Abstract
Background
Over 60 years ago clinical patterns resembling tick-borne rickettsioses have been described for the first time in Kazakhstan. Since 1995 the incidence of clinical cases of tick-borne rickettsioses in humans seems to be rising but studies on epidemiological data regarding the occurring etiological agents, tick vector species, prevalence and distribution throughout Kazakhstan are still scarce to date. The aim of the study was molecular investigation of ticks for spotted-fever group rickettsiae in the endemic Kyzylorda region and the so far considered as non-endemic Almaty region. A total of 2341 ticks was collected in the two regions in Kazakhstan and sorted in 501 pools: Ixodes persulcatus (243); Dermacentor marginatus (129); Haemaphysalis punctata (104); Hyalomma asiaticum (17); Dermacentor reticulatus (3); and Rhipicephalus turanicus (5). Pools were tested for Rickettsia spp. using real-time PCR. For positive samples multilocus sequence typing (MLST) was performed.
Results
The calculated minimum infection rate (MIR) for rickettsiae in the investigated ticks in Almaty region varied between 0.4–15.1% and 12.6–22.7% in the Kyzylorda region. At least four different Rickettsia species were identified in the two selected regions of Kazakhstan. Two of these are already known to science: Rickettsia raoultii and R. slovaca, the latter being reported for the first time in Almaty region One new form, “Candidatus R. yenbekshikazakhensis”, was described by MLST of six gene fragments in Almaty region and one new genotype, “genotype R. talgarensis” was detected using three gene fragments.
Conclusions
Kazakh physicians should be aware of rickettsioses after tick bites in both regions studied. Both, R. raoultii and R. slovaca should be included in the diagnostics. The role for human diseases has further to be investigated for the newly described rickettsiae, “Candidatus R. yenbekshikazakhensis” and “Genotype R. talgarensis”.
Similar content being viewed by others
Background
Bacteria in the genus Rickettsia are arthropod-transmitted pathogens of vertebrates [1]. Rickettsiae are intracellular parasites, and are symbionts in the broad sense as these have close relationships with their hosts. They are the causative agents of numerous diseases of humans [2] which can occur from subclinical to severe forms [1, 3]. According to recent data, Rickettsia spp. that cause infections in humans are divided into two major groups: the typhus group (Rickettsia prowazekii and Rickettsia typhii) and the spotted fever group (SFG) (Rickettsia rickettsii, Rickettsia slovaca, Rickettsia sibirica, Rickettsia raoultii, Rickettsia conorii, Rickettsia peacockii, Rickettsia honei, Rickettsia japonica, Rickettsia montanensis, Rickettsia massiliae, Rickettsia ripicephali, Rickettsia amblyommii, Rickettsia africae, Rickettsia parkeri, Rickettsia heilongjiangensis, Rickettsia phillipi). The major typhus group includes the typhus group itself and the “ancestral” group with R. bellii subgroup and R. canadensis subgroup. The major spotted fever group consists of the “classical” spotted fever group (R. rickettsia subgroup, R. conorii subgroup, R. australis subgroup) and two transitional groups, R. felis group and R. akari group. Rickettsiae are widespread among arthropods including lice, fleas and most species of ixodid ticks [4,5,6].
The knowledge on the tick-associated rickettsiae and their significance of inducing human diseases has been considerably enhanced in the past three decades The main reason for progress is that molecular methods such as multilocus sequence typing (MLST) or next-generation sequencing have helped to identify new and previously recognized rickettsiae in ticks [7]. MLST led to the description of several new “Candidatus” Rickettsia species by describing at least four or five gene fragments or new Rickettsia genotypes if less than four sequences are characterized [4, 8,9,10].
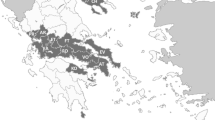
The clinical pictures of human cases of tick-born rickettsioses were first described in Kazakhstan during expeditions to Almaty region in 1949–1951 [11]. A few years later, clinical pictures of tick-borne rickettsioses were described further in five districts i.e. South Kazakhstan, West Kazakhstan, Pavlodar, North Kazakhstan and Akmola regions [12]. The causative agent of the North Asian tick-borne rickettsiosis (R. sibirica) was first described and isolated in 1961 by intra-abdominal infection of guinea pig males with homogenates containing Dermacentor marginatus and Haemaphysalis punctata ticks, which were collected in Yenbekshikazakh district of Almaty region [13]. Since 1995, clinical case definition criteria and a complement fixation test (CFT) with R. sibirica are used in Kazakhstan for diagnostics and consequently official registration of tick-borne rickettsiosis cases in humans. Currently, annual data exist for four regions in Kazakhstan (North Kazakhstan, Pavlodar, East Kazakhstan and Kyzylorda), which are currently considered as endemic regions for tick-borne rickettsioses. According to available statistical data, in total 3904 human cases of tick-borne rickettsiosis were officially registered in Kazakhstan from 1995 to 2016. In this period the incidence rate of this disease increased from 0.41 to 0.87 (per 100,000 inhabitants per year). The biggest increase was observed during this period in Kyzylorda region (incidence of 1.64–11.1 per 100,000 inhabitants per year) and Pavlodar region (incidence of 1.07–7.0 per 100,000 inhabitants per year). According to the currently available data, the Kyzylorda region is supposed to be an endemic area for tick-borne rickettsioses in Kazakhstan [14].
There exist some data on Rickettsia species that might circulate in the endemic regions of Kazakhstan. Rickettsia raoultii was reported in Dermacentor spp. and Ixodes spp. in three regions (Kyzylorda, Karaganda and East Kazakhstan) [15,16,17,18,19,20] and R. conorii caspia, R. raoultii and R. aeschlimannii were detected in ticks collected in the western, northern and central regions of Kazakhstan [21]. Rickettsia aeschlimannii was reported in Haemaphysalis punctata originating from Almaty region [17, 18]. Recently, Hay et al. (2016) [22] demonstrated the presence of R. conorii caspia in ticks engorged on four-striped grass rats (Rhabdomys pumilio) collected in the West Kazakhstan region. In 2017, Rickettsia asembonensis and Rickettsia felis/“Candidatus Rickettsia senegalensis” were detected in fleas collected in Almaty region [22, 23].
So far data concerning Rickettsia spp. from the spotted fever group circulating in the Almaty region are limited and there are no registered epidemiological data on human infections from this region [24]. Currently, there are still large gaps regarding the knowledge on circulating Rickettsia species in ticks and their geographical distribution in Kazakhstan. Here, we present data of a molecular study of ticks for SFG rickettsiae in two regions, the Almaty region, which is considered so far non-endemic but remains the most densely populated region in Kazakhstan, and in the endemic Kyzylorda region.
Methods
Tick sampling
Ticks were collected by flagging the vegetation in three districts of Almaty region (Talgar, Yeskeldy and Yenbekshikazakh districts) and Kyzylorda region (Syrdarya, Shyeli and Zhanakorgan districts), Kazakhstan, in May-June 2015.
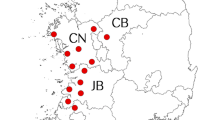
The sampling sites of the ticks of Almaty region have the following characteristics: Yeskeldi district (44°54′12″N, 78°29′42″E) with Tekeli city (44°49′48″N, 78°49′26″E) is located in Almaty region adjacent to the Peopleʼs Republic of China and is characterized by coniferous forests and open steppe vegetation. The region is mountainous with an altitude of 1400–2200 m above sea level (masl). More than 40% of the area is covered by forest, and the remaining parts constitute of pasture and agricultural land. Animal husbandry is practiced widely. The average annual precipitation has been reported as 250–300 mm [25]. Talgar district (43°18′55″N, 77°14′35″E) with Talgar city (43°18′0″N, 77°14′0″E) is 40 km away from Almaty city and comprises forested taiga, forested steppe and arid fields, the latter mainly covered by gramineous plants. Nearly 20% of its northern part is at an altitude of 1800–2400 masl. The annual precipitation is 200–300 mm [25]. Yenbekshikazakh district (43°21′0″N, 77°28′0″E) with Yesyk city (43°21′0″N, 77°28′0″E) has an altitude of 560–1300 masl and offers areas of maritime climate in summer and very cold temperatures (from -25 °C down to -50 °C) during winter. Average precipitation is reported as 200–700 mm/year. Plain steppe and meadows dominate most parts of this area [25].
In the Kyzylorda region, ticks were collected in three districts: Syrdarya (45°34′12″N, 65°36′0″E), Shyeli (44°10′0″N, 66°44′0″E) and Zhanakorgan (43°56′24″N, 67°13′12″E). Kyzylorda region (45°0′0″N, 64°0′0″E) is located in the south-western part of Kazakhstan, to the east of the Aral Sea in the lower reaches of the River Syrdarya, mainly within the Turan Lowland (altitude of 50–200 masl). The region borders the neighboring country Uzbekistan, as well as three other Kazakh regions: Aktobe region (to the west), Karaganda region (to the north), and South Kazakhstan (to the east). The climate is rather continental and extremely arid with prolonged hot and dry summers and with a comparatively warm, short and moderate winter. The amount of precipitation in the north-west near the Aral Sea coast is about 100 mm (the lowest in Kazakhstan) and up to 175 mm in the southeast, in the foothills of Karatau Mountain. A significant part of the region is occupied by sands, almost devoid of vegetation [26].
Sample preparation
The collected ticks were stored at -20 °C until further study. The laboratory study was conducted in batches. After thawing, all field ticks have been sorted by genus, species, stage and sex following the official guidelines for tick specification in Kazakhstan [27,28,29,30]. Next, the ticks were grouped into pools by genus, species, stage and sex (with a maximum of 5 adult ticks in a pool). Each pool has been homogenized using the TissueLyser II instrument, after adding ceramic granules and 1 ml medium Dulbecco’s Modified Eagle Medium (DMEM) (BioloT, Saint-Petersburg, Russia) to each tube. Following Kazakh guidelines for biosafety and biosecurity, aliquots containing tick homogenates were inactivated in a water bath at 56 °C for 30 min, before DNA extraction. DNA was extracted from 200 μl tick homogenates using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions.
PCR and species identification of rickettsiae
All PCRs were conducted in a three-room-regime. The presence of rickettsial DNA was determined by a real-time PCR assay targeting the pan-rickettsial citrate synthase gene using the primers PanRick_gltA_2_for (5′-ATA GGA CAA CCG TTT ATT T-3′), PanRick_gltA_2_rev (5′-CAA ACA TCA TAT GCA GAA A-3′), PanRick_gltA_2_taq (5′-6FAM-CCT GAT AAT TCG TTA GAT TTT ACC G-DB-3′) and Uracil-DNA-glycosylase (UDG) in order to eliminate carry-over contamination [31, 32] in a Rotor-Gene Q (Qiagen) machine.
In all samples yielding a positive pan-rickettsial citrate synthase gene signal, a multilocus sequence typing (MLST) targeting six gene fragments (partial fragments of ompB, ompAIV, 23S-5S interspacer region, 16S, sca4, gltA) was conducted for Rickettsia species identification [31,32,33,34]. First, for all these samples the partial outer membrane protein B (ompB) gene (RR 120–2788, cRR 120–3599) was amplified following published protocols [34]. Secondly, for the samples with positive sequences (depending on the sequence result) five additional fragments were investigated using previously published primers [31,32,33,34,35,36,37,38]: ompAIV (RR 190–5125, cRR 190–6013), 23S-5S interspacer region (23s for, 23s rev), 16S (Ric, Ric RT), sca4 (Rsca4_1707f, Rsca4_2837r) and partial gltA (Rh314, Rh654) [31,32,33,34].
Master mix solution for 23S-5S interspacer region PCR (50 µl including 5 µl DNA) were prepared with 0.2 mM dNTP Mix (Thermofisher-Invitrogen, Schwerte, Germany), 0.5 µM of each primer (23s for, 23s rev), 1.5 U Platinum® Taq DNA Polymerase High Fidelity (Thermofisher-Invitrogen), 1× PCR buffer (Thermofisher) and 2.0 mM MgSO4 (Thermofisher). The initial denaturation was performed for 3 min at 95 °C, 45 cycles of amplification each starting with denaturation for 20 s at 95 °C, followed by annealing for 30 s at 57 °C and elongation for 60 s at 68 °C, and a final elongation step at 68 °C for 10 min [35, 39].
Gene D (sca4) sequences were amplified with 0.2 mM dNTP Mix (Thermofisher-Invitrogen), 0.1 µM of each primer (Rsca4_1707f, Rsca4_2837r), 1.0 U Platinum® Taq DNA Polymerase High Fidelity (Thermofisher-Invitrogen), 1× PCR buffer, 3 mM MgSO4 and 2 µl DNA in a final reaction volume of 50 µl. After an initial denaturation for 3 min at 95 °C, 40 cycles with denaturation for 30 s at 95 °C, annealing for 35 s at 53 °C and elongation for 90 s at 68 °C, and a final extension step at 68 °C for 7 min [37, 39].
Partial 16S sequences were amplified using 0.2 mM dNTP Mix (Thermofisher-Invitrogen), 0.5 µM of each primer (Ric, Ric RT) with 1.5 U Platinum® Taq DNA Polymerase High Fidelity (Thermofisher-Invitrogen), 1× PCR buffer, 2.5 mM MgSO4 and 5 µl DNA in a final reaction volume of 50 µl. After an initial denaturation for 3 min at 95 °C, 45 cycles with denaturation for 30 s at 94 °C, annealing for 30 s at 63 °C, and elongation for 120 s at 68 °C were performed, followed by a final extension step at 68 °C for 7 min [36, 39].
The partial gltA gene was amplified with 0.2 mM dNTP Mix (Thermofisher- Invitrogen), 0.5 µM of each primer (Rh314, Rh654), 1.0 Unit Platinum® Taq DNA Polymerase High Fidelity (Thermofisher-Invitrogen), 1× PCR buffer, 2 mM MgSO4 and 5 µl DNA in a final reaction volume of 50 µl. After an initial denaturation for 2 min at 94 °C, 45 cycles with denaturation for 20 s at 94 °C, annealing for 30 s at 54 °C and elongation for 60 s at 68 °C were conducted, followed by a final extension step at 68 °C for 5 min [38, 39].
PCR products were visualized in a 1.5% agarose gel and purified using the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s recommendations. PCR product sequencing was carried out using the ABI Prism BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and 3500xl Genetic Analyzer (Hitachi, Japan) with the primers used for the PCR amplifications. pGEM-3Zf(+) control template was used as a sequencing control. Quantification of the PCR products was performed on a Fluorometer Qubit 2.0 (Invitrogen, USA). Sequence analyses were carried out with Chromas Lite 2.01 [40] and Bioedit 7.2.5. [41]. Obtained sequences were compared with sequences from GenBank using BLAST 2.2.32 [42, 43].
Phylogenetic trees were constructed using the Maximum Likelihood method based on the Tamura 3-parameter model [44] with the software package MEGA 6 [45]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log-likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
The binomial (Clopper-Pearson) ‘exactʼ method based on the beta distribution was used for the calculation of 95% confidence intervals (CI).
The minimum infection rate (MIR) was calculated as the ratio of the number of positive tick pools to the total number of ticks of the same species.
Results
Six tick species collected from 6 districts of Almaty and Kyzylorda regions in Kazakhstan were studied: Ixodes persulcatus (n = 1193; 243 pools); Dermacentor marginatus (n = 578; 129 pools); Haemaphysalis punctata (n = 470; 104 pools); Hyalomma asiaticum (n = 77; 17 pools); Dermacentor reticulatus (n = 14; 3 pools); and Rhipicephalus turanicus (n = 9; 5 pools). Ixodes persulcatus, D. marginatus and H. punctata were the most abundant tick species collected. Four tick species out of six (I. persulcatus, H. punctata, D. marginatus and D. reticulatus) were detected in Almaty region, with three of them (I. persulcatus, H. punctata and D. reticulatus) found only in this region. Three tick species out of six (Hy. asiaticum, Rh. turanicus, D. marginatus) were collected in Kyzylorda region, with two of them (Hy. asiaticum and Rh. turanicus) found only from this region. Dermacentor marginatus was collected more often in Kyzylorda region than in Almaty region (Table 1). Figure 1 provides an overview of collection sites and tick species studied.

(modified after [14])
Incidence of tick-borne rickettsiosis in Kazakhstan based on complement fixation test
The overall prevalence as per minimum infection rate (MIR) of rickettsial DNA in the tick species and in the collecting localities (Almaty and Kyzylorda regions) was 42.3% (212/501; 95% CI: 37.9–46.8%). The largest number of the Rickettsia partial gltA real-time PCR-positive tick pools was determined in Dermacentor spp. (128/132; 97.0%; 95% CI: 92.4–99.2%) and H. punctata (79/104; 76.0%; 95% CI: 66.6–83.8%) (Table 1) collected from three selected districts of Kyzylorda region (Rickettsia prevalence 56.8–100%) and in Yenbekshikazakh district (79/113; 69.9%; 95% CI: 60.6–78.2%) of Almaty region (Table 2). The smallest number of the Rickettsia partial gltA real-time PCR-positive tick pools was found in I. persulcatus (3/243;1.2%; 95% CI: 0.3–3.6) and H. asiaticum (1/17; 5.9%; 95% CI: 0.2–28.7%) (Table 1) collected from Almaty region (Rickettsia prevalence 1.9–11.5%) and in Zhanakorgan district (25/44; 56.8%; 95% CI: 41–71.7%) of Kyzylorda region (Table 3). All Rh. turanicus from Zhanakorgan district of Kyzylorda region were negative in the screening real-time PCR (Table 1).
Moreover, the MIR was calculated for each tick species and for both selected regions in this study (Tables 1 and 2). A high MIR of rickettsiae was detected in Dermacentor (MIR = 21.4–21.6%) and Haemaphysalis (MIR = 17.0%) ticks (Table 2) collected from Yenbekshikazakh district (Almaty region, MIR = 15.1%) and from the three districts of Kyzylorda region (MIR = 12.6–22.7%) (Table 3).
As a result of MLST, BLAST and phylogenetic analyses, rickettsiae could be determined to the species level for 209 of the 212 Rickettsia-positive samples. A total of four Rickettsia spp. were identified in the molecular study (Tables 4, 5, 6, Figs. 2, 3, 4, 5, 6, 7, 8). Two already known species, R. raoultii and R. slovaca, were identified. Rickettsia raoultii was confirmed in 124 samples (124/209; 59.3%; 95% CI: 52.3–66.1%) by sequencing partial ompB (n = 123), partial ompAIV (n = 9) and 23S-5S (n = 9), in D. marginatus, D. reticulatus and Hy. asiaticum from Kyzylorda region (n = 113) and in the vicinities of Tekeli city in Almaty region (n = 11). Further, R. slovaca (n = 3) by partial ompB (n = 2), partial ompAIV (n = 1), 23S-5S (n = 2) genes was detected in D. marginatus pools only around Tekeli city in Almaty region (3/209; 1.4%; 95% CI: 0.3–4.1%), (Tables 4, 5, 6, Figs. 3, 4, 5, 6, 7, 8).
Maximum Likelihood phylogenetic tree based on 226 partial ompB DNA sequences, with 203 sequences originating from amplificates from Kazakh tick DNA and 23 from the GenBank database. 124 sequences from Kazakh ticks were 100% identical to R. raoultii, two were 100% identical to R. slovaca, and 77 sequences formed a new cluster “Candidatus Rickettsia yenbekshikazakhensis” (76 sequences from Yenbekshikazakh district, 1 from Yeskeldy district-Tekeli city). The tree with the highest log-likelihood (-3541.6714) is shown. There were a total of 806 positions in the final dataset
Maximum Likelihood phylogenetic tree based on 62 partial ompAIV sequences, with 44 sequences originating from amplificates from Kazakh tick DNA and 20 from the GenBank database. 10 sequences from Kazakh ticks were 100% identical to R. raoultii, one sequence was identical to R. slovaca. 30 sequences formed a new cluster “Candidatus Rickettsia yenbekshikazakhensis” (29 sequences from Yenbekshikazakh district and 1 from Yeskeldy district around Tekeli city) and three a new cluster “genotype Rickettsia talgarensis” (1 sequence from Yenbekshikazakh district, 2 from Yeskeldy district-Tekeli city). There were a total of 864 positions in the final dataset. The tree with the highest log-likelihood (-1803.5066) is shown
Maximum Likelihood phylogenetic tree based on 40 partial 23S-5S sequences, with 22 sequences originating from Kazakh ticks and 18 from GenBank. Nine sequences from Kazakh ticks were 100% identical to R. raoultii, two sequences were identical to R. slovaca. Nine sequences Yenbekshikazakh district formed a new cluster “Candidatus Rickettsia yenbekshikazakhensis“. There were a total of 367 positions in the final dataset. The tree with the highest log-likelihood (-1572.3294) is shown
Maximum Likelihood phylogenetic tree based on partial 27 partial 16S sequences, with 8 sequences originating from Kazakh ticks and 19 from GenBank. Six sequences formed a new cluster “Candidatus Rickettsia yenbekshikazakhensis” (5 sequences from Yenbekshikazakh district, 1 from Yeskeldy district-Tekeli city) and two sequences from DNA of ticks from Tekeli the new cluster “genotype Rickettsia talgarensis”. There were a total of 717 positions in the final dataset. The tree with the highest log-likelihood (-1287.3794) is shown
Maximum Likelihood phylogenetic tree based on f partial 57 sca4 sequences with 34 sequences originating from Kazakh tick DNAs (33 from Yenbekshikazakh district, 1 from Yeskeldy district-Tekeli city) and 23 from GenBank. There were a total of 1.115 positions in the final dataset. The tree with the highest log-likelihood (-4809.7101) is shown
Maximum Likelihood phylogenetic tree based on partial 55 partial gltA sequences, with 35 sequences originating from Kazakh tick DNAs forming the new “Candidatus Rickettsia yenbekshikazakhensis” (34 from Yenbekshikazakh district, 1 from Yeskeldy district around Tekeli city) and 20 from GenBank. There were a total of 318 positions in the final dataset. The tree with the highest log-likelihood (-641.7358) is shown
The new “Candidatus R. yenbekshikazakhensis” was confirmed in 80 samples (80/209; 38.2%; 95% CI: 31.7–45.2%) and by MLST of the partial fragments of ompB (n = 77), ompAIV (n = 30), 23S-5S (n = 9), sca4 (n = 34) and 16S (n = 6), gltA (n = 35) genes (Tables 4, 5, 6, 7). “Candidatus Rickettsia yenbekshikazakhensis” was identified in 16.0% of all 501 tick pools studied (80/501; 95% CI: 12.9–19.5%). This rickettsia was detected with a high prevalence in H. punctata pools (80/104, 76.9%) from Yenbekshikazakh district (n = 78) and in the vicinities of Tekeli city (n = 1) in Almaty region, respectively. This “Candidatus” species was detected only in one tick species (H. punctata) collected from two districts (Yeskeldy and Yenbekshikazakh districts) of Almaty region (Tables 4, 5, 7, Figs. 3, 4, 5, 6, 7, 8).
We further detected the new “genotype R. talgarensis” in three samples by analysis of the partial ompAIV (n = 3), 23S-5S (n = 2) and 16S (n = 2) genes in the ticks (3/209, 1.5%) in 0.6% of all 501 tested tick pools (3/501; 95% CI: 0.1–1.7%) (n = 2341). This genotype was only present in I. persulcatus collected in two districts (Talgar and Yenbekshikazakh) in Almaty region (Tables 4, 5, 7, Figs. 3, 4, 5, 6, 7, 8).
Unfortunately, one sample could not be sequenced and in two samples a mixture of different Rickettsia species was detected by sequencing of gene fragments. Overlapping chromatograms indicating a mixture of sequences were found for partial ompAIV, gltA, 16S and sca4 sequences for sample Kyzylorda 061 (D. marginatus, Kyzylorda region, Shieli district) and in the ompB, ompAIV and gltA sequences for sample Tekeli 076 (D. marginatus, Almaty region, Yeskeldy district), respectively.
Discussion
To our knowledge, this study is the first large-scale, comprehensive investigation of rickettsiae of the spotted fever group conducted in two selected pilot regions of Kazakhstan. The difference in natural landscapes in both selected regions explains the variety of collected ticks. Dermacentor marginatus is the most abundant tick typically found at the collection sites in the desert and semi-desert landscape of Kyzylorda region [26] which mirrors the habitat of this tick species [46, 47]. In comparison, the three selected collection sites in the Almaty region are characterized by the presence of a mountainous landscape covered with forests which are the classical habitats for Ixodes spp. [46] exhibiting in Almaty region the highest abundance of all tick species (48.1%). Almaty region showed the wider variety of tick species with five out of the seven species identified in this study (I. persulcatus, H. punctata, D. marginatus and D. reticulatus).
The identification of the ticks investigated in this study was performed using morphological markers [27,28,29,30]. For D. marginatus and D. niveus, there is an ongoing discussion if these two species are conspecific. Genetic markes seem to give evidence for that despite a detailed comparison is still missing [48,49,50]. Herein, both morphologically different species were summed up and data presented as data for Dermacentor marginatus.
Our results show that five of the seven collected tick species are positive for Rickettsia spp. In general, in Kyzylorda region where Dermacentor spp. dominated, 56.8–100% of the ticks’ pools were Rickettsia-positive, and only R. raoultii was found in the two species of Dermacentor. Surprisingly at the three collection sites in the Almaty region, which has been considered so far as a non-endemic region, all four Rickettsia species detected in this study were found. Rickettsia raoultii was detected in 59% of the tick pools and R. slovaca was detected in three pools; both species are human pathogens. The present data indicate that the main vectors of these two pathogens are ticks of the genus Dermacentor, which is in line with data from neighboring countries, i.e. Russian Federation, Mongolia or northwestern China which is located close to the Almaty region of Kazakhstan [7, 51,52,53,54,55,56,57,58,59]. Of note, in our study R. raoultii was also for the first time detected in one Hy. asiaticum tick pool collected from Kyzylorda region of Kazakhstan.
Rickettsia. raoultii and R. slovaca are known human pathogens that cause the scalp eschar and neck lymph adenopathy after a tick bite (SENLAT), tick-borne lymphadenopathy (TIBOLA) or Dermacentor-borne necrosis erythema lymphadenopathy (DEBONEL) after a tick bite [7, 60]. The high MIR of R. raoultii in the ticks studied and a recent case study in China were 26 cases of R. raoultii infections with varying severity were described [61], indicate that R. raoultii should be included in the diagnosis of rickettsioses in Kazakhstan. The occurrence of R. slovaca was previously described in Melophagus ovinus, the sheep ked, collected in localities of Xinjiang Uygur Autonomous Region (northwestern China), that borders the Almaty region of Kazakhstan [58]. The detection of R. slovaca leads to the conclusion that further data on its natural foci in Kazakhstan as well as the role for human infections are needed.
We here report a new “Candidatus R. yenbekshikazakhensis” by performing a MLST of six gene fragments. For the ompB, 23S-5S, 16S and sca4 but not for the ompAIV and gltA it fulfills the criteria of Fournier et al. [8] to designate it as a new “Candidatus” species (Table 7). It has been suggested to taxonomically classify rickettsiae as new “Candidatus” if at least four or five sequences are newly described [4, 8,9,10]. The closest species is R. massiliae which is also known to be pathogenic to humans inducing a SENLAT syndrome [62, 63]. The new “Candidatus R. yenbekshikazakhensis” was detected in two regions and in 87.6% of all H. punctata ticks studied, which might therefore be its main vector.
Further, the “genotype R. talgarensis” was detected in three tick pools. The analysis of three gene fragments, ompAIV, 23S-5S and 16S could be performed showing a quite high divergence to all known rickettsiae (Table 7). The detected agent fulfills therefore, the criteria to be described as a new genotype [8]. For both, “Candidatus R. yenbekshikazakhensis” and “genotype R. talgarensis” the pathogenicity is still unknown and should be the aim of further studies.
Conclusions
The clinical cases of tick-borne rickettsioses, which were registered by using CFT over the past 20 years in Kazakhstan, are so far not confirmed by other serological methods such as ELISA and by pathogen detection (e.g. rickettsial DNA by PCR). With the rising evidence on the relevance of rickettsiae in human infections and for improving epidemiological data, routine laboratory diagnostic tools must be implemented in all reporting laboratories in Kazakhstan. Our data also indicate that clinicians should be aware of SENLAT syndrome which is caused by two confirmed pathogens (R. raoultii and R. slovaca) circulating in the territory of Almaty and Kyzylorda regions. The present data indicate that tick-borne rickettsiae and associated pathological conditions in humans should be further investigated in all regions of Kazakhstan to estimate the importance and clinical impact caused by all four described rickettsiae.
Abbreviations
- CFT:
-
complement fixation test
- DEBONEL:
-
Dermacentor-borne necrosis erythema lymphadenopathy
- MCL:
-
maximum composite likelihood
- MIR:
-
minimum infection rate
- MLST:
-
multilocus sequence typing
- PCR:
-
polymerase chain reaction
- SENLAT:
-
scalp eschars and neck lymphadenopathy
- TIBOLA:
-
tick-borne lymphadenopathy
References
Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10:694–719.
Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc Biol Sci. 2006;273:2097–106.
Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world, emerging diseases challenging old concepts. Clin Microbiol Rev. 2005;18:719–56.
Fournier PE, Raoult D. Current knowledge on phylogeny and taxonomy of Rickettsia spp. Ann N Y Acad Sci. 2009;1166:1–11.
Murray GG, Weinert LA, Rhule EL, Welch JJ. The phylogeny of Rickettsia using different evolutionary signatures: how tree-like is bacterial evolution? Syst Biol. 2016;65:265–79.
Shpynov SN, Fournier PE, Pozdnichenko NN, Gumenuk AS, Skiba AA. New approaches in the systematics of rickettsiae. New Microbes New Infect. 2018;23:93–102.
Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world, a geographic approach. Clin Microbiol Rev. 2013;26:657–702.
Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoul D. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–65.
Raoult D, Fournier PE, Eremeeva M, Graves S, Kelly PJ, Oteo JA, Sekeyova Z, Tamura A, Tarasevich I, Zhang L. Naming of rickettsiae and rickettsial diseases. Ann N Y Acad Sci. 2005;1063:1–12.
Merhej V, Angelakis E, Socolovschi C, Raoult D. Genotyping, evolution and epidemiological findings of Rickettsia species. Infect Genet Evol. 2014;25:122–37.
Bartoshevich EN. To the issue of rickettsioses. Health Care of Kazakhstan. 1952;3:20–4 (In Russian).
Kereyev NI. Human natural focal diseases in Kazakhstan. Alma-Ata. 1965:98–119 (In Russian).
Arkhangelskiy DS. Experimental study of tick-borne rickettsial pathogen in Almaty region. In: Collection of scientific papers of the Institute of Microbiology and Virology, Vol. 4. Physiology and ecology of microorganisms. Alma-Ata. 1961. p. 176–85 (In Russian).
Epidemiological situation of infectious diseases in the Republic of Kazakhstan from 2016. Annual report from Scientific Practical Center of Sanitary Epidemiological Expertise and Monitoring. Almaty: Kazakhstan; 2016. (In Russian).
Shpynov S, Parola P, Rudakov N, Samoilenko I, Tankibaev M, Tarasevich I, Raoult D. Detection and identification of spotted fever group rickettsiae in Dermacentor ticks from Russia and central Kazakhstan. Eur J Clin Microbiol Infect. Dis. 2001;20:903–5.
Shpynov SN, Rudakov NV, Tarasevich IV, Tankibayev MA. Genotyping of rickettsiae and erlichiae from Ixodes ticks in Russia and Kazakhstan. In: Gene diagnosis of infectious diseases. 4th all-Russian scientific and practical conference. Moscow, 2002. p. 256–57 (In Russian).
Shpynov S, Fournier PE, Rudakov N, Tankibaev M, Tarasevich I, Raoult D. Detection of a rickettsia closely related to Rickettsia aeschlimannii, “Rickettsia heilongjiangensis”, Rickettsia sp. strain RpA4, and Ehrlichia muris in ticks collected in Russia and Kazakhstan. J Clin Microbiol. 2004;42:2221–3.
Shpynov S, Rudakov N, Yastrebov V. Identification of new genotypes of rickettsia tick-borne spotted fever group in the south of the Ural, Siberia, Far East and Kazakhstan. Epidemiol Inf Dis. 2005;1:23–7 (In Russian).
Yegemberdiyeva R, Shapiyeva Zh. Clinical and epidemiological characteristic of tick-borne rickettsiosis in Kazakhstan. In: Abstract book of the international conference on zoonoses. Ulaanbaatar. 2008. p. 48–51 (In Russian).
Kyraubayev K, Shapiyeva Zh, Utegenova E, Zhandosov S, Beysenaeva M, Ziyadina L, et al. Study of Dermacentor marginatus ticks for rickettsiae in central Kazakhstan. In: Abstract Book of 114th general meeting of ASM. Boston. 2014. p. 139.
Rudakov NV, Shpynov SN, Samoilenko IE, Tankibaev MA. Ecology and epidemiology of spotted fever group rickettsiae and new data from their study in Russia and Kazakhstan. Ann N Y Acad Sci. 2003;990:12–24.
Hay J, Yeh KB, Dasgupta D, Shapieva Zh, Omasheva G, Deryabin P, et al. Biosurveillance in central Asia: successes and challenges of tick-borne disease research in Kazakhstan and Kyrgyzstan. Front Publ Health. 2016;4:1–6.
Sansyzbayev Y, Nurmakhanov T, Berdibekov A, Vilkova A, Yeskhodzhayev O, St John HK, et al. Survey for rickettsiae within fleas of great gerbils, Almaty oblast, Kazakhstan. Vector Borne Zoonotic Dis. 2017;17:172–8.
Yegemberdiyeva R, Dmitrovskiy A, Shapiyeva Zh, et al. Nozological structure and distribution of tick-borne infections in Kazakhstan. Natl Priorities Russia. 2016;4:30–3 (In Russian).
Almaty region of Kazakhstan. http://www.zhetysu.gov.kz/ru/o-regione. Accessed 10 May 2017 (In Russian).
Kyzylorda region of Kazakhstan. https://e-kyzylorda.gov.kz/?q=ru/content/prirodno-klimaticheskie-usloviya. Accessed 10 May 2017 (In Russian).
Pomerantsev BI. Ixodid ticks. Ixodidae. In: Fauna SSSR Fauna of the USSR. Arachnids, vol 4. Leningrad: USSR Academy of Sciences Publ.; 1950 (In Russian).
Philippova NA. Ixodid ticks. Ixodinae. In: Fauna SSSR Fauna of the USSR. Arachnids, 3rd edn. Moscow: Nauka; 1977 (In Russian).
Fedorova SJ. Key identification of Ixodidae at Kirghizstan. Issledovaniya Zhivoy Prirody Khirghizstana. 2013;1:47–54 (in Russian).
Gugliemone AA, Robbins RG, Apanaskevich DA, Petney TN, Estrada-Pena A, Horak IG, et al. The Argasidae, Ixodidae and Nuttalliellidae (Acari: Ixodida) of the world: a list of valid species names. Zootaxa. 2010;2528:1–28.
Woelfel R, Essbauer S, Dobler G. Diagnostics of tick-borne rickettsioses in Germany: a modern concept for a neglected disease. Int J Med Microbiol. 2008;29:368–74.
Schex S, Dobler G, Riehm J, Müller J, Essbauer S. Rickettsia spp. in wild small mammals in Lower Bavaria, south-eastern Germany. Vector Borne Zoonotic Dis. 2011;11:493–502.
Fournier PE, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48:839–49.
Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol. 2000;50:1449–55.
Jado I, Escudero R, Gil H, Jiménez-Alonso MI, Sousa R, García-Pérez AL, et al. Molecular method for identification of Rickettsia species in clinical and environmental samples. J Clin Microbiol. 2006;44:4572–6.
Nilsson K, Jaenson T, Uhnoo I, Lindquist O, Pettersson B, Uhlén M, et al. Characterization of a spotted fever group Rickettsia from Ixodes ricinus ticks in Sweden. J Clin Microbiol. 1997;35:243–7.
Matsumoto K, Inokuma H. Identification of spotted fever group Rickettsia species by polymerase chain reaction-restriction fragment length polymorphism analysis of the sca4 gene. Vector Borne Zoonotic Dis. 2009;9:747–9.
Nilsson K, Lindquist O, Liu A, Jaenson T, Friman G, Påhlson C. Rickettsia helvetica in Ixodes ricinus ticks in Sweden. J Clin Microbiol. 1999;37:400–3.
Essbauer S, Hofmann M, Kleinemeier C, Wölfel S, Matthee S. Rickettsia diversity in southern Africa: a small mammal perspective. Ticks Tick Borne Dis. 2018;9:288–301.
Aktas M, Özübek S, Altay K, Ipek ND, Balkaya İ, Utuk AE, Kırbas A, Şimsek S, Dumanlı N. Molecular detection of tick-borne rickettsial and protozoan pathogens in domestic dogs from Turkey. Parasit Vectors. 2015;8:157.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis programme for Windows 95/98/NT. Nucl Acids Symp. 1999;41:95–8.
Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14.
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;4:1757–64.
Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol. 1992;9:678–87.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Filippova NA. Ixodid ticks of subfamily Amblyomminae. Fauna of Russia and neighboring countries. St. Petersburg Nauka. 1997;4:244 (In Russian).
Ramezani Z, Chavshin AR, Telmadarraiy Z, Edalat H, Dabiri F, Vatandoost H, et al. Ticks (Acari: Ixodidae) of livestock and their seasonal activities, northwest of Iran. Asian Pacific J Trop Dis. 2014;4:S574–758.
Estrada-Pena A, Estrada-Pena R. Notes on Dermacentor ticks: redescription of Dermacentor marginatus with the synonymies of Dermacentor niveus and Dermacentor daghestanicus (Acari: Ixodidae). J Med Entomol. 1991;28:2–15.
Filippova NA, Plaksina MA. Some aspects of intraspecific variability of the closely related species of the Dermacentor marginatus complex (Acari: Ixodidae) as demonstration of microevolutionary process]. Parazitologiya. 2005;39:337–64 (In Russian).
Moshaverinia A, Shayan P, Nabian S, Rahbari S. Genetic evidence for conspecificity between Dermacentor marginatus and Dermacentor niveus. Parasitol Res. 2009;105:1125–32.
Shpynov S, Fournier PE, Rudakov N, Tarasevich I, Raoult D. Detection of members of the genera Rickettsia, Anaplasma, and Ehrlichia in ticks collected in the Asiatic part of Russia. Ann NY Acad Sci. 2006;1078:378–83.
Samoylenko I, Shpynov S, Raoult D, Rudakov N, Fournier PE. Evaluation of Dermacentor species naturally infected with Rickettsia raoultii. Clin Microbiol Infect. 2009;15:305–6.
Rudakov NV, Schpynov SN, Samoylenko IE, Fournier PE, Reschetnikova TA, Kumpan LV, Raoult D. Characterisation of the Omsk collection of rickettsial strains. Clin Microbiol Infect. 2009;15:298–9.
Speck S, Derschum H, Damdindorj T, Dashdavaa O, Jiang J, Kaysser P, et al. Rickettsia raoultii, the predominant Rickettsia found in Mongolian Dermacentor nuttalli. Ticks Tick Borne Dis. 2012;3:227–31.
Tian Z, Liu G, Shen H, Xie J, Luo J, Tian M. First report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasit Vectors. 2012;5:19.
Guo LP, Mu LM, Xu J, Jiang SH, Wang AD, Chen CF, et al. Rickettsia raoultii in Haemaphysalis erinacei from marbled polecats, China-Kazakhstan border. Parasit Vectors. 2015;8:461.
Liu D, Wang Y, Zhang H, Liu Z, Wureli H, Wang S, Tu C, Chen C. First report of Rickettsia raoultii and R. slovaca in Melophagus ovinus, the sheep ked. Parasit Vectors. 2016;9:1–6.
Boldbaatar B, Jiang RR, von Fricken ME, Lkhagvatseren S, Nymadawa P, Baigalmaa B, Wang YW, Anderson BD, Jiang JF, Gray GC. Distribution and molecular characteristics of rickettsiae found in ticks across Central Mongolia. Parasit Vectors. 2017;10:61.
Han R, Yang J, Niu Q, Liu Z, Chen Z, Kan W, Hu G, Liu G, Luo J, Yin H. Molecular prevalence of spotted fever group rickettsiae in ticks from Qinghai Province, northwestern China. Infect Genet Evol. 2018;57:1–7.
Ibarra V, Oteo JA, Portillo A, Santibáñez S, Blanco JR, Metola L, et al. Rickettsia slovaca infection: DEBONEL/TIBOLA. Ann N Y Acad Sci. 2006;1078:206–14.
Li H, Zhang PH, Huang Y, Du J, Cui N, Yang ZD, et al. Isolation and identification of Rickettsia raoultii in human cases: a surveillance study in three medical centers in China. Clin Infect Dis. 2018;66:1109–15.
Cascio A, Torina A, Valenzise M, Blanda V, Camarda N, Bombaci S, et al. Scalp eschar and neck lymphadenopathy caused by Rickettsia massiliae. Emerg Infect Dis. 2013;19:836–7.
Zaharia M, Popescu CP, Florescu SA, Ceausu E, Raoult D, Parola P, Socolovschi C. Rickettsia massiliae infection and SENLAT syndrome in Romania. Ticks Tick Borne Dis. 2016;7:759–62.
Acknowledgements
The authors gratefully acknowledge the tick collectors from sanitary epidemiological stations of Almaty and Kyzylorda regions. We thank the staff of Scientific Practical Center for Sanitary Epidemiological Expertise and Monitoring, Kazakh National Medical University and in regions for technical assistance. We also express our gratitude to the Exceed Program of the German Federal Ministry for Economic Cooperation and Development (BMZ) and the German Academic Exchange Services (DAAD) through the CIHLMU (Centre for International Health, Ludwig-Maximilians-Universität, Munich, Germany).
Funding
The study is supported by the German Biosecurity Programme of the German Federal Foreign Office. This data were gained in the project “Aufbau eines deutsch-kasachischen Netzwerkes zur Diagnostik von Infektionskrankheiten verursacht durch potentielle B-Agenzien” Kapitel 0502, Titel 68777 (Obj. Nr. AA: 02761868).
Availability of data and materials
All data collected and analyzed during this study are available from the corresponding author upon reasonable request. The newly generated sequences were submitted to the GenBank database under the accession numbers MG973754–MG973956 (ompB), MG973957–MG973991 (gltA), MG973992–MG974035 (ompA), MG9704036–MG974057 (23S-5S interspacer region), MG974058–MG974091 (sca4), and MG974092–MG974099 (16S).
Authors’ contributions
SE, SF, RY, AD, LY and ZS designed this research. ZS collected field data. NT, KA, LZ, JZ, AA, ZK and AO performed tick and molecular biological studies. NT conducted statistical analysis. NT and SE wrote the draft manuscript. MH, GF and GD are supervisors of NT and read the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Disclaimer
The opinions expressed by the authors contributing to this study do not necessarily reflect the opinions of the involved institutes.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Turebekov, N., Abdiyeva, K., Yegemberdiyeva, R. et al. Prevalence of Rickettsia species in ticks including identification of unknown species in two regions in Kazakhstan. Parasites Vectors 12, 197 (2019). https://doi.org/10.1186/s13071-019-3440-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3440-9