak zone of Santa Cruz, Bolivia. R7356 project summary. Centre for Tropical Veterinary Medicine, University of Edinburgh. Project completion summary on the Animal Health Research Programme, Project R7356. 2001. https://assets.publishing.service.gov.uk/media/57a08d6140f0b64974001818/R7356_FTR.pdf. Accessed 24 Dec 2018.
Mulandane FC, Fafetine J, Abbeele JVD, Clausen P, Hoppenheit A, Cecchi G, et al. Resistance to trypanocidal drugs in cattle populations of Zambezia. Parasitol Res. 2018;117:429–36.
Taioe MO, Motloang MY, Namangala B, Chota A, Molefe NI, Musinguzi SP, et al. Characterization of tabanid flies (Diptera : Tabanidae) in South Africa and Zambia and detection of protozoan parasites they are harbouring. Parasitology. 2017;144:1162–78.
Dias JATS. Tabanídeos (Diptera-Tabanidae) de Moçambique. Contribuição para o seu conhecimento. Estudos Gerais Universitários de Moçambique: Lourenço Marques (=Maputo); 1966.
Oldroyd H. The horse-flies of the Ethiopian Region (Diptera:Tabanidae). I. Haematopota and hippocentrum. London: British Museum (Natural History); 1952. p. 226.
Oldroyd H. The horse-flies (Diptera: Tabanidae) of the Ethiopian region. II. Tabanus and related genera. London: British Museum (Natural History); 1954. p. 341.
Oldroyd H. The horse-flies (Diptera: Tabanidae) of the Ethiopian region. III. Subfamilies Chrysopinae, Scepsidinae and Pangoniinae and a revised classification. London: British Museum (Natural History); 1957. p. 489.
Herczeg T, Blahó M, Száz D, Kriska G, Gyurkovszky M, Farkas R, et al. Seasonality and daily activity of male and female tabanid flies monitored in a Hungarian hill-country pasture by new polarization traps and traditional canopy traps. Parasitol Res. 2014;113:4251–60.
Krčmar S, Radolić V, Lajoš P, Lukačević I. Efficiency of colored modified box traps for sampling of tabanids. Parasite. 2014;21:67.
Brightwell R, Dransfield R. Survey of Tabanidae (horseflies) in southern England. Influential points. 2014. https://influentialpoints.com/pdfs/A_preliminary_survey_of_tabanidae_horseflies_in_southern_England_2014.pdf. Accessed 21 Oct 2018.
Nthiwa DM, Odongo DO, Ochanda H, Khamadi S, Gichimu BM. Trypanosoma infection rates in Glossina species in Mtito Andei Division, Makueni County, Kenya. J Parasitol Res. 2015;2015:607432.
Wondimu A, Reta E, Fekadu A. Tsetse density and bovine trypanosomosis in selected sites of Konta, Ethiopia. J Vet Med Anim Health. 2017;9:78–82.
Lendzele S, Abdoulmoumini M, Ayg L. Spatial repartition of tabanids (Diptera: Tabanidae) in different ecological zones of North Cameroon. Biodivers Int J. 2017;1:1–5.
Kappmeier K. A newly developed odour-baited “H trap” for the live collection of Glossina brevipalpis and Glossina austeni (Diptera: Glossinidae) in South Africa. Onderstepoort J Vet Res. 2000;67:15–26.
Esena RK. A new trap for Glossina morsitans as a strategy for quality improvement of African trypanosomiasis control. J Biol Agric Health. 2013;3:54–66.
Brightwell R, Dransfield RD, Kyorku C, Golder TK, Tarimo SA, Mungai D. A new trap for Glossina pallidipes. Trop Pest Manag. 1987;33:151–9.
Morlais I, Grebaut P, Bodo JM, Djoha S, Cuny G, Herder S. Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Trop. 1998;70:109–17.
Geysen D, Delespaux V, Geerts S. PCR-RFLP using SSU-rDNA amplification as an easy method for species-specific diagnosis of Trypanosoma species in cattle. Vet Parasitol. 2003;110:171–80.
Köppen W, Geiger R. Klimate der Erde. Gotha: Justus Perthes; 1928.
Peel MC, Finlayson BL, Mcmahon TA. Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci. 2007;11:1633–44.
MAE (Ministerio da Administração Estatal). Perfil do Distrito de Nicoadala, Província da Zambézia, Moçambique. 2005. http://www.portaldogoverno.gov.mz/por/content/download/2902/23562/version/1/file/Nicoadala.pdf. Accessed 21 Aug 2018.
Mihok S. The development of a multipurpose trap (the Nzi) for tsetse and other biting flies. Bull Entomol Res. 2002;92:85–403.
Barclay HJ, Hargrove JW. Probability models to facilitate a declaration of pest-free status, with special reference to tsetse (Diptera: Glossinidae). Bull Entomol Res. 2005;95:1–11.
Bouyer J, Ravel S, Guerrini L, Dujardin J-P, Sidibé I, Vreysen MJB, Solano P, De Meeûs T. Population structure of Glossina palpalis gambiensis (Diptera: Glossinidae) between river basins in Burkina Faso: consequences for area-wide integrated pest management. Infect Genet Evol. 2010;10:321–8.
Vale GA, Hall DR. The role of 1-octen-3-oQ, acetone and carbon dioxide to improve baits for tsetse flies. Bull Entomol Res. 1985;75:219–31.
FAO. Collection of entomological baseline data for tsetse area-wide integrated pest management programmes. FAO/AIEA: Rome; 2008. http://www.fao.org/3/i0535e/i0535e.pdf. Accessed 15 June 2016.
Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–13.
Ravel S, De Meeûs T, Dujardin JP, Zézé DG, Gooding RH, Dusfour I, et al. The tsetse fly Glossina palpalis palpalis is composed of several genetically differentiated small populations in the sleeping sickness focus of Bonon, Côte d’Ivoire. Infect Genet Evol. 2007;7:116–25.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
Katoh S. MAFFT multiple sequence alignement software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0. Mol Biol Evol. 2016;35:1547–9.
Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67.
Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop. New Orleans; 2010. p. 1–8.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3.
Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4.
Rambaut A, Suchard M, Xie D, Drummond AJ. Tracer v1.6. 2014. http://beast.community. Accessed 2 Aug 2018.
Shannon CE, Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press; 1949.
Maity A, Naskar A, Parui P, Homechaudhuri S, Banerjee D. Seasonal variation in diversity of horse flies (Diptera: Tabanidae) from the part of Central Himalaya. J Adv Zool. 2018;39:63–83.
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: community ecology package. R package version 2.5-2. 2018.
Cannon RM, Roe RT. Livestock disease surveys: a field manual for veterinarians. Canberra: Australian Government Publishing Service; 1982. p. 35.
Bruford MW, Hanotte O, Brookfield JFY, Burke T. Multilocus and single-locus DNA fingerprinting. In: Hoelzel AR, editor. Molecular genetic analysis of populations: a practical approach. 2nd ed. Oxford: IRL Press; 1998. p. 287–336.
TIBCO Software Inc. Statistica (data analysis software system), version 13. 2017.
Mugasa CM, Villinger J, Gitau J, Ndungu N, Ciosi M, Masiga D. Morphological re-description and molecular identification of tabanidae (Diptera) in East Africa. ZooKeys. 2018;769:117–44.
Rodrigues CM, Garcia HA, Rodrigues AC, Costa-Martins AG, Pereira CL, Pereira DL, et al. New insights from Gorongosa National Park and Niassa National Reserve of Mozambique increasing the genetic diversity of Trypanosoma vivax and Trypanosoma vivax-like in tsetse flies, wild ungulates and livestock from East Africa. Parasites Vectors. 2017;10:337.
Garcia HA, Rodrigues CMF, Rodrigues AC, Pereira DL, Pereira CL, Camargo EP, et al. Remarkable richness of trypanosomes in tsetse flies (Glossina morsitans morsitans and Glossina pallidipes) from the Gorongosa National Park and Niassa national Reserve of Mozambique revealed by fluorescent fragment length barcoding (FFLB). Infect Genet Evol. 2018;63:370–9.
Ahmed AB, Okiwelu SN, Samdi SM. Species diversity and seasonal occurrence of some biting flies in southern Kaduna, Nigeria. Afr J Biomed Res. 2005;8:113–8.
Koné N, N’goran EK, Sidibe I, Kombassere AW, Bouyer J. Spatio-temporal distribution of tsetse and other bitting flies in the Mouhoun River basin, Burkina Faso. Med Vet Entomol. 2011;25:156–68.
Chainey JE, Oldroyd H. Family Tabanidae. In: Crosskey RW, editor. Catalogue of the Diptera of the Afrotropical Region. London: British Museum (Natural History); 1980. p. 275–308.
Esterhuizen J. Seasonal abundance of horse flies (Diptera: Tabanidae) from two conservation areas in north eastern KwaZulu-Natal Province, South Africa. Afr Entomol. 2006;14:395–7.
Van den Bossche P, Staak C. The importance of cattle as a food source for Glossina morsitans morsitans Westwood (Diptera: Glossinidae) in Katete District, eastern Province, Zambia. Acta Trop. 1997;65:105–9.
Simukoko H, Marcotty T, Phiri I, Geysen G, Vercruysse J, Van den Bossche P. The comparative role of cattle, goats and pigs in the epidemiology of livestock trypanosomiasis on the plateau of eastern Zambia. Vet Parasitol. 2007;147:231–8.
Gondwe N, Marcotty T, Vanwambeke SO, De Pus C, Mulumba M, Van den Bossche P. Distribution and density of tsetse flies (Glossinidae: Diptera) at the game/people/livestock interface of the nkhotakota game reserve human sleeping sickness focus in Malawi. EcoHealth. 2009;6:260–5.
Kilemwa AM. Environmental impact of animal traction in Rukwa Region, Tanzania. In: Starkey P, Kaumbutho P, editors. Meeting the challenges of animal traction. A resource book of the animal traction network for eastern and southern Africa (ATNESA), Harare, Zimbabwe. London: Intermediate Technology Publications; 1999. p. 326.
Van den Bossche P, Rocque LS, Hendrickx G, Bouyer J. A changing environment and the epidemiology of tsetse transmitted livestock trypanosomiasis. Trends Parasitol. 2010;26:236–43.
Malele II, Nyingilili HS, Msangi N. Factors defining the distribution limit of tsetse infestation and the implication for livestock sector in Tanzania. Afr J Agric Res. 2011;6:2341–7.
Mweempwa C, Marcotty T, De Pus C, Penzhorn BL, Dicko AH, Bouyer J, et al. Impact of habitat fragmentation on tsetse populations and trypanosomosis risk in eastern Zambia. Parasites Vectors. 2015;8:406.
Torres A, Jaeger JAG, Alonso JC. Assessing large-scale wildlife responses to human infrastructure development. Proc Natl Acad Sci USA. 2016;113:8472–7.
MAE (Ministerio da Administração Estatal). Perfil do Distrito de Nicoadala, Província da Zambézia. Moçambique. 2014. http://www.maefp.gov.mz/wp-content/uploads/2017/04/Nicoadala.pdf. Accessed 28 Aug 2018.
Mulla AF, Rickman LR. How do African game animals control trypanosome infections? Parasitol Today. 1988;4:352–4.
Njiokou F, Laveissière C, Simo G, Nkinin S, Grébaut P, et al. Wild fauna as a probable animal reservoir for Trypanosoma brucei gambiense in Cameroon. Infect Genet Evol. 2006;6:147–53.
Enderson NE, Mubanga J, Fevre EM, Picozzi K, Eisler MC, Thomas R, et al. Characterisation of the wildlife reservoir community for human and animal trypanosomiasis in the Luangwa Valley, Zambia. PLoS Negl Trop Dis. 2011;5:e1211.
Auty H, Anderson NE, Picozzi K, Picozzi K, Eisler MC, Thomas R, et al. Trypanosome diversity in wildlife species from the Serengeti and Luangwa Valley ecosystems. PLoS Negl Trop Dis. 2012;6:e1828.
Teixeira, AHS, Valadão FG, Dias JATS. Anais dos Serviços de Veterinária. Secçaõ de economia da repartição técnica de zootecnia e fomento pecuário. Mozambique. 1960. Report Number 8. 202 pp.
WHO. Control and surveillance of human African trypanosomiasis. World Health Organ Tech Rep Ser. 2013;984:1–237.
Wiel K. The importance of wildlife as a reservoir for human and animal African Trypanosomiasis. M.Sc thesis. Faculty of Veterinary Medicine, Ghent University, Ghent; 2016.
Usher J. A review of the South African horse fly fauna (Diptera: Tabanidae). Ann Natal Museum. 1972;21:459–507.
Malele II, Ouma JO, Nyingilili HS, Kitwika HS, Malulu DJ, Magwisha HB, et al. Comparative performance of traps in catching tsetse flies (Diptera: Glossinidae) in Tanzania. Onderstepoort J Vet Res. 2016;83:1057.
Mamabolo MV, Ntantiso L, Latif A, Majiwa PAO. Natural infection of cattle and tsetse flies in South Africa with two genotypic groups of Trypanosoma congolense. Parasitology. 2009;136:425–31.
De Beer CJ, Venter GJ, Green KK, Esterhuizen J, Ntshangase J, Vreysen MJB, et al. An update of the tsetse fly (Diptera : Glossinidae) distribution and African animal trypanosomosis prevalence in north-eastern KwaZulu-Natal, South Africa. Onderstepoort J Vet Res. 2016;83:a1172.
Sutrisno H. The impact of human activities to dynamic of insect communities: a case study in Gunung Salak, West Java. HAYATI J Biosci. 2010;17:161–6.
Parra-Henao G, Alarcón-Pineda EP. Ecology and parasitological analysis of horse flies (Diptera: Tabanidae) in Antioquia, Colombia. Caldasia. 2008;30:179–88.
Barros ATM. Seasonality and relative abundance of Tabanidae (Diptera) captured on horses in the Pantanal, Brazil. Mem Inst Oswaldo Cruz. 2001;96:917–23.
Motloang M, Masumu J, Mans B, Van den Bossche P, Latif AA. Vector competence of Glossina austeni and Glossina brevipalpis for Trypanosoma congolense in KwaZulu-Natal, South Africa. Onderstepoort J Vet Res. 2012;79:353.
Reifenberg JM, Cuisance D, Frezil JL, Cuny G, Duvallet G. Comparison of the susceptibility of different Glossina species to simple and mixed infections with Trypanosoma (Nannomonas) congolense savannah and riverine forest types. Med Vet Entomol. 1997;11:246–52.
Salekwa L, Nnko H, Ngonyoka A, Estes AB, Agaba M, Gwakisa PS. Relative abundance of tsetse fly species and their infection rates in Simanjiro, northern Tanzania. Livest Res Rural Dev. 2014;26:213.
Shereni W, Anderson NE, Nyakupinda L, Cecchi G. Spatial distribution and trypanosome infection of tsetse flies in the sleeping sickness focus of Zimbabwe in Hurungwe District. Parasites Vectors. 2016;9:605.
Malele II, Magwisha HB, Nyingilili HS, Nyingilili HS, Mamiro KA, Rukambile EJ, et al. Multiple Trypanosoma infections are common amongst Glossina species in the new farming areas of Rufiji District, Tanzania. Parasites Vectors. 2011;4:217.
Bitew M, Amedie Y, Abebe A, Tolosa T. Prevalence of bovine trypanosomosis in selected areas of Jabi Tehenan District, West Gojam of Amhara regional state, Northwestern Ethiopia. Afr J Agric Res. 2011;6:140–4.
Jordan AM. Trypanosomiasis control and African rural development. London: Longman; 1986.
Leak SGA. Tsetse biology and ecology: their role in the epidemiology and control of trypanosomiasis. Wallingford: CABI Publishing; 1999.
Kubi C, Van Den Abbeele J, De Deken R, Marcotty T, Dorny P, Van den Bossche P. The effect of starvation on the susceptibility of teneral and non-teneral tsetse flies to trypanosome infection. Med Vet Entomol. 2006;20:388–92.
Akoda K, Van Den Abbeele J, Marcotty T, De Deken R, Sidibe I, Van den Bossche P. Nutritional stress of adult female tsetse flies (Diptera: Glossinidae) affects the susceptibility of their offspring to trypanosomal infections. Acta Trop. 2009;111:263–7.
Ndegwa PN, Irungu LW, Moloo SK. Effect of puparia incubation temperature: increased infection rates of Trypanosoma congolense in Glossina morsitans centralis, G. fuscipes fuscipes and G. brevipalpis. Med Vet Entomol. 1992;6:127–30.
Bouyer J, Koné N, Bengaly Z. Dynamics of tsetse natural infection rates in the Mouhoun river, Burkina Faso, in relation with environmental factors. Front Cell Infect Microbiol. 2013;3:47.
Fermino B, Paiva F, Viola LB, Rodrigues CMF, Garcia HA, Campaner M, et al. Shared species of crocodilian trypanosomes carried by tabanid flies in Africa and South America, including the description of a new species from caimans, Trypanosoma kaiowa n. sp. Parasites Vectors. 2019;12:225.
Bose R, Heister NC. Development of Trypanosoma (M.) theileri in tabanids. J Eukaryot Microbiol. 1993;40:788–92.
Kelly S, Ivens A, Mott GA, et al. An alternative strategy for trypanosome survival in the mammalian bloodstream revealed through genome and transcriptome analysis of the ubiquitous bovine parasite Trypanosoma (Megatrypanum) theileri. Genome Biol Evol. 2017;9:2093–109.
Schlafer DH. Trypanosoma theileri: a literature review and report of incidence in New York cattle. Cornell Vet. 1979;69:411–25.
Rodrigues AC, Paiva F, Campaner M, Stevens JR, Noyes HA, Teixeira MMG. Phylogeny of Trypanosoma (Megatrypanum) theileri and related trypanosomes reveals lineages of isolates associated with artiodactyl hosts diverging on SSU and ITS ribosomal sequences. Parasitology. 2006;132:215–24.
Martins JR, Leite RC, Doyle RL. Tripanosomatides like Trypanosoma theileri in the cattle tick Boophilus microplus. Rev Bras Parasitol Vet. 2008;17:113–4.
Sood NK, Singla LD, Singh RS, Uppal SK. Association of Trypanosoma theileri with peritonitis in a pregnant cross-bred cow: a case report. Vet Med. 2011;56:82–4.
Desquesnes M, Dia ML. Mechanical transmission of Trypanosoma congolense in cattle by the African tabanid Atylotus agrestis. Exp Parasitol. 2003;105:226–31.
Moore S, Shrestha S, Tomlinson KW, Vuong H. Predicting the effect of climate change on African trypanosomiasis: integrating epidemiology with parasite and vector biology. J R Soc Interface. 2012;9:817–30.
Lord JS, Hargrove JW, Torr SJ, Vale GA. Climate change and African trypanosomiasis vector populations in Zimbabwe’s Zambezi Valley: a mathematical modelling study. PLoS Med. 2018;15:e1002675.
Desquesnes M, Dia ML. Mechanical transmission of Trypanosoma vivax in cattle by the African tabanid Atylotus fuscipes. Vet Parasitol. 2004;119:9–19.
Abebe G, Jobre Y. Trypanosomosis: a threat to cattle production in Ethiopia. Rev Med Vet. 1996;147:897–902.
Jamal S, Sigauque I, Macuamule C, Neves L, Penzhorn BL, Marcotty T, et al. The susceptibility of Trypanosoma congolense isolated in Zambézia Province, Mozambique, to isometamidium chloride, diminazene aceturate and homidium chloride. Onderstepoort J Vet Res. 2005;72:333–8.
Specht EJK. Prevalence of bovine trypanosomosis in Central Mozambique from 2002 to 2005. Onderstepoort J Vet Res. 2008;75:73–81.
Acknowledgements
We would like to acknowledge the Biotechnology Center, Eduardo Mondlane University and the University of Pretoria, Department of Veterinary Tropical Diseases, particularly Miss Ilse Vorster, for all the laboratory assistance. FCM would like to acknowledge the Italian Agency for Cooperation and Development (AICS) for the financial aid for publication through the project AID11096. LPS would like to acknowledge a DST/NRF freestanding award (116711) as funding for his postdoctoral fellowship.
Funding
The present study was financed by the European Union through the EU funded TRYRAC project (TRYRAC/DCI-FOOD/2011/279-754).
Author information
Authors and Affiliations
Contributions
LN and FCM conceived and designed the study. FCM coordinated the fieldwork and molecular biology analysis. LPS conducted the taxonomic identification of the flies together with FCM, and was also responsible for the phylogenetic analysis. DB conducted the statistical analysis. JB, JMF, JVDB, MO and VD contributed to the revisions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
An uncorrected p-distance data-display network, using all characters recovered from SplitsTree using the Tabanidae cox1 data. Bootstrap (bs) support calculated from 1000 replicates is indicated for the various groupings. The double-digit numbers (intuitional voucher numbers) on the tips of the network represent the sequences from this study.
Additional file 2: Figure S2.
An uncorrected p-distance data-display network, using all characters recovered from SplitsTree using the Glossinidae cox1 data. Bootstrap (bs) support calculated from 1000 replicates is indicated for the various groupings. The double-digit numbers on the tips of the network represent the sequences from this study. Red arrows indicate the specimen sequences from this study.
Additional file 3: Table S1.
Metadata for all specimens analysed.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mulandane, F.C., Snyman, L.P., Brito, D.R.A. et al. Evaluation of the relative roles of the Tabanidae and Glossinidae in the transmission of trypanosomosis in drug resistance hotspots in Mozambique. Parasites Vectors 13, 219 (2020). https://doi.org/10.1186/s13071-020-04087-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04087-1