Abstract
Background
Glossina m. morsitans is the primary vector of the Trypanosoma brucei group, one of the causative agents of African trypanosomoses. The parasites undergo metacyclogenesis, i.e. transformation into the mammalian-infective metacyclic trypomastigote (MT) parasites, in the salivary glands (SGs) of the tsetse vector. Since the MT-parasites are largely uncultivable in vitro, information on the molecular processes that facilitate metacyclogenesis is scanty.
Methods
To bridge this knowledge gap, we employed tandem mass spectrometry to investigate protein expression modulations in parasitized (T. b. brucei-infected) and unparasitized SGs of G. m. morsitans. We annotated the identified proteins into gene ontologies and mapped the up- and downregulated proteins within protein-protein interaction (PPI) networks.
Results
We identified 361 host proteins, of which 76.6 % (n = 276) and 22.3 % (n = 81) were up- and downregulated, respectively, in parasitized SGs compared to unparasitized SGs. Whilst 32 proteins were significantly upregulated (> 10-fold), only salivary secreted adenosine was significantly downregulated. Amongst the significantly upregulated proteins, there were proteins associated with blood feeding, immunity, cellular proliferation, homeostasis, cytoskeletal traffic and regulation of protein turnover. The significantly upregulated proteins formed major hubs in the PPI network including key regulators of the Ras/MAPK and Ca2+/cAMP signaling pathways, ubiquitin-proteasome system and mitochondrial respiratory chain. Moreover, we identified 158 trypanosome-specific proteins, notable of which were proteins in the families of the GPI-anchored surface glycoproteins, kinetoplastid calpains, peroxiredoxins, retrotransposon host spot multigene and molecular chaperones. Whilst immune-related trypanosome proteins were over-represented, membrane transporters and proteins involved in translation repression (e.g. ribosomal proteins) were under-represented, potentially reminiscent of the growth-arrested MT-parasites.
Conclusions
Our data implicate the significantly upregulated proteins as manipulators of diverse cellular processes in response to T. b. brucei infection, potentially to prepare the MT-parasites for invasion and evasion of the mammalian host immune defences. We discuss potential strategies to exploit our findings in enhancement of trypanosome refractoriness or reduce the vector competence of the tsetse vector.
Similar content being viewed by others
Background
Trypanosoma brucei group causes African trypanosomoses, a group of neglected zoonotic tropical diseases endemic in 37 sub-Saharan African countries [1], against which there are no effective vaccines or drugs [2, 3]. Amongst the medically and agriculturally important tsetse fly species, Glossina morsitans morsitans is highly significant in the savannah woodlands and is the primary vector of the species of T. brucei group [4].
In order to complete their complex developmental cycles in the tsetse vector, trypanosomes face two critical replicative stage barriers: colonization of the midguts and establishment in the salivary glands (SGs). The pathway followed by the parasites between these two independent multi-replicative stages is time-dependent, irreversibly transient and eventually influences metacyclogenesis in the SGs (differentiation into the mammalian-infective metacyclic trypomastigote (MT) parasites) [5–9]. Critical replicative events occur ~3 days post-infective blood meal when only a small proportion (≤ 10 %) of the parasites is able to pass through the midgut barrier [8]. Upon establishment of infection, parasite numbers per fly gut remain remarkably constant [6, 8]. However, since the SG-derived MT-parasites are uncultivable in vitro, molecular mechanisms that promote metacyclogenesis remain to be investigated.
Whilst the SGs determine success of metacyclogenesis, there is limited knowledge on how trypanosomes adapt to and evade the host defence responses in the SGs [10]. It is however generally known that trypanosomes modulate SGs microenvironment [11], and that factors such as parasite genotypes, midgut antioxidant status, lectins and environmental stimuli influence parasite maturation [12, 13]. Most trypanosome research has focussed on the bloodstream and/or in vitro cultured procyclic forms, thus creating a knowledge gap with regard to metacyclogenesis. The availability of complete genomes of G. morsitans [14] and T. brucei [15] makes it possible to identify proteins involved in the mechanisms that facilitate metacyclogenesis. Such proteins are ideal candidates to develop improved strategies for tsetse and trypanosomosis control, especially via the sterile insect technique (SIT) programs [16], which have so far been employed in areas without active disease transmission [2].
Vector competence is one of the key pillars in the SIT programs, which involves mass release of sterile males into wild populations of the target species. More importantly, the sexually sterilized males are still competent trypanosome vectors, thus increasing risks of disease transmission when millions of sterile males are released into trypanosome-infested areas. Attempts have been previously made to make the sterile males vector incompetent via drug-supplemented blood meals, an approach now known to be inefficient [17]. Thus, alternative and/or complementary approaches are necessary, especially with the risk of trypanosomes developing resistance to the trypanocidal drug supplements. In this regard, metacyclogenesis and transmission of the mammalian-infective MT-parasites potentially represent vulnerable and attractive intervention points to enhance the natural trypanosome refractoriness or reduce the vectorial competence of the sterile males used in the SIT campaigns.
We hypothesized that T. b. brucei manipulates expression of proteins involved in pathways that specifically prepare the MT-parasites for successful transmission to and infection of susceptible mammalian hosts. To test this hypothesis, we employed tandem mass spectrometry (LC-MS/MS) to determine T. b. brucei-induced protein expression modulations in parasitized SGs of G. m. morsitans compared to unparasitized SGs. We also aimed at highlighting major metacyclic T. b. brucei-specific proteins, which are potentially critical for parasite survival in the SGs and transmission to susceptible mammalian hosts. We discuss our findings from the perspective of potential approaches to enhance trypanosome refractoriness in tsetse as an anti-vector strategy against African trypanosomosis.
Methods
Tsetse flies and parasites infections
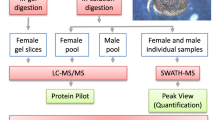
Male G. m. morsitans were obtained from the Institute of Tropical Medicine (Antwerp, Belgium) and infected with a highly transmissible T. b. brucei strain (EATRO 1125 AnTaR 1) [11]. Male flies were used because they mature significantly more midgut infections than females [5]. For the infections, teneral flies (24–48 h post-adult eclosion) received their first blood meals supplemented with ~12 μg of parasites/fly [11]. Fully-engorged flies were selected, reared for 28 days post-infection (dpi) in controlled insectaria (65 % relative humidity; 26 °C) and subsequently fed on clean (trypanosome-free) defibrinated horse blood (in vitro; 2–3 times/week) [18]. Control flies were prepared from males of the same batch, age and feeding history as the parasitized flies. The control flies were maintained on clean blood meals and handled as described for their parasitized counterparts. All assays consisted of biological triplicates (20 flies per group) in small holding cells (3.5 cm in diameter × 6 cm height).
Parasite scoring and SG dissections
Forty-eight h after the last blood meal, the flies were scored for the presence of the MT-parasites in the SGs using phase-contrast microscopy (×400) as previously described [11]. Briefly, the SGs were considered parasitized if stuffed with trypanosomes (i.e. the fly’s spit full of the parasites). Fly’s spit (from the control group) completely devoid of trypanosomes were considered unparasitized. For mass spectrometry, 10 intact pairs of SGs were selected from each of the replicated fly groups and immediately preserved at -20 °C in 150 μl sterile saline supplemented with complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany).
SG extracts preparation and SDS-PAGE
SGs were individually homogenized using a glass/Teflon homogenizer and ultra-sonicated (Sonifier cell disruptor, Branson Instruments Co., Stanford, Connecticut, USA). Homogenates were freeze-thawed and clarified three times by centrifugation (7500× g; 10 min; 4 °C) to completely remove cell debris. Supernatants were pooled and proteins quantified using the standard BCA method (Bio-Rad, Hercules, California, USA). Equal protein quantities (600 ng) were resolved in 12 % SDS-PAGE gels and stained (CBB stain; NuPAGE Novex; Invitrogen Life Technologies, Carlsbad, California, USA). The middle sections of entire gel lanes were longitudinally excised (from top to bottom), divided into five equal fractions (covering the entire gel lanes) and cut into small pieces (~1 mm3) as previously described [19].
LC-MS/MS measurements and protein identification
Tryptic peptides for subsequent LC-MS/MS measurements were prepared as previously described [19]. Briefly, the gel pieces were washed with 50 mM ammonium bicarbonate (ABC) buffer and ABC buffer/50 % acetonitrile (ACN) and proteins reduced and alkylated using dithiothreitol and iodoacetamide. Gels were washed with ABC/ABC-ACN buffer, followed by in-gel trypsin digestions and LC-MS/MS measurements [20]. Proteins were identified by searching the MS/MS spectra (using MaxQuant/Andromeda [21, 22]) against G. m. morsitans and T. brucei databases (downloaded from UniProt), a decoy database (constructed by reversing all the protein sequences) and a database of common contaminants (available from MaxQuant). MaxQuant search parameters included fixed carbamidomethylation (C), oxidation (M), acetylation and deamidation (N and Q). Two peptides (at least one unique and unmodified) matching the same protein were required for protein identification at a maximum false discovery rate (FDR) of ≤ 0.01. The unique identifiers of the proteins downloaded from the UniProt databases were used to identify and classify the LC-MS/MS peptides as specific to the host (G. m. morsitans) and the parasite (T. b. brucei). LC-MS/MS peptide hits to the decoy database and hits with modified peptides only were deleted from the final list of protein groups.
Protein quantification and normalization
Unique and ‘razor’ (non-unique) peptides were used for peptide assignments and protein quantification [21, 23]. For each of the above-described five gel fractions, peptides were matched across different MS/MS runs based on mass and retention time (‘match between runs’ of 2 min). Label-free quantification (LFQ) was enabled. To minimize technical variations and to easily compare abundances of the same proteins (parasitized vs unparasitized), Log10 normalized LFQ was used across the biological triplicates. To compare levels of different proteins from the same samples (parasitized and unparasitized), Log10 iBAQ (intensity-based absolute quantitation) was used [24]. To determine up- and downregulated proteins (parasitized vs unparasitized SGs), t-tests were performed on Log10 LFQ using the MaxQuant’s Perseus module. Proteins were considered to be up- or downregulated when their Log10 iBAQ ratios (parasitized vs unparasitized) were larger or smaller than zero, respectively, and significantly upregulated when the FDR was ≤ 0.05.
Gene ontology (GO) annotations and PPI network analyses of upregulated SG proteins
Blast2GO v. 3.2 [25] was used to classify the identified proteins along three biological aspects, i.e. biological process (BP), molecular function (MF) and cellular component (CC) Gene Ontology (GO) terms. It should be noted that the GO terms are descriptions of the different protein functional classes and how they relate to each other. To provide a broader overview of the ontologies, the GO classes were grouped into GO-slim terms using CateGOrizer [26]. To correctly place the significantly upregulated proteins within signaling pathways and networks, computational predictions of protein-protein interactions (PPIs) were inferred using interolog mapping [27]. For this, human orthologs to the identified G. m. morsitans proteins were obtained from Ensembl [28] and used to generate an exhaustive list of possible interacting protein combinations using custom in-house Python scripts. Putative interactions for these combinations were then determined using FpClass [29] (probability ≥ 0.2) and mapped back onto the SG protein datasets obtained from the above-mentioned LC-MS/MS measurements. The resulting significant interactome was rendered in Cytoscape v3.30 [30].
Results
Parasitized SG proteome of G. m. morsitans
Analysis of the LC-MS/MS data resulted in 5469 and 4366 total and unique peptides, respectively. These peptides mapped to 874 protein groups. Removal of the common contaminants, applying extra filter steps such as removing single peptide hits (thereby decreasing FDR to below 0.01) and hits to the decoy database resulted in 523 non-redundant (nr) protein groups (Additional file 1: Table S1). Of these, 363 protein groups had specific peptide hits to the tsetse vector (G. m. morsitans) proteins (Additional file 1: Table S2) and 158 protein groups had specific peptide hits to the parasite (T. b. brucei) proteins (Additional file 1: Table S3). We also obtained peptide hits to four proteins specific to the bacterial endosymbionts Wigglesworthia glossinidia (n = 3) and Sodalis glossinidius (n = 1) (Additional file 1: Table S4). These symbiont proteins have been reported in the genome of G. m. morsitans [14]. It should be noted that there were no host and/or parasite and/or symbiont proteins that grouped together in one protein group. Figure 1 shows the abundance distribution of the proteins identified in parasitized SGs compared to unparasitized SGs.
Distribution of proteins in parasitized G. m. morsitans SG proteome. Scatter plot distribution of the protein expression measurements for the identified G. m. morsitans proteins (red; n = 361), Trypanosoma proteins (green; n = 158) and bacterial endosymbiont proteins (purple; n = 4). Selections of the most notable proteins are named in the figure. See text for abbreviations
Table 1 presents the topmost abundant 25 proteins, amongst which were the following blood feeding-associated proteins: tsetse salivary gland proteins-1 & 2 (Tsal1/2), tsetse salivary growth factors-1 & 2 (TSGF-1/2), salivary antigen-5-protein (TAg5), 5′-nucleotidase-related saliva protein (5′Nuc), adenine deaminase (ADA) and 5′-nucleotidase-related SG protein-3 (Sgp3). Also abundant were proteins related to cellular proliferation/differentiation (adenosine deaminase-related growth factors, muscle LIM protein at 84B), cytoskeletal proteins of the actin and myosin families, lectins, molecular chaperones (e.g. 14-3-3 protein zeta family chaperones) and enzymes (e.g. trehalose-6-phosphate synthase, isovaleryl-CoA dehydrogenase, and aspartate aminotransferase).
Amongst the least abundant proteins that we identified included dihydrolipoamide transacylase α-ketoacid dehydrogenase (DBT) complex proteins (for amino acid metabolism), GTP-binding proteins, Ras-related protein and clathrin adaptor complex proteins (for endocytotic trafficking), 26S proteasome regulatory complex proteins (for protein turnover), bifunctional ATP/sulfurylase-adenosine 5′-phosphosulfate (APS) kinase (for uptake/assimilation of inorganic sulphate) and translocon-associated (TRAP) complex proteins (for endoplasmic reticulum (ER)-targeting of nascent polypeptides) (see Additional file 1: Table S2).
The parasitisation of G. m. morsitans SGs appears not to have affected the expression of at least four host proteins, including bifunctional methylene-tetrahydrofolate dehydrogenase (MTHF-folD) (for thymidylate/methionine/purine synthesis) and 25-/28-kDa glutathione S-transferase 1 (GST1) proteins (for detoxification and lipophilic compound transport) (Additional file 1: Table S8).
Preferentially expressed proteins in parasitized SGs and T. b. brucei-induced changes
Compared to the unparasitized SGs, 276 G. m. morsitans proteins were found to be upregulated in the parasitized SGs (Table S5); of these 32 proteins were significantly upregulated (Table 2). Topmost of the upregulated proteins included Ca2+/calmodulin-dependent protein kinase (CaMK), tetraspanin 42Ed (Tsp42Ed), β-integrin, stress-associated ER protein-2 (Serp-2), small ubiquitin-related modifier-3 (SUMO), a homolog to uracil-DNA degrading factor-like (UDE) protein and various subunits of the vacuolar ATPases and chaperonin containing t-complex polypeptide-1 (TcP-1). Others included amino acid metabolism-related proteins such as methylglutaconyl-CoA hydratase (AUH), glutamine synthetase (GS), δ-1-pyrroline-5-carboxylate dehydrogenase (P5CDH), aspartate aminotransferase (AspAT) and isovaleryl-CoA dehydrogenase (IVD).
Several proteins associated with the ubiquitin-proteasome system (UPS) were also upregulated (Additional file 1: Tables S5 and S6), including ubiquitin C-terminal hydrolase (uCHL), ubiquitin-40S ribosomal protein S27a fusion protein (RpS27A), ubiquitin activating enzyme (uBA1), ubiquitin-protein ligase (E3) and various subunits of the proteasome regulatory complex proteins α-/β-types (PSMA/PSMB). According to our protein identification criteria, the repertoire of the downregulated proteins did not have any UPS-associated proteins.
Nine proteins known to be involved in Glossina immunity were upregulated. Among these were thioredoxin peroxidases (TrxP), alkyl hydroperoxide reductase (AhpC), Serp-2, cathepsin B-like cysteine proteinase (CatB), a truncated nitric oxide synthase (NOS) and transferrin. Of these, only Serp-2 and CatB were significantly upregulated (Additional file 1: Tables S5 and S6). Other putative immunity-related proteins included integrin-β, Tsp42Ed and adaptor protein downstream of receptor kinase (Drk), all of which were significantly upregulated. Top BLASTp (bitscore >75; E-value ≤1.0E-4) of these immunity-related proteins against the Insect Innate Immunity Database (IID) v 2.2.26 [31] resulted in significant homologies to immunity proteins that have been reported in the Acyrthosiphon pisum (pea aphid), Drosophila melanogaster (common fruit fly), Apis mellifera (western honeybee) and Anopheles gambiae (malaria mosquito) (Table 3).
Gene ontology (GO) and PPI network analyses of parasitized SGs during T. b. brucei infection
All of the significantly upregulated proteins were associated with at least one GO term for a total of 9323 term occurrences. Using the CateGOrizer, the GO terms were trimmed down and grouped into 127 Go-Slim terms associated with biological processes (BP; 55.1 %), molecular functions (MF; 25.9 %) and cellular components (CC; 19 %) ontologies (Fig. 2). Proteins involved in metabolism and cell- or organelle reorganization were present amongst the key BP ontologies (Additional file 1: Table S9), while the majority of MF ontologies represented proteins associated with catalytic, binding and hydrolase activities (Additional file 1: Table S10). Topmost in the CC ontology were proteins localized in the cellular, intracellular and cytoplasmic compartments of the SGs (Additional file 1: Table S11).
The predictions of protein-protein interaction network of the upregulated host proteins resulted in 10,861 putative interacting protein pairs, of which 225 pairs had significant interacting probabilities (Additional file 1: Table S12). A single high-scoring network was obtained consisting of 88 nodes, a clustering coefficient of 0.107 and an average number of neighbours of 5.0 (Fig. 3). Eighteen of the significantly upregulated proteins formed nine of the main PPI network hubs (Fig. 3). Seven of these proteins formed network nodes with the most edges including SUMO, TcP-1s, mitochondrial NADH-ubiquinone oxidoreductase (nuo-24), V-ATPase-D, Drk and GS.
The most significant PPI network model for the upregulated proteins in parasitized SG proteome of G. m. morsitans. Highlighted are the main hubs formed by the proteins that were found to be significantly upregulated in the parasitized SGs. The PPI was visualized in Cytoscape. The significantly upregulated proteins SUMO, TcP-1, nuo-24, V-ATPase-D, Drk and GS occupy central positions in the PPI network
Expression of abundant proteins is downregulated in parasitized SGs
A total of 81 proteins were downregulated in parasitized SGs. However, only salivary secreted adenosine (ADA; previously annotated as adenosine deaminase-related growth factor-3; ADGF-3) was significantly downregulated (Additional file 1: Table S7). Despite their high abundance, the expression patterns of TAg5, 5′-Nuc, Tsal1/2, TSGF-1/2, ADA, ADGF-C, Spg3 and TRAP was downregulated. Further, > 20 % (n = 18) of the downregulated proteins were ribosomal protein (RP) components (Additional file 1: Table S7), including several proteins of the 40S and 60S subunits, glycine/glutamate-rich protein (Sgp1), proline-rich protein (Sgp2), mu-clathrin adaptor complex proteins, subunits of the vesicle coat complex II (COPII), ADGF-C and lectins.
Expression of metacyclic trypomastigote-specific proteins
Table 4 presents major clusters of trypanosome-specific proteins identified in this study. Notable abundant proteins included the glycosylphosphatidyl inositol (GPI)-anchored proteins bloodstream stage alanine-rich proteins (BARPs) and variant/invariant surface glycoproteins (VSGs/ISGs), calpains/small kinetoplastid calpain-related proteins (CALPs/SKCRPs), retrotransposon hot spot proteins (RHS), kinetoplastid membrane protein-11 (KMP-11), tryparedoxins (TXNs) and membrane transporters. We also identified several trypanosome-specific RPs, with an over-representation of the 40S RP compared to the 60S RP families (Table 4). Also identified were paraflagellar rod proteins (PFRA and PFRC), flagellar attachment zone protein 1 (FAZ1), flagellar calcium-binding protein (Tb-44A) and C-terminal motor kinesin (Table 4). Here, it is important to note that FAZ and C-terminal motor kinesin are important in the adjustments of the flagellar positions/sizes [32], depending on the parasite life-cycle stages.
Twenty-five of the parasite protein groups were of unknown functions (denoted as ‘uncharacterized proteins’ in Additional file 1: Table S3). Top-BLASTp analyses (bitscore >100; E-value ≤ 1.0E-6) of these proteins against the nr-NCBI protein database yielded eight hits to proteins with known functions in flagellate protozoans (Table 5). Six of these were hits to proteins reported in the recently sequenced genome of the African crocodilian trypanosome, T. grayi (vectored by G. palpalis), which is more closely related to T. cruzi than T. brucei [33]. Other homologies included proteins associated with antigenic variations (Pro-Glu/polymorphic GC-rich repeat (PE-PGRS) protein [34] and acyl-CoA-binding protein [35]), parasite proliferation (auxin-induced in root cultures 9 (AIR9)-like protein [36]) and the chemosensational intraflagellar transport, osmotic avoidance abnormal protei3 (OSM3)-like kinesin [37].
Discussion
A robust immune system in tsetse midguts makes the flies naturally Trypanosoma-refractory [38]. Following an infected blood meal, the absolute parasite numbers drastically drop at the midgut barrier (days 1–3), then proliferates (day 4) and stably colonizes the midguts where the established population reaches approximately 5 × 105 trypanosomes [8, 39]. Trypanosomes then migrate to the ectoperitrophic space (day 5) [39], congregate within the proventriculus (days 6–8) and subsequently colonize and complete metacyclogenesis in the SGs (days 12–18) [8]. The MT-parasites then detach from the SG epithelium into the lumen and are uniquely adapted to infect and survive in susceptible mammalian host [32]. The entire process takes approximately 3–5 weeks [40], implying that at the time of the SG dissections (i.e. 28 dpi), MT-parasites were continuously produced.
Trypanosoma b. brucei suppresses expression of abundant host proteins to promote MT-parasite transmission
Previous transcriptomic analyses on G. m. morsitans SGs reported seven protein families: (i) Tsal1/2; (ii) 5′Nuc/apyrase; (iii) ADA; (iv) TAg5 family-related proteins; (v) Glycine-glutamate (GE)-rich proteins; (vi) C-type lectins; and (vii) housekeeping genes [13, 41]. We identified these protein families with similar abundance profiles as reported across different tsetse species [11, 41–46]. Among these, the expression of tag5, tsal1/2, tsgf1/2, 5′-nuc and spg3 genes were previously reported to be decreased in parasitized flies [11]; these previous results agree with our results (see Table 1 and Additional file 1: Table S7). We however did not detect the anticoagulant tsetse thrombin inhibitor (TTI), echoing reports by Capello et al. [47] and Telleria et al. [13], who noted moderate down-regulation of TTI in parasitized SGs. It is notable that the differentially modulated proteins are involved in key processes such as blood feeding which, despite their high abundance, were downregulated in the parasitized SGs compared to unparasitized SGs (Table 1). Reduction in the expression of these proteins could reduce fly feeding performance, which in turn increases tsetse biting frequencies to achieve full engorgement [11]. One could conclude that an increase in biting frequencies promotes the competence of the tsetse vector in trypanosome transmission.
Trypanosoma b. brucei induces upregulation of host proteins essential for parasite differentiation and survival
Cellular proliferation and homeostasis are important conditions for parasite survival. We identified several proteins, whose upregulation may be advantageous to T. b. brucei (see Table 2). For instance, CaMK is one of the most important regulators of stage-specific morphological differentiation of Trypanosoma, Leishmania and Plasmodium parasites [48]. Upregulation of Serp-2 is also notable owing to its central roles in ER stress, an important cue for Plasmodium parasites to switch to transmissible sexual stages in mosquitoes [49]. Further, one of the parasitic survival tactics is to alter intra-cellular locations, binding partners and/or functions of specific proteins and antagonizing other protein modifications. One of the quickest ways to achieve these changes is via sumoylation [50, 51], a posttranslational modification mediated by protein SUMO. Notable also is the upregulation of proteins in the family of the ubiquitous and highly conserved V-ATPases; at least eight different V-ATPase subunits were variously upregulated in parasitized SGs (see Additional file 1: Table S5). Mosquito cells over-expressing V-ATPases are reported to be preferentially invaded by Plasmodium parasites [52]; perhaps trypanosomes has a similar invasion preference for V-ATPase expression in SGs. Homeostasis-associated proteins such as ArgK are also important for parasite survival; dsRNA-mediated silencing of ArgK significantly reduced Plasmodium parasite loads in Anopheles gambiae [53]. In our study, ArgK was not only the most abundant protein (Table 1), but also amongst the upregulated proteins (see Additional file 1: Table S5).
Glossina m. morsitans overexpresses immunity-related proteins in responses to SG parasitisation
Overcoming host immune responses is one of the most challenging parts of trypanosome life-cycle because immunity counter-defence provides a nutritious and equable environment. Lehane et al. [54] reported that out of the 68 putative immunity-related genes, 15 genes were actually overexpressed in the midguts of T. b. brucei-infected G. m. morsitans. Midgut expression of 12 of the 15 genes in T. b. gambiense-infected G. p. gambiensis was recently quantified by Hamidou et al. [55], who noted a time-dependent and variable high expression of five out of the 12 genes. Eight of these previously reported immune-related proteins were upregulated in parasitized SGs (see Table 3), a result in agreement with another transcriptome-based study on parasitized SGs of G. m. morsitans [13]. The only exemption was that only Serp-2 and CatB were significantly upregulated in parasitized SGs; CatB was not reported by Hamidou et al., [55]. These differences potentially indicate tissue-specific differential expression patterns of these proteins, for instance in the midgut versus the SGs.
In addition to the above-mentioned previously reported tsetse immunity genes, we also identified other proteins implicated in the immunity of other insects (see Table 3). These included Rac/Rab GTPases (for immune surveillance against pathogens and parasites in Drosophila [56]), ubiquitin protein ligase (for Drosophila humoral systemic immune response [57]), a homolog to the Pro-Glu/polymorphic GC-rich repeat (PE-PGRS) protein (for antigenic variations in intracellular parasites [34]), Tsp42Ed and integrins (for activation of immune signaling Plasmodium-infected mosquitoes [53, 58]) and Drk (implicated in Drosophila hemocyte immunity [59]). Finally, loosely associated to insect immunity is amino acid metabolism; some of the upregulated proteins we identified are related to amino acid metabolism (see Table 1). However, a link between upregulation of proteins involved in amino acid metabolism and parasite infection in the SGs is unclear. This notwithstanding, it is rather obvious that following a blood meal, increase in amino acid metabolism is important for insect immune responses (via activation of proteolytic immunity cascades such as prophenoloxidase) and detoxification (removal of amino acid metabolites). Although likely to occur in the hemolymph, these processes are reportedly present in the SGs of locusts, leafhoppers and aphids [60]. Similar scenario cannot be totally ruled out in the case of tsetse immune responses to trypanosome infections in the SGs.
Trypanosoma b. brucei controls the efficiency of protein turnover in the SGs of T. b. brucei
It is notable that > 20 % of the downregulated host-specific proteins in parasitized SGs were RPs. This modulation of RP expression is in agreement with recent transcriptome-based studies in the differential expression of the RP genes in T. b. gambiense-infected G. p. gambiensis midguts [61]. Downregulation of RPs implicates protein translational regulation to prepare mammalian-infective MT-parasites, or reflects the fly’s adaptive response to SG parasitisation. Notably, the downregulation of RPs appears to be accompanied by upregulation of at least 17 proteins related to the UPS pathway, suggesting that metacyclogenesis requires protein translation and/or degradation efficiency. Moreover, the importance of protein turnover/folding/modifications during parasite infections is underscored by differential modulations of the UDE homolog [62], TcP-1 components [63] and various molecular chaperones identified in the current study.
Trypanosoma b. brucei induces upregulation of key regulators (host proteins) of diverse pathways
The identification of the significantly upregulated SUMO as one of the top interacting proteins in the predicted PPI network (Fig. 3) is interesting given its central roles in the regulation of various cellular processes including transcription, replication, chromosome segregation and DNA repair. SUMO is required for activation of the Ras/MAPK pathway in Drosophila S2 cells [64]; Drk is one of the essential components of the pathway. Additionally, the 14-3-3 protein family, which constitute a multitude of functionally diverse signalling proteins related to behaviour, is also a SUMO substrate. Sumoylation of 14-3-3 proteins results in modulation of diverse cellular processes, including cell cycle regulation, metabolism control, apoptosis and gene transcription control [65]. Furthermore, sumoylation and ubiquitin-proteasome systems are known to selectively modify the functions, sub-cellular locations and half-life of proteins in a very specific manner to maintain cellular homeostasis [66]. According to our PPI predictions (see Fig. 3 and Additional file 1: Table S12) sumoylation targets include several proteins of the TcP-1 complex, members of the 70-kDa heat shock protein family (Hsp70-4 and Hsp70-5), translation initiation factors and several enzymes (see Fig. 3).
On the other hand, members of the NADH-ubiquitin oxidoreductases (in our case the nuo-24 protein; significantly upregulated; Table 2) are important for energy metabolism and nucleotide synthesis; these proteins are also the initial enzymes in mitochondrial transport chain (mtECT). In fact, mtECT-related proteins were reported to be upregulated during the invasion of mosquito SGs by Plasmodium parasites [67]. Following a blood meal, GS (a key enzyme in glutamine biosynthesis) is upregulated in mosquito midguts [68], which agrees with the results shown in Additional file 1: Table S5. Glutamine is eventually used for production of chitin, which is also produced in other tissues including the SGs. Perhaps the upregulation of GS is related to chitin biosynthesis in Glossina, requiring modulation during parasite infection as can be observed in our PPI predictions, i.e. Serp-4 and ubiquitin-related proteins were first neighbours of GS (see Fig. 3). Finally, the central hub occupied by the V-ATPases in the predicted PPI network is noteworthy because these electrolyte/solute trans-epithelia transporters are critical in saliva secretion in other insects such as the blowflies [69]. Trypanosoma. b. brucei may therefore modulate the expression of V-ATPases to facilitate saliva-mediated transmission to the mammalian host, which possibly explains the upregulation of various V-ATPases (Additional file 1: Table S5). Taken together, the central hubs occupied by the significantly upregulated proteins in the predicted PPI network potentially indicate manipulations of diverse cellular processes in response to trypanosome infection. These processes require strict control of the participating proteins and enzymes.
Trypanosoma. b. brucei MT-parasites express specific proteins in preparation to invade mammalian host
During metacyclogenesis, the MT-parasites re-acquire the VSG/ISG coat and enter the SG lumens in preparation for invading susceptible mammalian hosts [70]. Expressions of VSG and the VSG-shielded ISGs are activated in the SGs to mediate the parasite’s evasion of and/or escape the immune surveillance of the host, and successful establishment of mammalian infections [71, 72]. Trypanosomes have other VSG-shielded proteins or proteins that are ‘hidden’ within the flagellar pocket, where they mediate numerous host-parasite interactions. For instance BARPs are predominantly used by the parasites to attach to the SG plasma membranes and promote the release of VSGs [32, 73]. In fact, mutations of BARPs are reported to block cytokinesis in T. b. brucei [74]. Further, expression site-associated gene (ESAG) proteins are widely distributed amongst different Trypanosoma species albeit with low expression [72, 75], probably explaining the detection of only few ESAG variants in our study (see Table 4 and Additional file 1: Table S3). Notably, to enhance adaptive immunity, T. brucei switches the expression of VSGs with compatible ESAG variants [76]. Moreover, the identification of members of the diverse and rapidly evolving RHS family in our study is noteworthy (see Table 4). Since RHS proteins are localized in polymorphic subtelomic regions [77], they are likely to confer the MT-parasites with selective advantages (via antigenic variations). Additionally, detoxification is also important in parasite metabolism, which underscores the importance of the abundance of TXNs, members of the peroxiredoxin AhpC-type family (Table 4). Abundant in all T. brucei life stages, TXNS are crucial in nucleotide synthesis and adaptive defence (hyperoxide detoxification) [78, 79].
KMP-11 and CALP/SKCRP are well-conserved heterodimeric proteins with strict stage-specific expression patterns in kinetoplastids [80–82]. Stebeck et al. [82] reported that KM-11 is differentially expressed in various trypanosome life-cycle stages. CALP/SKCRP are abundantly expressed in mammalian-infective MT-parasites, and are involved in various cellular Ca2+-regulated processes such as signal transduction, differentiation, virulence, cytoskeletal rearrangement, and protein-membrane interactions with host structures [80, 83]. Out of the 18 (12 CALPs; six SKCRPs) family members known in T. brucei [81], we identified eight including CAP5.5 V, a variant of the insect stage-specific CAP5.5 (see Table 4). Finally, one of the uncharacterized proteins that we identified (C9ZWF1; Table 5) was homologous to the T. grayi calpain-like cysteine peptidase, an important protein in parasite cell cycle progression [84].
The interchange between the functionally and morphologically distinct forms of trypanosomatids (from epimastigotes to trypomastigotes) is accompanied by efficient regulation of gene expression, which is largely posttranslational [85]. Among other strategies, this regulation can be through the control of the expression of translation factors; translation of RPs is reduced during differentiation of trypanosomatids into the MT-parasites [86]. The possibility of this form of translational repression partially explains our detection of only few RPs (see Table 4), potentially because the MT-parasites are essentially growth-arrested. Our results echo studies on the on differential expression of RPs in the closely related Chagas disease causative agent, T. cruzi [86].
During active replication, trypanosomes heavily rely on membrane transporters for nutrient and metabolite salvage from the host. The main transporters include purine/pyrimidine transporters (trypanosomes are incapable of de novo purine synthesis) [87], hexose/sugar transporters (to meet parasite’s nutritional requirements) [88], amino acid transporters (for incorporation into proteins by the parasite) [89, 90], aquaporins (for osmotic protection and metabolism) [91, 92] and fatty acids transporters (for preservation of organelle vitality and expansion of vacuole sizes to the accommodate growing parasite progenies) [93]. Trypanosoma brucei contains a repertoire of 305 transporters clustered in 23 families [94]. We identified only three transporters, all with low abundances (see Table 4). Potentially, the growth-arrested MT-parasites may not require expression of many transporters; expression of the transporter genes is probably activated in the next life-cycle stage, i.e. proliferating bloodstream forms in the mammalian host.
Potential approaches to enhance trypanosome refractoriness in tsetse
Factors such as the lack of effective vaccines, resistance to pesticides/drugs and inefficient public health infrastructures make African trypanosomosis increasingly important in sub-Saharan Africa. The fact that sterile males are still competent trypanosome vectors reduces the efficacy of tsetse and trypanosomosis control via the SIT programs. One of the strategies to solve this problem is to enhance the natural trypanosome refractoriness or reduce the competence of the tsetse vector. This can be achieved by identifying candidate genes and creating transgenic sterile males that are incapable of transmitting trypanosomes to mammalian hosts. Ideally, the transgenic sterile males released into wild tsetse populations could eventually block trypanosome transmission. This would be important especially in the event that the areas into which these males are released have active trypanosome circulation.
Although a proteomic approach is far better (in protein identification and observed quantitative expression changes) than a transcriptomic approach, the former approach is often not precise enough to directly make valid biological inferences, especially due to the post-translational modifications that play important roles in protein functions. Nevertheless, our proteomics and pathway analyses are important milestones in the identification and characterization of SG proteins that potentially contribute to the trypanosome infection status (refractoriness) in the tsetse vector. Having confirmed the protein abundances and expression patterns (parasitized vs unparasitized SGs), we are now designing bioassays to functionally appraise the biological consequences (by immunoblotting, qPCR, RNAi gene silencing) of the protein level changes of some of the proteins identified in our proteomics data sets. After validation, the next objective is to use modern tools such as the piggyBac transposon or CRISPR/Cas9 systems [95], or paratransgenesis [96] to enhance trypanosome refractoriness of the sterile males that are used in SIT programs. Previous studies have provided proof of principle that these methods are feasible in creation of parasite-refractory insect vectors. For instance, using the piggyBac system, Ito et al. [97] created Plasmodium-refractory anopheline mosquitoes by over-expressing a blood-inducible anti-plasmodium gene (salivary gland/midgut-binding peptide 1; SM1) in a tissue-specific manner. Recently, Gantz et al. [98] used the CRISPR/Cas9 system to create Plasmodium-resistant mosquitoes by expression of blood-inducible and tissue-specific single chain variable fragment antibodies (scFvs) against Plasmodium proteins. In the case of tsetse flies, De Vooght et al. [96] demonstrated that transformed Sodalis can secrete significant amounts of functional Nanobodies against Trypanosoma VSG epitopes. The concept of paratransgenesis has also been demonstrated in the control of Leishmania transmission by sand flies [99]. Taken together, our data take us a step closer towards improved anti-vector methods against tsetse and African trypanosomiasis. We have made significant progress towards this direction in that the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture has brought together research experts in different disciplines in a collaborative coordinated research project to explore ways to enhance trypanosome refractoriness in tsetse [2].
Conclusions
Our data suggest that T. b. brucei modifies SG protein composition and functions (suppression of abundant SG proteins) and induces SG cellular proliferation (upregulation of immunity, stress, homeostasis and translatome-related proteins). Further, the repertoire of T. b. brucei-specific proteins largely consisted of proteins reminiscent of non-replicative MT-parasites (suppression of RPs and transporters), and proteins critical for preparing the trypomastigotes for invasion and evasion of mammalian host immune responses (over-representation of immunity, signal transduction and virulence-related proteins). In response to T. b. brucei infection of G. m. morsitans SGs, divergent cellular processes appear to be manipulated via modulations of proteins involved in various pathways, which is accompanied by modulations of proteins involved in the control of protein turnover. Similar to the tsetse midgut barrier, SG micro-environment is a critical bottleneck with key determinants to T. b. brucei life-cycle transitions. Our proteomic data provide evidence that these genes are not only transcribed (as evidenced by previous transcriptome-based studies), but are also translated into (potentially functional) proteins. These proteins, especially the immunity-related proteins, present an attractive platform to enhance trypanosome refractoriness as an anti-vector strategy to control tsetse and African trypanosomosis. However, as we have discussed above, we are aware of the need to experimentally validate our proteomics data, a process which is now on-going.
Abbreviations
5′ Nuc, 5′-nucleotidase-related saliva protein; ABC, ABC ammonium bicarbonate; ACN, acetonitrile; ADA, salivary adenosine deaminase; ADGF, adenosine deaminase growth factor; AhpC, alkyl hydroperoxide reductase; AIR9, auxin-induced in root cultures 9; APS, bifunctional ATP/sulfurylase-adenosine 5′-phosphosulfate; ArgK, arginine kinase; AspAT, aspartate aminotransferase; AUH, methylglutaconyl-CoA hydratase; BARP, bloodstream stage alanine-rich proteins; BLAST, basic local alignment search tool; BP, biological process; CALP, calpain; CaMK, Ca2+/calmodulin-dependent protein kinase; CatB, cathepsin B-like cysteine proteinase; CBB, colloidal brilliant blue; CC, cellular component; CRISPR/Cas9, Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR associated protein 9; DBT, dihydrolipoamide transacylase α-ketoacid dehydrogenase; Drk, downstream of receptor kinase; ESAG, expression site-associated gene; FAZ, flagellar attachment zone protein; FDR, false discovery rate; GE-rich, glycine-glutamate-rich protein; gGAPDH, glycosomal glyceraldehayde-3-phosphate dehydrogenase; GO, gene ontology; GPI, glycosylphosphatidyl inositol; GS, glutamine synthetase; GST1, glutathione S-transferase 1; iBAQ, intensity-based absolute quantitation; IID, insect innate immunity database; IPR, iron regulatory protein; ISG, invariant surface glycoprotein; IVD, isovaleryl-CoA dehydrogenase; KMP-11, 11-kDa kinetoplastid membrane protein-11; LC-MS/MS, liquid chromatography coupled to electrospray and tandem mass spectrometry; LFQ, label-free quantification; MF, molecular function; MT, mammalian-infective metacyclic trypomastigote; mtECT, mitochondrial transport chain; MTHF-folD, bifunctional methylene-tetrahydrofolate dehydrogenase; NOS, nitric oxide synthase; nuo-24, mitochondrial NADH-ubiquinone oxidoreductase; OSM3, osmotic avoidance abnormal protei3; P5CDH, δ-1-pyrroline-5-carboxylate dehydrogenase; PFR, paraflagellar rod protein; PPI, protein-protein interaction; PSMA/PSMB, proteasome regulatory complex proteins α-/β-types; Ras-MAPK, Ras-mitogen-activated protein kinase; RH, relative humidity; RHS, retrotransposon hot spot protein; RpS27A, ubiquitin-40S ribosomal protein S27a fusion protein; scFvs, single chain variable fragment antibodies; SDS-PAGE, sodium dodecyl sulphate polyacrylamide gel electrophoresis; Serp-2, stress-associated endoplasmic reticulum protein 2; SG, salivary gland; Sgp3, 5′ nucleotidase-related SG protein 3; SIT, sterile insect technique; SKCRP, small kinetoplastid calpain-related protein; SUMO, small ubiquitin-related modifier 3; TAg5, antigen-5-related allergen; TcP-1, Chaperonin containing t-complex polypeptide 1; TRAP, translocon-associated complex protein; TrxP, thioredoxin peroxidases; Tsal1/2, tsetse salivary gland proteins 1 and 2; Tsp, tetraspanin; TXN, tryparedoxin; uBA1, ubiquitin activating enzyme; uCHL, ubiquitin C-terminal hydrolase; UDE, uracil-DNA degrading factor-like protein; UPS, ubiquitin-proteasome system; V-ATPases, vacuolar ATPases; VSG, variant surface glycoprotein
References
Steverding D. The history of African trypanosomiasis. Parasit Vectors. 2008;1:3.
Abbeele VDJ, Bourtzis K, Weiss B, Cordón-Rosales C, Miller W, Abd-Alla AMM, et al. Enhancing tsetse fly refractoriness to trypanosome infection: a new IAEA coordinated research project. J Invertebr Pathol. 2013;112:S142–7.
Anene BM, Onah DN, Nawa Y. Drug resistance in pathogenic African trypanosomes: what hopes for the future? Vet Parasitol. 2001;96:83–100.
Krafsur ES. Tsetse flies: genetics, evolution and role as vectors. Infect Genet Evol. 2009;9:124–41.
Dale C, Welburn SC, Maudlin I, Milligan PJ. The kinetics of maturation of trypanosome infections in tsetse. Parasitology. 1995;111:187–91.
Welburn SC, Maudlin I. Control of Trypanosoma brucei brucei infections in tsetse, Glossina morsitans. Med Vet Entomol. 1997;11:286–9.
Sharma R, Gluenz E, Peacock L, Gibson W, Gull K, Carrington M. The heart of darkness: growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol. 2009;25:517–24.
Abbeele VDJ, Claes Y, van Bockstaele D, Le Ray D, Coosemans M. Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology. 1999;118:469–78.
Rotureau B, Abbeele VDJ. Through the dark continent: African trypanosome development in the tsetse fly. Front Cell Infect Microbiol. 2013;3:53.
Roditi I, Lehane MJ. Interactions between trypanosomes and tsetse flies. (Special Section: Host-microbe interactions-parasites). Curr Opin Microbiol. 2008;11:345–51.
Abbeele VDJ, Caljon G, De Ridder K, De Baetselier P, Coosemans M. Trypanosoma brucei modifies the tsetse salivary composition, altering the fly feeding behavior that favors parasite transmission. PLoS Pathog. 2010;6:1–9.
MacLeod ET, Darby AC, Maudlin I, Welburn SC. Factors affecting trypanosome maturation in tsetse flies. PLoS One. 2007;2:1–5.
Telleria EL, Benoit JB, Zhao X, Savage AF, Regmi S, Alves e Silva TL, et al. Insights into the trypanosome-host interactions revealed through transcriptomic analysis of parasitized tsetse fly salivary glands. PLoS Negl Trop Dis. 2014;8:e2649.
International Glossina Genome Initiative. Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science. 2014;344:380–6.
Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–22.
Vreysen MJB, Seck MT, Sall B, Bouyer J. Tsetse flies: their biology and control using area-wide integrated pest management approaches. J Invertebr Pathol. 2013;112(Supplement 1):S15–25.
Bouyer J. Does isometamidium chloride treatment protect tsetse flies from trypanosome infections during SIT campaigns? Med Vet Entomol. 2008;22:140–3.
Feldmann U. Guidelines for the rearing of tsetse flies using the membrane feeding technique. In: Ochieng’-Odero JPR, editor. Techniques of insect rearing for the development of integrated pest and vector management strategies. Nairobi: ICIPE Science Press; 1994. p. 449–71.
Kariithi HM, Ince IA, Boeren S, Murungi EK, Meki IK, Otieno EA, et al. Comparative analysis of salivary gland proteomes of two Glossina species with differential hytrosavirus pathologies. Front Microbiol. 2016;7:89.
Lu J, Boeren S, de Vries SC, van Valenberg HJ, Vervoort J, Hettinga K. Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J Proteomics. 2011;75:34–43.
Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72.
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805.
Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods. 2011;53:453–9.
Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42.
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6.
Hu Z-L, Bao J, Reecy JM. CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J Bioinform. 2008;9:108–12.
Nguyen PV, Srihari S, Leong HW. Identifying conserved protein complexes between species by constructing interolog networks. BMC Bioinform. 2013;14:S8.
Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–9.
Kotlyar M, Pastrello C, Pivetta F, Lo SA, Cumbaa C, Li H, et al. In silico prediction of physical protein interactions and characterization of interactome orphans. Nat Methods. 2015;12:79–84.
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2.
Brucker RM, Funkhouser LJ, Setia S, Pauly R, Bordenstein SR. Insect Innate Immunity Database (IIID): an annotation tool for identifying immune genes in insect genomes. PLoS One. 2012;7:e45125.
Langousis G, Hill KL. Motility and more: the flagellum of Trypanosoma brucei. Nat Rev Microbiol. 2014;12:505–18.
Kelly S, Ivens A, Manna PT, Gibson W, Field MC. A draft genome for the African crocodilian trypanosome Trypanosoma grayi. Sci Data. 2014;1:140024.
Banu S, Honore N, Saint-Joanis B, Philpott D, Prevost MC, Cole ST. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol. 2002;44:9–19.
Milne KG, Ferguson MA. Cloning, expression, and characterization of the acyl-CoA-binding protein in African trypanosomes. J Biol Chem. 2000;275:12503–8.
May SF, Peacock L, Meida Costa CI, Gibson WC, Tetley L, Robinson DR, et al. The Trypanosoma brucei AIR9-like protein is cytoskeleton-associated and is required for nucleus positioning and accurate cleavage furrow placement. Mol Microbiol. 2012;84:77–92.
Cole DG. Intraflagellar transport: keeping the motors coordinated. Curr Biol. 2005;15:R798–801.
Maudlin I. Transmission of African trypanosomiasis: interactions among tsetse immune system, symbionts, and parasites. In: Harris K, editor. Advances in disease vector research. New York: Springer; 1991. p. 117–48.
Gibson W, Bailey M. The development of Trypanosoma brucei within the tsetse fly midgut observed using green fluorescent trypanosomes. Kinetoplastid Biol Dis. 2003;2:1.
Vickerman K, Tetley L, Hendry KA, Turner CM. Biology of African trypanosomes in the tsetse fly. Biol Cell. 1988;64:109–19.
Alves-Silva J, Ribeiro JMC, Abbeele VDJ, Attardo G, Hao Z, Haines LR, et al. An insight into the sialome of Glossina morsitans morsitans. BMC Genomics. 2010;11:213.
Caljon G, De Ridder K, De Baetselier P, Coosemans M, Van Den AJ. Identification of a tsetse fly salivary protein with dual inhibitory action on human platelet aggregation. PLoS One. 2010;5:e9671.
Li S, Aksoy S. A family of genes with growth factor and adenosine deaminase similarity are preferentially expressed in the salivary glands of Glossina m. morsitans. Gene. 2000;252:83–03.
Li S, Kwon J, Aksoy S. Characterization of genes expressed in the salivary glands of the tsetse fly, Glossina morsitans morsitans. Insect Mol Biol. 2001;10:69–76.
Zhao X, Alves e Silva TL, Cronin L, Savage AF, O’Neill M, Nerima B, et al. Immunogenicity and serological cross-reactivity of saliva proteins among different tsetse species. PLoS Negl Trop Dis. 2015;9:e0004038.
Abbeele VDJ, Caljon G, Dierick JF, Moens L, De Ridder K, Coosemans M. The Glossina morsitans tsetse fly saliva: general characteristics and identification of novel salivary proteins. Insect Biochem Mol Biol. 2007;37:1075–85.
Cappello M, Li S, Chen X, Li CB, Harrison L, Narashimhan S, et al. Tsetse thrombin inhibitor: bloodmeal-induced expression of an anticoagulant in salivary glands and gut tissue of Glossina morsitans morsitans. Proc Natl Acad Sci U S A. 1998;95:14290–5.
Ogueta SB, Macintosh GC, Téllez-Inon MT. Stage-specific substrate phosphorylation by a Ca2+/calmodulin-dependent protein kinase in Trypanosoma cruzi. J Eukaryot Microbiol. 1998;45:392–6.
Chaubey S, Grover M, Tatu U. Endoplasmic reticulum stress triggers gametocytogenesis in the malaria parasite. J Biol Chem. 2014;289:16662–74.
Békés M, Drag M. Trojan horse strategies used by pathogens to influence the small ubiquitin-like modifier (SUMO) system of host eukaryotic cells. J Innate Immun. 2012;4:159–67.
Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–9.
Shahabuddin M, Pimenta PF. Plasmodium gallinaceum preferentially invades vesicular ATPase-expressing cells in Aedes aegypti midgut. Proc Natl Acad Sci U S A. 1998;95:3385–9.
Jaramillo-Gutierrez G, Rodrigues J, Ndikuyeze G, Povelones M, Molina-Cruz A, Barillas-Mury C. Mosquito immune responses and compatibility between Plasmodium parasites and anopheline mosquitoes. BMC Microbiol. 2009;9:154.
Lehane MJ, Aksoy S, Gibson W, Kerhornou A, Berriman M, Hamilton JV, et al. Adult midgut expressed sequence tags from the tsetse fly Glossina morsitans morsitans and expression analysis of putative immune response genes. Genome Biol. 2003;4:R63.
Hamidou SI, Tchicaya B, Chuchana P, Geiger A. Midgut expression of immune-related genes in Glossina palpalis gambiensis challenged with Trypanosoma brucei gambiense. Front Microbiol. 2014;5:609.
Xavier MJ, Williams MJ. The Rho-family GTPase Rac1 regulates integrin localization in Drosophila immunosurveillance cells. PLoS One. 2011;6:e19504.
Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–74.
Zhuang S, Kelo L, Nardi JB, Kanost MR. An integrin-tetraspanin interaction required for cellular innate immune responses of an insect, Manduca sexta. J Biol Chem. 2007;282:22563–72.
Ragab A, Buechling T, Gesellchen V, Spirohn K, Boettcher AL, Boutros M. Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J. 2011;30:1123–36.
Wells KL. The effects of immune challenge on phenoloxidase activity in locust salivary glands in vitro. Biosci Horizons. 2008;1:122–7.
Hamidou Soumana I, Klopp C, Ravel S, Nabihoudine I, Tchicaya B, Parrinello H, et al. RNA-seq de novo assembly reveals differential gene expression in Glossina palpalis gambiensis infected with Trypanosoma brucei gambiense versus non-infected and self-cured flies. Front Microbiol. 2015;6:1259.
Bekesi A, Pukancsik M, Haasz P, Felfoldi L, Leveles I, Muha V, et al. Association of RNA with the uracil-DNA-degrading factor has major conformational effects and is potentially involved in protein folding. FEBS J. 2011;278:295–315.
Kubota H, Hynes G, Willison K. The chaperonin containing t-complex polypeptide 1 (TCP-1): multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol. Eur J Biochem. 1995;230:3–16.
Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One. 2009;4:e5905.
Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2005;2005:re10.
Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ Res. 2007;100:1276–91.
Rosinski-Chupin I, Briolay J, Brouilly P, Perrot S, Gomez SM, Chertemps T, et al. SAGE analysis of mosquito salivary gland transcriptomes during Plasmodium invasion. Cell Microbiol. 2007;9:708–24.
Smartt CT, Chiles J, Lowenberger C, Christensen BM. Biochemical analysis of a blood meal-induced Aedes aegypti glutamine synthetase gene. Insect Biochem Mol Biol. 1998;28:935–45.
Baumann O, Bauer A. Development of apical membrane organization and V-ATPase regulation in blowfly salivary glands. J Exp Biol. 2013;216:1225–34.
Matthews KR. The developmental cell biology of Trypanosoma brucei. J Cell Sci. 2005;118:283–90.
Horn D. Antigenic variation in African trypanosomes. Mol Biochem Parasitol. 2014;195:123–9.
Jackson DG, Windle HJ, Voorheis HP. The identification, purification, and characterization of two invariant surface glycoproteins located beneath the surface coat barrier of bloodstream forms of Trypanosoma brucei. J Biol Chem. 1993;268:8085–95.
Urwyler S, Studer E, Renggli CK, Roditi I. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol Microbiol. 2007;63:218–28.
Nolan DP, Jackson DG, Biggs MJ, Brabazon ED, Pays A, Van LF, et al. Characterization of a novel alanine-rich protein located in surface microdomains in Trypanosoma brucei. J Biol Chem. 2000;275:4072–80.
Son HJ, Cook GA, Hall T, Donelson JE. Expression site-associated genes of Trypanosoma brucei rhodesiense. Mol Biochem Parasitol. 1989;33:59–66.
Cully DF, Ip HS, Cross GA. Coordinate transcription of variant surface glycoprotein genes and an expression site associated gene family in Trypanosoma brucei. Cell. 1985;42:173–82.
Bringaud F, Biteau N, Melville SE, Hez S, El-Sayed NM, Leech V, et al. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot Cell. 2002;1:137–51.
Castro H, Romao S, Carvalho S, Teixeira F, Sousa C, Tomas AM. Mitochondrial redox metabolism in trypanosomatids is independent of tryparedoxin activity. PLoS One. 2010;5:e12607.
Trujillo M, Budde H, Piñeyro MD, Stehr M, Robello C, Flohé L, et al. Trypanosoma brucei and Trypanosoma cruzi tryparedoxin peroxidases catalytically detoxify peroxynitrite via oxidation of fast reacting thiols. J Biol Chem. 2004;279:34175–82.
Branquinha MH, Marinho FA, Sangenito LS, Oliveira SSC, Gonçalves KC, Ennes-Vidal V, et al. Calpains: potential targets for alternative chemotherapeutic intervention against human pathogenic trypanosomatids. Curr Med Chem. 2013;20:3174–85.
Ersfeld K, Barraclough H, Gull K. Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J Mol Evol. 2005;61:742–57.
Stebeck CE, Beecroft RP, Singh BN, Jardim A, Olafson RW, Tuckey C, et al. Kinetoplastid membrane protein-11 (KMP-11) is differentially expressed during the life cycle of African trypanosomes and is found in a wide variety of kinetoplastid parasites. Mol Biochem Parasitol. 1995;71:1–13.
Liu W, Apagyi K, McLeavy L, Ersfeld K. Expression and cellular localisation of calpain-like proteins in Trypanosoma brucei. Mol Biochem Parasitol. 2010;169:20–6.
Atkinson HJ, Babbitt PC, Sajid M. The global cysteine peptidase landscape in parasites. Trends Parasitol. 2009;25:573–81.
De Gaudenzi JG, Noe G, Campo VA, Frasch AC, Cassola A. Gene expression regulation in trypanosomatids. Essays Biochem. 2011;51:31–46.
Smircich P, Eastman G, Bispo S, Duhagon MA, Guerra-Slompo EP, Garat B, et al. Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi. BMC Genomics. 2015;16:443.
Landfear SM, Ullman B, Carter NS, Sanchez MA. Nucleoside and nucleobase transporters in parasitic protozoa. Eukaryot Cell. 2004;3:245–54.
Dean P, Major P, Nakjang S, Hirt RP, Embley TM. Transport proteins of parasitic protists and their role in nutrient salvage. Front Plant Sci. 2014;5:153.
Sherman IW. Amino acid metabolism and protein synthesis in malarial parasites. Bull World Health Organ. 1977;55:265–76.
Sherman IW. Transport of amino acids and nucleic acid precursors in malarial parasites. Bull World Health Organ. 1977;55:211–25.
Beitz E. Aquaporins from pathogenic protozoan parasites: structure, function and potential for chemotherapy. Biol Cell. 2005;97:373–83.
Mandal G, Orta JF, Sharma M, Mukhopadhyay R. Trypanosomatid aquaporins: roles in physiology and drug response. Diseases. 2014;2:3–23.
Ehrenman K, Sehgal A, Lige B, Stedman TT, Joiner KA, Coppens I. Novel roles for ATP-binding cassette G transporters in lipid redistribution in Toxoplasma. Mol Microbiol. 2010;76:1232–49.
Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–9.
Dong S, Lin J, Held NL, Clem RJ, Passarelli AL, Franz AW. Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PLoS One. 2015;10:e0122353.
De Vooght L, Caljon G, Stijlemans B, De Baetselier P, Coosemans M, Van Den Abbeele J. Expression and extracellular release of a functional anti-trypanosome Nanobody® in Sodalis glossinidius, a bacterial symbiont of the tsetse fly. Microb Cell Factories. 2012;11:1–11.
Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417:452–5.
Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015;112:E6736–43.
Hurwitz I, Hillesland H, Fieck A, Das P, Durvasula R. The paratransgenic sand fly: a platform for control of Leishmania transmission. Parasit Vectors. 2011;4:82.
Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–56.
Acknowledgements
The authors acknowledge Prof. Jan Van Den Abbeele of the Institute of Tropical Medicine Antwerp, Belgium, for preparing parasite infections. We thank our colleagues in the Joint FAO/IAEA coordinated research project (CRP No.: D4.20.15) for insights and discussions that informed several ideas discussed in this article.
Funding
This work was funded by the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, IAEA (CRP No.: D4.20.15) Vienna, Austria.
Availability of data and material
The mass spectrometry proteomics data (Project Name: Proteomics of Tsetse salivary glands before and after infection with Trypanosoma parasite; Project accession: PXD004577) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [100].
Authors’ contributions
Conceived and designed experiments: HMK, JMV, AMMA. Performed experiments: HMK, SB. Analysed data: HMK, EKM, SB. Wrote paper: HMK. All authors read and approved final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Table S1.
523 non-redundant protein groups identified by LC-MS/MS. Table S2. 362 protein groups specific to the host vector. Table S3. 158 protein groups specific to the parasite identified in parasitized SG of G. m. morsitans. Table S4. Four bacterial endosymbiont proteins identified in the G. m. morsitans by LC-MS/MS. Table S5. 276 protein groups upregulated in the parasitized SGs of G. m. morsitans. Table S6. 32 protein groups significantly upregulated in the parasitized SG of G. m. morsitans. Table S7. 81 protein groups downregulated in parasitized SGs. Table S8. Four host proteins not affected by parasite infection. Table S9. Distribution of GO terms associated with biological processes. Table S10. GO terms associated with molecular functions. Table S11. GO terms associated with cellular compartments. Table S12. 225 protein pairs with significant interacting probabilities (≥0.2) determined by FpClass. (XLSX 352 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kariithi, H.M., Boeren, S., Murungi, E.K. et al. A proteomics approach reveals molecular manipulators of distinct cellular processes in the salivary glands of Glossina m. morsitans in response to Trypanosoma b. brucei infections. Parasites Vectors 9, 424 (2016). https://doi.org/10.1186/s13071-016-1714-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1714-z