Abstract
Background
Cystic echinococcosis (CE) is a zoonosis caused by cestodes of Echinococcus granulosus (sensu lato) complex. In Nigeria, reports on the prevalence of CE, although limited, have been found to vary with location and host with higher prevalence and fertility rate observed in camels than other livestock. Until now, information regarding the molecular characteristics, genetic population structure, and genotypes of Echinococcus is lacking. Therefore, this study was aimed at addressing these gaps in knowledge.
Methods
We describe the genetic status of 31 Echinococcus isolates collected from slaughtered livestock (camels, cattle and goats) based on the full-length mitochondrial cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase subunit 1 (nad1) genes.
Results
The resulting nucleotide sequences via the NCBI BLAST algorithm and Bayesian phylogeny of cox1 and cox1–nad1 genes using MrBayes v.3.1.2 showed that all isolates were clearly E. canadensis (G6/G7) and were 99–100% identical to previously reported G6/G7 haplotypes across Europe, Asia, North and East Africa.
Conclusions
Although, the G1 genotype is believed to be responsible for the majority of global CE burden, reports from a number of West African countries including Nigeria suggest that E. canadensis G6/G7 genotype could be the major causative agent of CE in the subregion. This study provides for the first time insight into the genetic population structure of Echinococcus species as well as implications for CE control in Nigeria.
Similar content being viewed by others
Background
Cystic echinococcosis (CE) is caused by larval stage metacestodes of Echinococcus granulosus (sensu lato) of which canids are definitive hosts while a wide range of domestic ungulates act mainly as intermediate hosts. Globally, the economic losses due to the burden of CE have been estimated as reaching billions of US dollars annually [1, 2]. CE is common in Africa, especially but not limited to northern and eastern African countries [2,3,4,5,6,7]. Additionally, high genetic diversity of Echinococcus species has been observed [7]. In most West African countries, data on the prevalence and genetic diversity of Echinococcus species is broadly lacking as information of the genotypes is only available for a few countries [8,9,10,11]. Species of E. granulosus (s.l.) have shown considerable variation within their mitochondrial DNA resulting in categorization into E. granulosus (sensu stricto) (G1, G3), E. equinus (G4), E. ortleppi (G5), E. canadensis (G6-G10), as well as E. felidis [12]. However, there are pending controversies regarding the taxonomy of E. canadensis group as some authors have suggested that genotypes G6/G7 be categorised as E. intermedius, while genotypes G8 and G10 as E. canadensis [12,13,14].
Therefore, knowledge of the identity of species and their genotypes from a range of hosts in a given location could prove useful in understanding and appreciating the disease dynamics; this could guide towards designing effective control and prevention schemes [15, 16].
In Nigeria, reports during the last three decades have shown a high prevalence of CE in livestock, particularly in the northern region [17, 18], with the highest prevalence and fertility rate in camels [18]. The seeming impact of CE infection on livestock production is yet to be evaluated in Nigeria. Therefore, in furthering this course, information regarding species variation and genotypes responsible for infection could be crucial. To our knowledge, we thus provide for the first time an insight into the genetic population structure and phylogenetic relationship of Echinococcus species in Nigeria.
Methods
Study area
Nigeria is in West Africa, has a population of over 180 million, and comprises 36 states and a Federal Capital Territory (Abuja). These states are grouped into six geopolitical zones (North-East, North-Central, North-West, South-East, South-South and South-West) based on ethnicity and common history/ancestry. The vegetation cover is mostly rainforest in the south and savannah in the north. Owing to its favorable climate, it supports large biodiversity and is thus endemic for a number of parasitic zoonoses including cystic echinococcosis. Sokoto and Maiduguri are capital cities located in the North-West and North-East zones of Nigeria, respectively, while Benin-city and Yenagoa are situated in the South-South zone (Fig. 1).
Parasite material
A total of 1592 heads of livestock [camels (n = 118), cattle (n = 856), sheep (n = 300) and goats (n = 318); Table 1] were investigated post-mortem for infection with Echinococcus spp. within a two-month period (November and December 2018) in the following locations: Sokoto modern abattoir, Sokoto state; Maiduguri abattoir, Borno state; Aduwawa cattle market in Benin-city, Edo state; and Yenagoa abattoir in Bayelsa state. Furthermore, faecal samples from privately-owned dogs (n = 51) within the city metropolis, and lions (n = 3) and a hyena (n = 1) from a zoological garden in Benin-city (Table 1) were examined for Echinococcus eggs using a modified sucrose flotation method and polymerase chain reaction after DNA extraction from faeces (QIAamp Fast DNA Stool Mini-kit; Qiagen, Hilden, Germany).
DNA extraction, amplification, and sequencing of isolates
Prior to DNA extraction, collected cysts were first cleaned with 75% ethanol and germinal layers were removed and repeatedly washed with phosphate-buffered solution (PBS). Protoscoleces from fertile cysts were also washed in PBS and stored until use. DNA was extracted from germinal layers and protoscoleces. Briefly, a portion of the germinal layer from each isolate was crushed in liquid nitrogen followed by total genomic DNA extraction using a Qiagen Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. Afterwards, PCR was conducted in a 25 µl final volume using 2 mM MgCl2, 0.2 mM dNTPs, 5 µl of 5× Taq buffer, 10 pmol of each primer, 0.5 µl of Ex Taq DNA polymerase (5 U/µl; TaKaRa, Kusatsu, Japan), 0.5 μl of genomic DNA extract (~20–200 ng) and RNAse free water to make up the final volume. The PCR conditions were as follows: initial denaturation at 95 °C for 5 min; 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 40 s and elongation at 72 °C for 60–90s; and a final extension step at 72 °C for 10 min.
Amplification of the complete mitochondrial nad1 gene (894 bp) using forward primer (5′-ATT ATA GAA AAT TTT CGT TTT ACA CGC-3′) and reverse primer (5′-ATT CAC AAT TTA CTA TAT CAA AGT AAC C-3′), and complete cox1 gene (1608 bp) using forward primer (5′-ATT ATA GAA AAT TTT CGT TTT ACA CGC-3′) and reverse primer (5′-AAG CAT GAT GCA AAA GGC AAA TAA ACC-3′) [19] was carried out for all isolates. In addition to the above primers, previously designed primers [20] were also used to detect the presence of Echnococcus in faecal samples. PCR products were detected in a 1.5% (w/v) agarose gel stained with GelRedTM. Five microliters of the amplicon was used for visualization while the rest was sequenced in an ABI3730xl DNA Analyser (Beijing Tsingke Biotechnology Co., Ltd., Beijing, China).
Molecular analysis
DNA sequences were viewed and manually corrected for any misread nucleotides using BioEdit software [21]. Resulting gene sequences were aligned with BioEdit [21] and the identity of each isolate was confirmed with their nucleotide sequence in the GenBank database using the NCBI BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Nucleotide and haplotype diversity indices were estimated in DnaSP v.6 [22]. Median-joining network [23] was inferred based on the sequences of mitochondrial cox1, nad1, and cox1–nad1 genes using PopART (http://popart.otago.ac.nz). Population neutrality indices; Tajima’s D [24] and Fu’s Fs [25] were calculated using DnaSP v.6 [22]. Bayesian phylogeny was inferred based on the cox1 and cox1–nad1 dataset using MrBayes v.3.1.2. Markov Chain Monte Carlo (MCMC) sampling was used to assess the posterior distribution of parameters with a chain length of 2,000,000 states, and 25% was discarded as burn-in. Parameters were logged every 1000 states. TreeView v.1.6.6. (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to display tree.
Results
Overall, of livestock examined, 19.49% (23/118) of camels, 0.47% (4/856) of cattle, 0.31% (1/318) of goats and 0% (0/300) of sheep were positive of 32 hydatid cysts from lungs, liver and spleen (Table 1). No infection was detected in all faecal samples examined. Out of the 32 isolates, 31 were successfully amplified for cox1 and nad1 mitochondrial genes and were identified as E. canadensis G6/G7 genotype using the NCBI BLAST algorithm with 99–100% identity. Analysis of the resulting sequences showed a total of 4 mutation sites (cox1= 3, nad1= 1) (Table 2) with 3 parsimony informative sites (nad1= 1, cox1= 2) (Table 3). No insertions or deletions were observed.
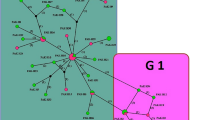
Haplotype network of Echinococcus canadensis
Among the 31 isolates, 4 and 2 haplotypes were found for cox1 and nad1 genes, respectively (Fig. 2a, b). The hosts and geographical origins of the isolates are shown in Table 1 (see Additional file 1: Figure S1 for the haplotype geographical location). Analysis of the concatenated cox1–nad1 (2502 bp) sequences showed 5 distinct haplotypes (Fig. 2c) with haplotype H2 found in both zones (North-West and North-East), constituting about 67.74% of the entire E. canadensis population. Additionally, 3/4 of the cattle isolates (Sokoto, North-West zone) formed a distinct haplotype (H5) (Fig. 2c). Representative cox1 and nad1 haplotype sequences from this study have been deposited in the GenBank database under the accession numbers MN025261–MN025264 (cox1), and MN025265 and MN025266 (nad1).
Neutrality and diversity indices
The diversity and neutrality indices for the entire E. canadensis population were calculated based on the sequences of cox1, nad1, and cox1–nad1 genes and are presented in Table 3. Overall, cox1–nad1 haplotype diversity (Hd) and nucleotide diversity (π) were 0.529 and 0.00024, respectively, while Tajima’s D and Fu’s Fs were negative and insignificant for the entire population (Table 3).
Phylogenetic analysis
The Bayesian phylogeny based on sequences of cox1 and cox1–nad1 mitochondrial genes placed all Nigerian E. canadensis G6/G7 isolates in the same clade with other G6/G7 genotype from different hosts and countries retrieved from GenBank (Fig. 3a, b).
Bayesian phylogeny of Nigerian Echinococcus canadensis G6/G7 inferred from the cox1 (1608 bp) gene (a) and cox1–nad1 (2502 bp) (b) concatenation. Red= Echinococcus canadensis G6/G7 cluster. Posterior probability values are depicted at the nodes. *Indicates haplotypes representing isolates from this study: cox1 haplotypes: MN025261–MN025264 (H1–H4); cox1–nad1 haplotypes: MN025261, MN025266 (H1) MN025261, MN025265 (H2) MN025262, MN025265 (H3) MN025263, MN025265 (H4) MN025264, MN025265 (H5)
Discussion
In Africa, E. canadensis (G6/G7) is the second leading cause of CE after E. granulosus (s.s.) and seems to be the major cause of CE in some countries [26, 27]. Its prevalence is also high in countries like Sudan and Egypt [26,27,28,29,30], with documented evidence in Mali and Mauritania [9, 10] where the population of the suitable intermediate host is high [31]. Similarly, the G6/G7 genotype has also been reportedly common in Middle East countries and Europe [32,33,34]. The peculiarity of this genotype to these countries could possibly have resulted from the ancient trade relationship with countries in northern Africa when camelid hosts served as major means of overland transportation. Although pigs also serve as suitable intermediate host for the G6/G7 genotype, the dearth of data on this intermediate host across Africa limits a clear description of the role of pigs in the epidemiology of the G6/G7 genotype.
The CE prevalence reported in this study showed some degree of variation among intermediate hosts as camels had a higher rate of infection. Previous studies have reported similar results implicating camels as most susceptible. In some areas, prevalence has been found to reach as high as 70% in camels and 40% in other livestock [18, 35, 36], although lower prevalence has also been documented in other locations [37, 38]. The high fertility rate observed in camels suggests that they play a significant epidemiological role in the transmission of CE in Nigeria, especially in the North-West and North-East zones. On the other hand, the situation in the South-West and South-South cannot be completely described at the moment due to the dearth of data. Previously, studies on CE in the Niger Delta areas (South-South) conducted in the last two decades reported prevalences ranging between 24.4–55.9% among livestock and over 80% in dogs [39]. Since then, follow-up studies have not been carried out to update CE epidemiological data in this region. A similar situation is the case for the South-West region where previous studies within the said period estimated CE prevalence at 28% in sheep and goats [40]. Currently, reports from the aforementioned region are seroprevalence estimations [41] which makes the CE status in both region unclear and difficult to describe because of the inability of current serological tools to discriminate between strains/genotypes and/or cross-reactivity with other Taenia species [42,43,44]. The absence of CE in the examined livestock from two South-South cities as reported in this study may not completely reflect the CE situation in the region. However, a possible explanation for this observation could relate to (i) the absence or extremely low population of camels of which they are potentially known to maintain the infection; (ii) the fact that majority of slaughtered animals for consumption in the southern part of Nigeria are reared in the north as only seemingly healthy livestock are transported to the south (since vendors need to be sure that such livestock can withstand the stress of road transportation); and (iii) the different climatic conditions and vegetation cover between northern and southern Nigeria which could impact egg survival. Nonetheless, a detailed longitudinal study in the south compared to the north will be vital in appraising the climatic and transboundary effect on CE prevalence across zones.
In this study, we confirm the presence of the ‘camel’ strain of E. granulosus in camelid and non-camelid hosts (cattle and goats) from the northern part of Nigeria. This report is also in line with previous observations regarding the preponderance of E. canadensis G6/G7 in the West African region. For example, the ‘camel’ strain has been implicated in camels, cattle and humans from Mauritania [8, 9, 11], with a similar finding in a dog from Mali [10].
The low nucleotide and haplotype diversity observed in this study is comparable to previous reports of E. canadensis G6/G7 population in some countries [33, 34, 45]. Additionally, a lower number of haplotypes was found among the 31 isolates using cox1, nad1 and cox1–nad1 genes when compared to haplotypes recorded in Mongolia [46], but similar to the number of haplotypes reported from Hungary [45]. Our result is also in conformity with previous observations of low polymorphism frequently exhibited by E. canadensis G6/G7 populations in most parts of the world [33, 34]. The haplotype and nucleotide diversity based on the 2502 bp cox1–nad1 genes when compared to global G6/G7 clusters recently reported by Addy et al. [34], were more similar to the French G6/7 population than those of northern and eastern African countries and other geographical locations.
The network analysis based on cox1–nad1 concatenated sequences showed that the major haplotype (H2) constituted 67.74% of the entire E. canadensis population and had a 100% homology with other widespread haplotypes in camels from Mauritania, Sudan and Iran [47], goat isolates from Sudan and Argentina [47], and human isolates from Kenya [47]. It was also identical to the Gmon haplotype of human origin from Mongolia [47] and the cox1 human isolates from Russia [48]. The same haplotype, when compared to the recent global G6/G7 clusters, showed 99.78–99.94% similarity to haplotypes from Kenyan camels, goats, dogs and humans; Sudanese camels and humans; French pigs; and Iranian camels and humans [34]. However, the cox1 of H3 and nad1 of H4 were found to be 100% identical to the haplotypes of camel origin from Kenya: Ec04 (GenBank: KX010833) and Ec01 (GenBank: KX010873), respectively.
The similarity observed between Nigerian isolates and those from elsewhere agrees with the initial proposition of non-geographical distinction among G6/G7 haplotypes [34]. This observation was further corroborated by the clustering of the Nigerian isolates with isolates from other hosts and locations as evident in the Bayesian phylogeny of the cox1 and cox1–nad1 concatenated sequences.
Furthermore, to understand the role played by wildlife in the transmission and maintenance of CE, we examined captive lions and hyena for the possibility of infection with Echinococcus since they fed on carcasses of goats/sheep. Clearly, no positive case was seen. However, it will be important in the future to carry out an extensive investigation on both captive and untamed wildlife so as to establish/understand the possible nature of the interface between domestic and wild animals in the transmission of CE across the country.
Conclusions
Our findings confirm camels as the major intermediate host responsible for the maintenance of CE in Nigeria. Although the G1 genotype is responsible for the majority of global CE burden, the case in the West African region following existing reports and our study suggest that the E. canadensis G6/G7 genotype due to the large involvement of camels may be the dominant species responsible for CE in the region. To our knowledge, we provide for the first time insights on the genotype/species of Echinococcus infecting livestock and identify E. canadensis G6/G7 genotype as the major cause of CE infection as well as establish their phylogenetic relationship with other isolates from different hosts/locations. This information could serve as a baseline for further studies. Regardless of the limited sample size analysed in this study, it has enriched CE data for Nigeria and indeed West Africa. We thus suggest that in the future, molecular studies should cover states in Nigeria that are yet to be investigated in order to provide robust data for control.
Availability of data and materials
All data supporting the conclusions of this article are included in the article and its additional file. Representative nucleotide sequences of cox1 and nad1 genes from the present study are available in the GenBank database under the Accession Numbers MN025261–MN025266.
Abbreviations
- WHO:
-
World Health Organization
- bp:
-
base pair or base pairs
- PCR:
-
polymerase chain reaction
- DNA:
-
deoxyribonucleic acid
- cox1:
-
cytochrome c oxidase subunit 1
- nad1:
-
NADH dehydrogenase subunit 1
- CE:
-
cystic echinococcosis
- MCMC:
-
Markov Chain Monte Carlo
- Hd:
-
haplotype diversity
References
WHO. Sustaining the drive to overcome the global impact of neglected tropical diseases: second WHO report on neglected diseases. Geneva: World Health Organization; 2013.
WHO. Echinococcosis. Fact sheet no. 377. Geneva: World Health Organization; 2015.
Aboelhadid SM, El-Dakhly KM, Yanai T, Fukushi H, Hassanin KM. Molecular characterization of Echinococcus granulosus in Egyptian donkeys. Vet Parasitol. 2013;193:292–6.
Azlaf R, Dakkak A. Epidemiological study of the cystic echinococcosis in Morocco. Vet Parasitol. 2006;137:83–93.
Bekele J, Butako B. Occurrence and financial loss assessment of cystic echinococcosis (hydatidosis) in cattle slaughtered at Wolayita Sodo municipal abattoir, southern Ethiopia. Trop Anim Health Prod. 2011;43:221–8.
Getaw A, Beyene D, Ayana D, Megersa B, Abunna F. Hydatidosis: prevalence and its economic importance in ruminants slaughtered at Adama municipal abattoir, Central Oromia, Ethiopia. Acta Trop. 2010;113:221–5.
Romig T, Deplazes P, Jenkins D, Giraudoux P, Massolo A, Craig PS, et al. Ecology and life cycle patterns of Echinococcus species. Adv Parasitol. 2017;95:213–314.
Bart JM, Bardonnet K, Elfegoun MC, Dumon H, Dia L, Vuitton DA, et al. Echinococcus granulosus strain typing in North Africa: comparison of eight nuclear and mitochondrial DNA fragments. Parasitology. 2004;128:229–34.
Farjallah S, Busi M, Mahjoub MO, Slimane BB, Said K, D’Amelio S. Molecular characterization of Echinococcus granulosus in Tunisia and Mauritania by mitochondrial rrnS gene sequencing. Parassitologia. 2007;49:239–46.
Mauti S, Traoré A, Crump L, Zinsstag J, Grimm F. First report of Echinococcus granulosus (genotype G6) in a dog in Bamako, Mali. Vet Parasitol. 2016;217:61–3.
Boue F, El-Berbri I, Hormaz V, Boucher JM, El Mamy AB, Traore A, et al. Use of FTA® card methodology for sampling and molecular characterization of Echinococcus granulosus sensu lato in Africa. Exp Parasitol. 2017;173:29–33.
Lymbery AJ. Phylogenetic pattern, evolutionary processes and species delimitation in the genus Echinococcus. Adv Parasitol. 2017;95:111–45.
Laurimäe T, Kinkar L, Moks E, Romig T, Omer RA, Casulli A, et al. Molecular phylogeny based on six nuclear genes suggests that Echinococcus granulosus sensu lato genotypes G6/G7 and G8/G10 can be regarded as two distinct species. Parasitology. 2018;145:1929–37.
Nakao M, Lavikainen A, Hoberg E. Is Echinococcus intermedius a valid species? Trends Parasitol. 2015;31:342–3.
Carmena D, Cardona GA. Echinococcosis in wild carnivorous species: epidemiology, genotypic diversity, and implications for veterinary public health. Vet Parasitol. 2014;202:69–94.
Wahlers K, Menezes CN, Wong ML, Zeyhle E, Ahmed ME, Ocaido M, et al. Cystic echinococcosis in sub-Saharan Africa. Lancet Infect Dis. 2012;12:871–80.
Dada BJO. Taeniasis, cysticercosis and echinococcosis/hydatidosis in Nigeria: III. prevalence of bovine and porcine cysticercosis, and hydatid cyst infection based on joint examination of slaughtered food animals. J Helminthol. 1980;54:293–7.
Dada BJO, Adegboye DS, Mohammed AN. The epidemiology of Echinococcus infection in Kano State, Nigeria. Ann Trop Med Parasitol. 1980;74:515–7.
Wu YT, Li L, Zhu GQ, Li WH, Zhang NZ, Li SN, et al. Mitochondrial genome data confirm that yaks can serve as the intermediate host of Echinococcus canadensis (G10) on the Tibetan Plateau. Parasit Vectors. 2018;11:166.
Liu CN, Lou ZZ, Li L, Yan HB, Blair D, Lei MT, et al. Discrimination between E. granulosus sensu stricto, E. multilocularis and E. shiquicus using a multiplex PCR assay. PLoS Negl Trop Dis. 2015;9:0004084.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8.
Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao Librado P, Ramos-Onsins SE, Sanchez-Gracia A. DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol. 2017;34:3299–302.
Bandelt H, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Bio Evol. 1999;16:37–48.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95.
Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–25.
Omer RA, Dinkel A, Romig T, Mackenstedt U, Elnahas AA, Aradaib IE, et al. A molecular survey of cystic echinococcosis in Sudan. Vet Parasitol. 2010;169:340–6.
Ahmed ME, Salim B, Grobusch MP, Aradaib IE. First molecular characterization of Echinococcus granulosus (sensu stricto) genotype 1 among cattle in Sudan. BMC Vet Res. 2018;14:36.
Abdel Aaty HE, Abdel-Hameed DM, Alam-Eldin YH, El-Shennawy SF, Aminou HA, Makled SS, et al. Molecular genotyping of Echinococcus granulosus in animal and human isolates from Egypt. Acta Trop. 2012;121:125–8.
Amer S, Helal IB, Kamau E, Feng Y, Xiao L. Molecular characterization of Echinococcus granulosus sensu lato from farm animals in Egypt. PLoS ONE. 2015;10:e0118509.
Alam-Eldin YH, Abdel Aaty HE, Ahmed MA. Molecular characterization of cystic echinococcosis: first record of G7 in Egypt and G1 in Yemen. Acta Parasitol. 2015;60:662–5.
Food and Agriculture Organization of the United Nations (FAO). FAOSTAT: Statistics Division. Production/live animals Nigeria. 2014. http://www.fao.org/faostat/en/#data/QA/visualize. Accessed 8 Apr 2019.
Ebrahimipour M, Sadjjadi SM, Yousofi Darani H, Najjari M. Molecular studies on cystic echinococcosis of camel (Camelus dromedarius) and report of Echinococcus ortleppi in Iran. Iran J Parasitol. 2017;12:323–31.
Karamian M, Haghighi F, Hemmati M, Taylor WR, Salehabadi A, Ghatee MA. Heterogenity of Echinococcus canadensis genotype 6 - the main causative agent of cysticechinococcosis in Birjand, Eastern Iran. Vet Parasitol. 2017;245:78–85.
Addy F, Wassermann M, Kagendo D, Ebi D, Zeyhle E, Elmahdi IE, et al. Genetic differentiation of the G6 / 7 cluster of Echinococcus canadensis based on mitochondrial marker genes. Int J Parasitol. 2017;47:923–31.
Dada BJ, Adegboye DS, Mohammed AN. The epidemiology of Echinococcus infection in Kaduna State, Nigeria. Vet Rec. 1979;104:312–3.
Magaji AA, Onwuegbunam CU, Sonfada ML, Salihu MD. Prevalence of hydatidosis in camels slaughtered in Sokoto Central Abattoir, Sokoto, Nigeria. Bull Anim Health Prod Afr. 2012;60:219–24.
Tijjani AO, Musa HI, Atsanda NN, Mamman B. Prevalence of hydatidosis in sheep and goats slaughtered at Damaturu abattoir, Yobe state, Nigeria. Nig Vet J. 2010;31:71–5.
Igwenagu E, Onyiche ET, Saidu AM, Chahari AM, Waziri A, Kayeri BK. Prevalence of hydatidosis and fertility of hydatid cyst in slaughtered camels in Maiduguri, Nigeria. Ife J Sci. 2018;20:299–303.
Arene FO. Prevalence of hydatid cysts in domestic livestock in the Niger Delta. Trop Anim Health Prod. 1985;17:3–5.
Anyanwale FO, Dipeolu OO, Esuruoso GC. Incidence of Echinococcus infection in dogs, sheep and goats slaughtered in Ibadan, Nigeria. Int J Zoonoses. 1982;9:65–7.
Adediran OA, Kolapo TU, Uwalaka EC. Echinococcus granulosus prevalence in dogs in southwest Nigeria. J Parasitol Res. 2014;2014:124358.
Craig PS, Rickard MD. Studies on the specific immunodiagnosis of larval cestode infections of cattle and sheep using antigens purified by affinity chromatography in any enzyme-linked immunosorbent assay (ELISA). Int J Parasitol. 1981;11:441–9.
Sangaran A, Bino Sundar ST, Latha BR. Antigen based detection of cystic echinococcosis in buffaloes using ELISA and Dot-EIA. J Parasitic Dis. 2016;41:128–30.
Tamarozzi F, Covini I, Mariconti M, Narra R, Tinelli C, De Silvestri A, et al. Comparison of the diagnostic accuracy of three rapid tests for the serodiagnosis of hepatic cystic echinococcosis in humans. PLoS Negl Trop Dis. 2016;10:e0004444.
Dán Á, Rónai Z, Széll Z, Sréter T. Prevalence and genetic characterization of Echinococcus spp. in cattle, sheep, and swine in Hungary. Parasitol Res. 2018;117:3019–22.
Ito A, Dorjsuren T, Davaasuren A, Yanagida T, Sako Y, Nakaya K, et al. Cystic echinococcoses in Mongolia: molecular identification, serology and risk factors. PLoS Negl Trop Dis. 2014;8:e2937.
Laurimae T, Kinkar L, Romig T, Omer RA, Casulli A, Umhang G, et al. The benefits of analysing complete mitochondrial genomes: deep insights into the phylogeny and population structure of Echinococcus granulosus sensu lato genotypes G6 and G7. Infect Genet Evol. 2018;64:85–94.
Konyaev SV, Yanagida T, Ingovatova GM, Shoikhet YN, Nakao M, Sako Y, et al. Molecular identification of human echinococcosis in the Altai region of Russia. Parasitol Int. 2012;61:711–4.
Acknowledgments
We would like to thank the following for their assistance during sample collection: Dr Attahiru Ahmad Rufa’i, Dr Muhammad Ahmad Ibrahim and the staff of Sokoto Central Abattoir, Sokoto State Veterinary Service, Sokoto, Nigeria; Dr Nura Abubakar, Sokoto State Veterinary Clinic, Sokoto, Nigeria; and Dr Saulawa MA, Veterinary Council of Nigeria, Abuja. We are also grateful to Dr Etinosa O. Igbinosa, Head of Applied Microbial Processes & Environmental Health Research Group, University of Benin, Nigeria for allowing us to access his laboratory facilities.
Funding
This study was supported by Central Public-Interest Scientific Institution Basal Research Fund (1610312017001; 1610312016012), National Key Basic Research Programme (973 Programme) of China (2015CB150300) and National Key Research and Development Plan (2017YFD0501301), while John Asekhaen Ohiolei’s PhD scholarship was provided by the Chinese Government Scholarship Programme of the People’s Republic of China.
Author information
Authors and Affiliations
Contributions
JAO, HY and WZJ conceived and designed the experiments. JAO, JL, MEO and AAM collected the samples. JAO, GZ, LL, MAA, YTW, HBY and RJM performed the experiments and the data analyses. JAO wrote the first draft of the manuscript, HBY, CI, BQF and WZJ improved paragraphs and provided constructive suggestions for revisions. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file

Additional file 1: Figure S1.
cox1–nad1 haplotypes geographical distribution. H2 was found in both zones whereas other haplotypes were present only in Sokoto, northwestern Nigeria.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ohiolei, J.A., Yan, HB., Li, L. et al. Cystic echinococcosis in Nigeria: first insight into the genotypes of Echinococcus granulosus in animals. Parasites Vectors 12, 392 (2019). https://doi.org/10.1186/s13071-019-3644-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3644-z