Abstract
Background
The yeast Starmerella bombicola is renowned for its highly efficient sophorolipid production, reaching titers and productivities of (over) 200 g/L and 2 g/(L h), respectively. This inherent efficiency has led to the commercialization of sophorolipids. While the sophorolipid biosynthetic pathway has been elucidated a few years ago, in this study, it is revisited and true key intermediates are revealed.
Results
Recently, Starmerella bombicola strains developed and evaluated in the past were reevaluated unveiling unexpected findings. The AT enzyme encoded in the sophorolipid biosynthetic gene cluster is the only described enzyme known to acetylate sophorolipids, while the SBLE enzyme encoded by the SBLE gene is described to catalyze the conversion of (acetylated) acidic sophorolipids into lactonic sophorolipids. A double knockout of both genes was described to result in the generation of bolaform sophorolipids. However, new experiments performed with respective S. bombicola strains Δsble, Δat Δsble, and ∆at revealed inconsistencies with the current understanding of the SL pathway. It was observed that the ∆sble strain produces mainly bolaform sophorolipids with higher acetylation degrees instead of acidic sophorolipids. Furthermore, the ∆at strain produces predominantly bolaform sophorolipids and lactonic sophorolipids with lower acetylation degrees, while the ∆at ∆sble strain predominantly produces bolaform sophorolipids with lower acetylation degrees. These results indicate that the AT enzyme is not the only enzyme responsible for acetylation of sophorolipids, while the SBLE enzyme performs an intramolecular transesterification reaction on bolaform glycolipids instead of an esterification reaction on acidic sophorolipids. These findings, together with recent in vitro data, led us to revise the sophorolipid biosynthetic pathway.
Conclusions
Bolaform sophorolipids instead of acidic sophorolipids are the key intermediates in the biosynthetic pathway towards lactonic sophorolipids. Bolaform sophorolipids are found in very small amounts in extracellular S. bombicola wild type broths as they are very efficiently converted into lactonic sophorolipids, while acidic sophorolipids build up as they cannot be converted. Furthermore, acetylation of sophorolipids is not exclusively performed by the AT enzyme encoded in the sophorolipid biosynthetic gene cluster and acetylation of bolaform sophorolipids promotes their transesterification. These findings led to the revision of the industrially relevant sophorolipid biosynthetic pathway.
Similar content being viewed by others
Background
The non-pathogenic yeast Starmerella bombicola is renowned for its exceptional sophorolipid (SL) production capacity, reaching titers (over) 200 g/L [1, 2]. The ‘wild type’ glycolipids typically consist of a sophorose disaccharide linked to a hydroxylated fatty acid and exhibit structural diversity with variations in acetylation, lactonization, and fatty acid composition. The remarkable production capability of S. bombicola has led to the commercialization of SLs in eco-friendly cleaning solutions and diverse industrial formulations, showcasing its significance in producing high-yield, structurally diverse SLs with wide-ranging applications [3]. The individual steps of the SL biosynthetic pathway were previously elucidated and reported upon by our research group [4, 5]. A schematic overview of this pathway is displayed in Supplementary Fig. 1, Additional File 1.
The current understanding of the SL biosynthetic pathway of S. bombicola involves five steps with di-acetylated (diAc) lactonic SLs as the final product. The first step consists of the (sub)terminal hydroxylation of a fatty acid (either coming from a hydrophobic substrate or de novo production) by the action of a cytochrome P450 monooxygenase (CYP52M1) [6]. Subsequent glycosylation of the hydroxy fatty acid involves two UDP-glucosyltransferases. The first one (UGTA1) [7] is responsible for the transfer of a glucose molecule from UDP-glucose to the hydroxylated fatty acid yielding a glucolipid (GL) and UDP. The second glucosylation step is catalyzed by UGTB1 [8], which connects a second glucose molecule from UDP-glucose to the glucose of the GL with a β(1,2) glycosidic bond. As such, an acidic SL is formed consisting of the disaccharide sophorose connected to a hydroxy fatty acid of which the structure is displayed in Fig. 1A. The acidic SLs are subsequently acetylated by the action of an acetyltransferase (AT) [9] and secreted by a specific SL transporter [4, 10]. Lastly, the secreted acidic SLs can be converted into acetylated lactonic SLs by the action of the S. bombicola lactone esterase (SBLE) [11, 12]. The chemical structure of a lactonic SL is presented in Fig. 1B. Due to the presence of a secretion signal on the SBLE protein, the lactonization step is performed extracellularly [11]. All genes encoding the enzymes that perform the intracellular steps of the SL biosynthetic pathway are located in a subtelomeric cluster on the second chromosome, together with the MDR gene [4]. The SBLE gene, in contrast, is located outside of this cluster close to the other end of chromosome two and was found to be differently regulated from the genes in the SL cluster [11].
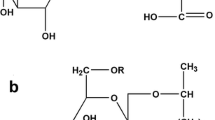
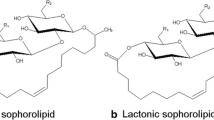
General structure of the described sophorolipids (SLs). A Acidic SL, B lactonic SL, C bolaform SL. The SLs can be acetylated on each of the glucose units and can thus occur as either non-acetylated (R = –H) or acetylated (R = –COCH3) with an acetylation degree up to two (for acidic and lactonic SLs) or four (for bolaform SLs)
Since 2011, our research group has developed several engineered S. bombicola strains to further elucidate the SL pathway. In a first article by Saerens et al. [9] a single deletion of the AT gene was described to produce mostly non-acetylated (nAc) lactonic SLs, in addition to minor amounts of nAc acidic SLs. This indicated the importance of the described acetyltransferase in SL acetylation, and the enzyme was considered to be the sole enzyme responsible for SL acetylation. The Δsble strain in which the SBLE gene had been deleted was reported to solely produce non-, mono- and di-acetylated acidic SLs [11]. Roelants et al. [13] conducted production experiments with this strain as well and reported the production of the acetylated and nAc congeners of acidic SLs. In addition, a strain overexpressing the SBLE gene was shown to almost uniformly produce lactonic SLs. Ciesielska et al. [5] later investigated the mode of action of the SBLE enzyme through in vitro enzyme assays and reported it to catalyze the intramolecular esterification (lactonization) of acetylated acidic SLs in an aqueous environment into acetylated lactonic SLs. No lactonization activity was reported to be observed for nAc acidic SLs, so acetylation was deemed to be essential for the intramolecular esterification catalyzed by the SBLE enzyme.
Subsequently, a strain combining these two deletions (i.e. the S. bombicola Δat Δsble strain) was described by Van Bogaert et al. [14]. Based on the previous findings and the proposed biosynthetic pathway, this strain was expected to produce solely nAc acidic SLs. However, it was found that in addition to these anticipated nAc acidic SLs, the modifications unexpectedly resulted in the biosynthesis of nAc bolaform sophorolipids (bola SLs), of which the general formula is shown in Fig. 1C. More specifically, the authors reported that 74% of the produced SLs were of the nAc bola SL congener [14, 15], which was also described in subsequent works [16, 17]. These bola SLs contain an additional sophorose molecule linked to the carboxyl function of the acidic SLs as confirmed by LC–MS and NMR analysis [14]. This analysis further confirmed variation in the incorporated fatty acid chain length and position of the hydroxyl group on the fatty acid (as is also the case for wild type SLs). Bola SL biosynthesis was proven to be attributed to the promiscuous activity of both UDP-glucosyltransferases (UGTA1 and UGTB1) on nAc acidic SLs, as they were found to display activity towards the carboxyl group of these SLs as well. The absence of acetyl groups was hypothesized to promote the formation of bola SL compounds starting from nAc acidic SLs as they were found by the authors to be produced by the Δat Δsble strain and not by the Δsble strain. The authors also hinted this as the potential reason why these bola SLs are only detected in marginal amounts (< 0.1% of produced SLs) in cultures of the wild type S. bombicola strain [18] as the presence of the AT enzyme in the wild type strain is efficiently giving rise to acetylated SLs, which would thus hamper the formation of bola SLs. Recently, Kobayashi et al. [19] confirmed the presence of trace amounts of bola SLs in the production profile of the wild type S. bombicola.
Because this was an unexpected finding, the glycolipid mixtures produced by both single deletion strains described above were investigated again. Therefore, a modified glycolipid extraction protocol was applied that was adjusted for the isolation of more hydrophilic compounds such as bola SLs. The Δsble strain was confirmed to only produce acidic SLs by the authors [14], while the Δat strain was found to also produce nAc bola SLs in addition to the previously reported nAc acidic and lactonic SLs [14]. The authors thus suggested again that the absence of acetylation seems to be a key factor triggering bola SL synthesis and suggested that this effect is enhanced by the absence of lactonic SLs (where the carboxyl group is not freely available anymore) as better production efficiencies of bola SLs seem to be obtained with the double deletion strain. nAc glycolipid compounds were hypothesized to allow a certain conformational orientation in the UGTA1 and UGTB1 enzymes, which would not be possible for their acetylated equivalents, thus resulting in further glucosylation of nAc acidic SLs resulting in nAc bola SLs.
This manuscript provides a reevaluation of the production spectra of the aforementioned S. bombicola deletion strains: ∆sble, ∆at and ∆at ∆sble. These in vivo experiments, along with the recently performed in vitro experiments with the SBLE enzyme, which are described elsewhere [20], provide new insights into the biosynthesis of SLs. The study demonstrates that S. bombicola strains lacking the SBLE gene primarily yield (acetylated) bolaform glycolipids. As such, this manuscript introduces a revised biosynthetic pathway for SLs, which more accurately reflects the functionality of the SBLE enzyme and thus clarifies its role in SL biosynthesis.
Methods
Strains and cultivation methods
Cloning and plasmid maintenance were performed with One Shot™ TOP10 Chemically Competent Escherichia coli cells (ThermoFisher Scientific). E. coli cells were grown at 37 °C in Luria–Bertani medium (LB; 10 g/L tryptone, 5 g/L yeast extract, 5 g/L sodium chloride and if required 15 g/L agar; Sigma-Aldrich) supplemented with 0.1 g/L ampicillin (LB + Amp; MP Biomedicals) when applicable. Wild type S. bombicola (ATCC 22214) and an URA3 auxotrophic mutant strain (PT36) were used during this study to serve as base strains to generate a set of novel strains described below [21]. Three existing S. bombicola strains developed in the past were also included: the single deletion strain Δsble [11], the double deletion strain Δat Δsble [14, 15] and the single deletion strain ∆at [9]. Solid synthetic dextrose with complete supplement mixture without uracil (SD CSM-URA; 6.7 g/L yeast nitrogen base without amino acids (Sigma-Aldrich), 20 g/L glucose (Cargill), 20 g/L Agar Noble (Difco), 0.77 g/L complete supplement mixture without uracil (MP biomedicals)) and yeast extract peptone dextrose (YPD; 20 g/L glucose (Cargill), yeast extract (DSM), 20 g/L bactopepton (BD biosciences), agar (Biokar Diagnostics)) supplemented with 1 g/L of hygromycin B (Sigma-Aldrich) were used for selection after transformation with the URA3 auxotrophic marker or the hygromycin resistance marker (hph gene of E. coli), respectively.
For the glycolipid production experiments, the production medium as described by Lang et al. [22] was used. Precultures of 5 mL were inoculated from cryovials (1%) and incubated for 48 h at 30 °C (200 rpm). Subsequently, shake flasks (n = 3) containing 100 mL production medium were inoculated (1%) from precultures. Shake flasks were incubated for 240 h at 30 °C (200 rpm). 37.5 g/L oleic acid (Sigma-Aldrich) was supplemented after 48 h of cultivation.
Analytical techniques
Cell dry weight (CDW) was measured by centrifuging 1 mL of shake flask culture (5 min, 14,000 rpm) and the supernatant was removed. The biomass pellet was resuspended in a 0.9% (w/v) NaCl solution and centrifuged once again for 5 min at 14,000 rpm. Finally, the supernatant was discarded and the biomass was placed in an oven at 60 °C for at least 50 h until constant weight in order to remove residual moisture. The net dry biomass was measured gravimetrically and the total CDW was expressed in g dry biomass/L. The pH of shake flask culture samples was measured with a Five easy F20 Mettler Toledo pH/mV meter with two point calibration.
For the detection of sophorose, the undiluted supernatant as obtained after the first centrifugation step of the broth sample (see above), was filtered through a PES filter (0.2 µm, Sartorius) prior to analysis. The supernatant samples were analyzed together with a commercial standard of glucose and non-acetylated sophorose (Sigma-Aldrich).
Glycolipid sample preparation was performed on broth samples of 0.5 mL which were obtained from shake flask cultures. First, two volumes of ethanol absolute (ChemLab) were added to the sample and the solution was vigorously vortexed for 5 min. Subsequently, samples were centrifuged (5 min, 14,000 rpm) and the obtained ‘supernatant’ (an aqueous-ethanol solution in which the glycolipid compounds are solubilized) served as the glycolipid sample for further analysis, which were diluted appropriately and filtered through a PES filter prior to analysis. In-house sophorolipid standards (C18:1 di-acetylated lactonic SLs, C18:1 non-acetylated acidic SLs, C18:1 acetylated acidic SLs (mix) and C18:1 acetylated bola SLs (mix of non- and mono-acetylated)) were analyzed together with the glycolipid samples.
Supernatant and glycolipid samples were analyzed by ultra performance liquid chromatography (UPLC) on an Acquity H-Class UPLC system (Waters) coupled with an Acquity Evaporative Light Scattering Detector (ELSD; Waters).
For the sugar analysis of the supernatant samples, the UPLC-ELSD system was equipped with an Acquity UPLC BEH amide column (130 Ä, 1.7 μm, 2.1 mm × 100 mm; Waters). The column was kept at 55 °C and samples were analyzed at a flow rate of 0.5 mL/min for 8 min/sample. A binary gradient elution system was applied, consisting of 1% triethyl amine in mQ water (eluent A) and 100% acetonitrile (eluent B). The gradient profile was as follows: during the first 3 min, the concentration of eluent A increases from 10 to 25%, which is kept at this level for 0.5 min. Next, the concentration of eluent A decreases again to 10% in 1 min and is maintained at this level for the remaining 3.5 min of the method.
Glycolipid samples were analyzed on the same UPLC-ELSD system in combination with an Acquity UPLC CSH C18 column (130 Ä, 1.7 μm, 2.1 mm × 50 mm; Waters). The column was kept at 30 °C and samples were analyzed at a flow rate of 0.6 mL/min for 10 min/sample. A binary gradient elution system was applied, consisting of 0.5% acetic acid in mQ water (eluent A) and 100% acetonitrile (eluent B). The gradient profile was as follows: during the first 6.8 min, the concentration of eluent B increases from 5 to 95%, after which it decreases again to 5% in 1.8 min and it was maintained at this level for the remaining 1.4 min.
Glycolipid samples were also analyzed by ultra performance liquid chromatography–high resolution mass spectrometry (UPLC–HRMS; Thermo Scientific™ Exactive™ Orbitrap Mass Spectrometer). Products were separated by UPLC according to Van Renterghem et al. [17] with a 2 µL injection volume. The column was kept at 30 °C. MS detection occurred in negative ionization mode with a heated electrospray ionization source operated with a sheath, auxiliary and sweep gas flow rate of 50, 20, and 2 arbitrary units (a.u.), respectively. The spray voltage and needle temperature were set to 4 kV and 40 °C, respectively. The capillary temperature and capillary, tube lens and skimmer voltage were set to 300 °C, 90 V, 60 V and 20 V, respectively. The mass detection range was set from 215 to 1800 m/z. The MS was operated at 50,000 resolving power, a duty cycle of 500 ms and single microscans, the automatic gain was 1000,000. Prior to analysis, the instrument was evaluated (and if required, calibrated) in positive and negative ionization mode using the manufacturer’s calibration reagents. Data processing was performed using the XCalibur (Version 4.1) software.
Molecular techniques
The plasmids carrying the deletion cassettes were cloned and maintained in E. coli, for which the cloning steps are described below. Three different deletion cassettes were constructed for gene deletion in S. bombicola, as shown in Supplementary Figs. 2–4 in Additional File 1.
Plasmids consisted of homologous regions of approximately 1000 bp originating from the S. bombicola genome, the URA3 auxotrophic marker under control of its own promoter or the hygromycin B resistance marker (hph gene of E. coli) under control of the promoter of the glyceraldehyde-3-phosphate dehydrogenase gene (pGAPD), the terminator of the Herpes simplex virus tyrosine kinase (tTK) and the pJET vector backbone (pJET; Thermo scientific) and were used as described by Lodens et al. [21]. In order to construct the deletion cassettes, fragments were first amplified by means of polymerase chain reaction (PCR) using the Primestar® HotStart DNA polymerase (Takara) according to the manufacturer’s instructions. Circular polymerase extension cloning (CPEC) was applied for plasmid assembly using the Q5® Hifi DNA polymerase (New England Biolabs) according to the manufacturer’s instructions and as described in Quan and Tian [23]. CPEC products were transformed into One Shot™ TOP10 Chemically Competent E. coli cells (ThermoFisher Scientific). Positive colonies were selected on LB + Amp plates and verified by colony PCR. Sanger sequencing of the assembled plasmids was performed by Macrogen inc.
Linear deletion cassettes were amplified with the Primestar® HotStart DNA polymerase (Takara). S. bombicola was transformed via electroporation according to Lodens et al. [21] and selected on the appropriate medium as described above. S. bombicola colony PCRs were performed to analyze the 5ʹ, 3ʹ and full overlap of the integration of the deletion cassette in the genome and were performed as described by Lodens et al. [21].
Results
Reevaluation of existing S. bombicola strains
The previously described S. bombicola strains Δsble [11, 12], Δat Δsble [14, 15] and ∆at [9] were reevaluated in a production experiment. This resulted in two unexpected observations in contradiction with former findings.
The first observation relates to the surprising detection of masses corresponding to (acetylated) bola SLs up to an acetylation degree of four in the samples from the Δsble strain, which is thus in strong contrast to these previous observations and reports by Ciesielska et al. [11], Roelants et al. [13] and Van Bogaert et al. [12] where only acidic SLs were described to be produced. The second observation similarly relates to the surprising detection of acetylated (bola) SLs up to an acetylation degree of two in the samples from the experiments with the Δat strain and the Δat Δsble strain, as opposed to all previous publications in which only non-acetylated compounds were reported to be present [9, 14, 15]. The novel findings are based on more thorough analytical analyses (i.e. the detection range in MS analysis) and an extraction method suitable for more hydrophilic compounds produced by the engineered strains (i.e. ethanol solubilization instead of ethyl acetate extraction), as described under the materials and methods section.
Both observations are thus in contradiction with current literature and unexpected as the Δsble strain had been described to exclusively produce (acetylated) acidic SLs and diAc acidic SLs have been described to be the substrate of the SBLE enzyme converting these into diAc lactonic SLs [5, 11]. The Δat Δsble strain and Δat had been described to produce only nAc (bola) SLs, due to mutation of the AT gene, which had been described as the only enzyme responsible for acetylation of SLs so far. The production of acetylated bola SLs has never been described before. As these findings were highly unexpected, additional experiments were performed.
As the abovementioned strains were developed a couple of years ago using restriction enzyme mediated methods, parts of the open reading frames (ORFs) of the respective genes were still present in the modified strains, potentially resulting in residual enzyme activity. Novel S. bombicola strains were thus generated with full deletion of the ORFs/coding sequences, as described under materials and methods, and their glycolipid production profile was evaluated.
Creation and evaluation of novel S. bombicola strains
The deletion cassettes described under materials and methods and in Supplementary Figs. 2–4 in Additional File 1 were amplified from their respective plasmids and used for transformation into S. bombicola strains. The SBLE and AT deletion cassettes with the URA3 marker were used for the creation of the S. bombicola strains containing a single gene deletion, i.e. ∆sble_full and ∆at_full, respectively. After successful verification of the gene deletions, the AT deletion cassette with the hygromycin resistance gene was used for the deletion of AT in the ∆sble_full strain.
The newly developed strains were evaluated for their production characteristics in shake flask experiments together with the wild type S. bombicola strain as described under materials and methods section. A similar pH decline was observed in all shake flasks, starting at a pH of approximately 5.8, decreasing rapidly to a pH of about 3 around 48 h after inoculation, at which value it remained. A similar growth was observed for all strains with a maximum cell dry weight (CDW) of 19 g/L after 84 h of inoculation. Afterwards, the CDW remained constant for all strains except for the wild type strain. This is mainly due to the fact that solid lactonic SLs remain with the cell pellet for the wild type causing a biased CDW determination. These lactonic SLs were clearly visible as a separate layer in the centrifuged broth samples gathered from the wild type strain from 84 h till 240 h of production, while no such layer was detected in the analogous samples from the ∆at, ∆sble and ∆at ∆sble S. bombicola strains. Production samples obtained at 180 h after inoculation were subjected to UPLC–HRMS and UPLC-ELSD analysis. The results are described in the text below and summarized in Fig. 2.
Ultra performance liquid chromatography-evaporative light scattering detector (UPLC-ELSD) chromatograms of production experiments with the A Wild type, B ∆at_full, C ∆sble_full and D ∆at_full ∆sble_full S. bombicola strains. Peaks correspond to the following sophorolipid (SL) compounds with respective masses as determined by ultra performance liquid chromatography–high resolution mass spectrometry (UPLC–HRMS): 1, C18:1 non-acetylated (nAc) bola SL, 945; 2, C18:1 mono-acetylated (mAc) bola SL, 987; 3, C18:0 mAc bola SL, 989; 4, C18:1 di-acetylated (diAc) bola SL, 1029; 5, C18:1 nAc acidic SL, 621; 6, C18:1 tri-acetylated (triAc) bola SL, 1071; 7, C18:0 triAc bola SL, 1073; 8, C18:1 tetra-acetylated (tetraAc) bola SL, 1113; 9, C18:0 nAc acidic SL, 623; 10, C18:1 mAc acidic SL, 663; 11, C18:1 nAc lactonic SL, 603; 12, C18:0 mAc acidic SL, 665; 13, C18:0 nAc lactonic, 605; 14, C18:1 diAc acidic SL, 705; 15, C18:1 diAc lactonic SL, 687
The wild type S. bombicola produces predominantly C18:1 diAc lactonic SL as reported earlier [24]. The novel ∆sble_full strain was evaluated, and the SL spectrum consists primarily of bola and acidic SLs with higher acetylation degrees (Fig. 2B). This shows again that just like the ∆sble strain in which the SBLE gene was partially deleted (data not shown), also the novel SBLE deletion strain with the full SBLE deletion produces (acetylated) bola SLs in contrast to the previous observations that acidic SLs are produced in ∆sble [13, 14]. Moreover, fully acetylated bola SLs (tetraAc C18:1 bola SLs) are produced in quite abundant amounts. This was completely unexpected, as Van Bogaert et al. [14] explicitly reported the absence of bola SLs for the Δsble strain and stated that it is in fact the absence of acetyl groups that triggers the formation of bola SLs starting from acidic SLs.
Upon analysis of the novel ∆at_full strain, in which the AT gene from the SL biosynthetic gene cluster had been completely deleted, indeed again acetylated glycolipid compounds were detected just like in the ∆at strain described by Saerens et al. [9] in which the AT gene was partially deleted. However, lower acetylation degrees as described above for the ∆sble strain were observed. The production spectrum of the ∆at_full strain consists mainly of nAc bola SLs as well as mAc bola SLs and acidic and lactonic SLs with lower degrees of acetylation, confirming the new results obtained with the partially deleted Δat strain described above. As the acetyltransferase from the SL biosynthetic gene cluster was fully removed in the novel ∆at_full strain and acetylated compounds were still observed, this indicates the activity of (an)other unknown acetyltransferases active on SLs.
Lastly, the ∆at ∆sble_full S. bombicola strain was generated. In this strain both the SBLE and the AT genes were fully deleted. The SL production spectrum was evaluated and found to predominantly consist of nAc bola and acidic SLs as well as bola and acidic SLs with lower acetylation degrees. No lactonic SLs were found to be present in the product of this strain, again confirming the new results obtained with the partially deleted Δat ∆sble strain described above. The presence of the low-acetylated compounds is also a new observation compared to what had been previously reported by Van Bogaert et al. [14].
Discussion
In general, it is observed that lactonic SLs are only observed for the wild type strain containing a functional SBLE gene. Furthermore, the ∆at strain produces predominantly bola SLs and lactonic SLs with lower acetylation degrees (non- and mono-acetylated), while the ∆sble strain produces mainly bola SLs with higher acetylation degrees (di-, tri- and tetra-acetylated). The ∆at∆ sble strain predominantly produces bola SLs with a lower degree of acetylation (non- and mono-acetylated). This indicates that the SBLE enzyme has a preference to perform a transesterification reaction on acetylated bolaform glycolipids.
These findings are in contrast with what has been described previously in literature [12, 14, 16, 17]; namely bola SLs can solely be produced as completely nAc molecules, because deletion of the AT gene was described to be required to generate bola SLs. The AT enzyme was moreover described to be the only enzyme acetylating (bola) glycolipids in S. bombicola [9, 14], so the bola SLs in the previously described literature did not contain any acetyl groups.
The aforementioned in vivo experiments, together with the in vitro experiments performed by Diao et al. [20] lead to the necessity to revise the SL biosynthetic pathway. As bola SLs with different degrees of acetylation are formed in the SBLE deletion strains, it is more likely for the bola SLs to be the secreted product rather than for the acidic SLs. The acidic SLs are then an intermediate in this pathway and secreted before full conversion to tetraAc bola SLs can happen and therefore present in the wild type S. bombicola and aforementioned strains. Moreover, a hydrolyzing action of the SBLE enzyme on nAc bola SLs was also proposed and evaluated by Diao et al. [20]. Hence, the authors propose the following revised SL biosynthetic pathway, presented in Fig. 3.
Revised pathway for sophorolipid (SL) biosynthesis by S. bombicola. A All genes, except for the extracellularly bound SBLE, are located in a biosynthetic gene cluster on the second chromosome. B In a first step, a fatty acid (either exogenous or endogenous) is hydroxylated on the terminal (ω) or subterminal (ω-1) position by a P450 monooxygenase CYP52M1 (1). The UDP-glucosyltransferases UGTA1 and UGTB1 consecutively attach one glucose to the free hydroxyl to form a sophorose unit linked to a fatty acyl chain, known as acidic SLs. Through the subsequent action of the same glucosyltransferases, a sophorose unit is attached on the carboxylic end to form a glycosidic ester linkage (2, 3). It must be noted that a simplified version of these glucosylation steps is displayed in this figure, since the exact order is still unknown. These bolaform SLs (bola SLs) can be acetylated to varying degrees (n = 1–4) (4). Through the action of an MDR transporter, bola SLs are secreted in the extracellular space (5). SBLE performs a transesterification reaction resulting in ring closure and the formation of (acetylated) lactonic SL with the release of (acetylated) sophorose (6). For low-acetylated bola SLs (n = 1–2), a hydrolysis route is the preferred mode of action of SBLE and (acetylated) acidic SLs are formed with the release of (acetylated) sophorose
The first step in SL biosynthesis entails the (sub)terminal hydroxylation of the fatty acid at the ω or ω-1 positions by CYP52M1, using the cofactor NADPH and oxygen. The highest efficiency is obtained for the conversion of oleic acid where hydroxylation occurs preferentially at the penultimate position [6, 25]. This results in the formation of ω-1 hydroxylated fatty acids. This is in contrast with, for example, the CYP52M1 of Starmerella kuoi, which solely performs ω-hydroxylation [18].
Subsequently, two glycosyltransferases are responsible for the glycosylation of the hydroxy fatty acids. The first one, UGTA1, transfers a glucose molecule from a UDP-glucose to the two sides of the hydroxy fatty acid, giving rise to a GL and/or bola GL, whereas the second one, UGTB1, transfers a second glucose molecule from UDP-glucose to the (bola) GL molecule, giving rise to an acidic SL and/or bola SL [7, 8]. The first glucose residue is linked to the hydroxylated fatty acid through its 1ʹ position, whereas the 1″ position of the second glucose molecule is linked to the 2’ positions of the first glucose residue [26]. This provides the stepwise formation of the β-1,2 glycosidic bond of the sophorose moiety.
The sophorose groups of the nAc bola SLs can subsequently be acetylated up to an acetylation degree of 4 at the 6ʹ and/or 6″ position of the glucose moieties by the acetyltransferase enzyme AT, which uses acetyl-CoA as the acetyl donor [9]. However, as acetylated compounds can still be detected in the strains containing at least a deletion of the AT gene, (an)other enzyme(s) is/are apparently responsible for part of the acetylation of SLs.
It is important to note that the exact order of glycosylations and acetylations is not certain or that there might not even be a fixed order. In Fig. 3, two glucose moieties are first attached at one end of the fatty acid and then afterwards again two at the other end to make the bolaform SL, but it is also plausible that the synthesis first goes through an intermediate with one glucose attached to each end of the fatty acid, i.e. a bola GL. The same applies to the acetylations: it is unclear in which order the glucose moieties get acetylated and if this only happens on the tetraglycosyl bolaform SL or if it is already possible on the intermediates before. More research is needed to confirm the exact order or the possibility of interchangeable steps leading to tetraAc bolaform SLs, like for example measuring the kinetic parameters of the glycosyltransferases and acetyltransferases on different substrates.
The non-, mono-, di-, tri- or tetra-acetylated bola SLs are secreted to the extracellular space by the MDR transporter protein using ATP [13, 27]. Recently a second transporter of the ABC family was identified with high similarity (72.59% on amino acid level) to the transporter present in the SL biosynthetic gene cluster [10]. Surprisingly, knocking out either one or both transporters gives rise to reduced SL production, indicating that both transporters are required for transport of SLs. These two transporters possibly assemble as oligomers with each other and as such ensure optimal transport.
After transport, the SLs can be converted to acetylated lactonic SLs in the extracellular space through a transesterification reaction by the SBLE enzyme. As bolaform SLs are formed in the SBLE deletion strains, bola SLs are thus most probably the correct substrate of the SBLE enzyme rather than acidic SLs, as stated in literature [11, 12]. Instead of lactonizing acidic SLs, SBLE thus performs a transesterification reaction on bola SLs, converting them to lactonic ones and releasing a sophorose molecule [20].
In addition to the detection of sophorose being released during the in vitro enzyme reactions with SBLE as described by Diao et al. [20], the presence of sophorose in the medium during a production experiment with wild type S. bombicola was demonstrated by means of UPLC-ELSD analysis of the culture supernatant (see chromatograms in Supplementary Fig. 5, Additional File 1). Non-acetylated sophorose was detected in the supernatant of wild type S. bombicola two days after addition of the hydrophobic substrate, whereas this was not the case for the Δsble_full knockout strain. Sophorose does not accumulate in the medium, as the signal decreased to non-detectable levels over the course of the production experiment. It is thus probably taken up by the cells again or split into the two constituting glucose molecules prior to uptake by the cells, as several glycosidases were identified in the exoproteome of S. bombicola [11]. This would enable the cells to recycle the glucose groups to make new bola molecules with a sophorose leaving group for the SBLE to perform the transesterification to lactonic SLs. In addition, large amounts of extracellular sophorose would also negatively influence the thermodynamic equilibrium of the transesterification reaction and would be wasteful for the cells, definitely taken into account SLs are produced in the stationary phase as a nutrient storage and for niche protection in the natural habitat of S. bombicola [28].
In a recent article by Kobayashi et al. [29] significant amounts of sophorolipid glycerides were also detected next to the bola and acidic SLs. The fact that small amounts of bola SLs were detected in their experiments with wild type S. bombicola, as well as reported by Price et al. [18] for the wild type strain, again strengthens the proof that the pathway goes through bola SLs as intermediate. In the experiments performed in this study no glycerides were detected, but this can be explained by Kobayashi et al. [29] feeding with rapeseed oil instead of oleic acid.
With the origin of bola and lactonic SLs cleared out, it is hypothesized that the acidic SLs present in the product mixture of the described S. bombicola strains originate from two different production routes. A first option is also through the conversion of bola SLs by the SBLE. This hydrolytic activity mainly happens for bola SLs with lower acetylation degrees whereas bola SLs with a higher degree of acetylation are converted to lactonic SLs by the same enzyme [20]. This would explain why in the wild type S. bombicola mainly diAc lactonic SLs are detected, and to a lesser extent also nAc and mAc acidic SLs, but almost no bola SLs, whilst in the Δsble strain bola SLs and acidic SLs are the most prevalent products. The observation that the Δsble and Δat Δsble S. bombicola strains in which the SBLE gene had been removed, are still capable of producing acidic SLs indicates that the hydrolysis of low-acetylated bola SLs by the SBLE cannot be the only source of acidic SLs. Thus, it is proposed that part of the extracellularly present acidic SLs are intermediate compounds of the SL biosynthetic pathway that get exported out of the cell before full conversion to tetraAc bola SLs can happen. Strikingly, other intermediate glycolipids of the biosynthetic pathway, such as acidic GLs and triglucolipids, are almost absent in the extracellular space. This is presumably related to the affinity of the transporter proteins for acidic and bola SLs, yet more research is needed to confirm this hypothesis.
Conclusions
Reevaluation of the production spectra of S. bombicola strains ∆sble, ∆at and ∆at ∆sble, together with recently performed in vitro experiments with the SBLE enzyme [20], provided new insights in the biosynthetic pathway of SLs. Rather than acidic SLs, bola SLs are the key intermediate towards the production of lactonic sophorolipids and the degree of acetylation is the determining factor steering the SBLE towards their transesterification into lactonic SLs (for higher acetylated bola SLs) or towards their hydrolysis into acidic SLs (for non- or low-acetylated bola SLs). Moreover, acetylation of SLs is not solely performed by the acetyltransferase encoded by the AT gene of the SL biosynthetic cluster. These findings led to the revision of what was assumed to be the SL biosynthetic pathway.
Availability of data and materials
No datasets were generated or analysed during the current study.
Abbreviations
- bola SL:
-
Bolaform sophorolipid
- CDW:
-
Cell dry weight
- CPEC:
-
Circular polymerase extension cloning
- diAc:
-
Di-acetylated
- E. coli :
-
Escherichia coli
- GL:
-
Glucolipid
- LB:
-
Luria–Bertani medium
- mAC:
-
Mono-acetylated
- nAc:
-
Non-acetylated
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- S. bombicola :
-
Starmerella bombicola
- SBLE:
-
Starmerella bombicola Lactone esterase
- SL(s):
-
Sophorolipid(s)
- tetraAc:
-
tetra-acetylated
- tetraAc:
-
tri-acetylated
- UPLC-ELSD:
-
Ultra performance liquid chromatography-evaporative light scattering detector
- UPLC–HRMS:
-
Ultra performance liquid chromatography–high resolution mass spectrometry
References
Daniel HJ, Reuss M, Syldatk C. Production of sophorolipids in high concentration from deproteinized whey and rapeseed oil in a two stage fed batch process using Candida bombicola ATCC 22214 and Cryptococcus curvatus ATCC 20509. Biotechnol Lett. 1998;20(12):1153–6.
Spencer JFT, Gorin PAJ, Tulloch AP. Torulopsis bombicola sp. n. Antonie Van Leeuwenhoek. 1970;36(1):129–33.
Banat IM, Franzetti A, Gandolfi I, Bestetti G, Martinotti MG, Fracchia L, et al. Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol. 2010;87(2):427–44.
Van Bogaert INA, Holvoet K, Roelants SLKW, Li B, Lin YC, Van de Peer Y, et al. The biosynthetic gene cluster for sophorolipids: a biotechnological interesting biosurfactant produced by Starmerella bombicola. Mol Microbiol. 2013;88(3):501–9.
Ciesielska K, Roelants SLKW, Van Bogaert INA, De Waele S, Vandenberghe I, Groeneboer S, et al. Characterization of a novel enzyme—Starmerella bombicola lactone esterase (SBLE)—responsible for sophorolipid lactonization. Appl Microbiol Biotechnol. 2016;100(22):9529–41.
Van Bogaert INA, Demey M, Develter D, Soetaert W, Vandamme EJ. Importance of the cytochrome P450 monooxygenase CYP52 family for the sophorolipid-producing yeast Candida bombicola. FEMS Yeast Res. 2009;9(1):87–94.
Saerens KMJ, Roelants SLKW, Van Bogaert INA, Soetaert W. Identification of the UDP-glucosyltransferase gene UGTA1, responsible for the first glucosylation step in the sophorolipid biosynthetic pathway of Candida bombicola ATCC 22214. FEMS Yeast Res. 2011;11(1):123–32.
Saerens KMJ, Zhang J, Saey L, Van Bogaert INA, Soetaert W. Cloning and functional characterization of the UDP-glucosyltransferase UgtB1 involved in sophorolipid production by Candida bombicola and creation of a glucolipid-producing yeast strain. Yeast. 2011;28(4):279–92.
Saerens KMJ, Saey L, Soetaert W. One-step production of unacetylated sophorolipids by an acetyltransferase negative Candida bombicola. Biotechnol Bioeng. 2011;108(12):2923–31.
Claus S, Jezierska S, Elbourne LDH, Van Bogaert I. Exploring the transportome of the biosurfactant producing yeast Starmerella bombicola. BMC Genomics. 2022;23(1):22.
Ciesielska K, Van Bogaert IN, Chevineau S, Li B, Groeneboer S, Soetaert W, et al. Exoproteome analysis of Starmerella bombicola results in the discovery of an esterase required for lactonization of sophorolipids. J Proteomics. 2014;26(98):159–74.
Van Bogaert I, Ciesielska K, Devreese B, Soetaert W, Roelants S. A lactonase derived from Candida bombicola and uses thereof. WO 2013/092421. 2013.
Roelants SLKW, Ciesielska K, De Maeseneire SL, Moens H, Everaert B, Verweire S, et al. Towards the industrialization of new biosurfactants: biotechnological opportunities for the lactone esterase gene from Starmerella bombicola. Biotechnol Bioeng. 2016;113(3):550–9.
Van Bogaert INA, Buyst D, Martins JC, Roelants SLKW, Soetaert WK. Synthesis of bolaform biosurfactants by an engineered Starmerella bombicola yeast. Biotechnol Bioeng. 2016;113(12):2644–51.
Soetaert W, Bogaert IV, Roelants S. Methods to produce bolaamphiphilic glycolipids. WO 2015/028278. 2015.
Soetaert W, Roelants S, Stevens C, Delbeke E, Luyten G, Pala M, et al. Efficient synthesis of omega-glycosides and alkyl glycosides. WO 2021/229017. 2021.
Van Renterghem L, Roelants SLKW, Baccile N, Uyttersprot K, Taelman MC, Everaert B, et al. From lab to market: An integrated bioprocess design approach for new-to-nature biosurfactants produced by Starmerella bombicola. Biotechnol Bioeng. 2018;115(5):1195–206.
Price NPJ, Ray KJ, Vermillion KE, Dunlap CA, Kurtzman CP. Structural characterization of novel sophorolipid biosurfactants from a newly identified species of Candida yeast. Carbohydr Res. 2012;348:33–41.
Kobayashi Y, Li Q, Ushimaru K, Hirota M, Morita T, Fukuoka T. Updated component analysis method for naturally occurring sophorolipids from Starmerella bombicola. Appl Microbiol Biotechnol. 2024;12(108):296.
Diao Z, Roelants SLKW, Luyten G, Goeman J, Vandenberghe I, Van Driessche G, et al. Revision of the sophorolipid biosynthetic pathway in Starmerella bombicola based on new insights in the substrate profile of its lactone esterase. Biotechnol Biofuels Bioprod. 2024;17:89.
Lodens S, De Graeve M, Roelants SLKW, De Maeseneire SL, Soetaert W. Transformation of an exotic yeast species into a platform organism: a case study for engineering glycolipid production in the yeast Starmerella bombicola. In: Braman JC, editor. Synthetic biology: methods and protocols. New York: Humana Press; 2018. p. 95–123.
Lang S, Brakemeier A, Heckmann R, Spockner S, Rau U. Production of native and modified sophorose lipids. Chim Oggi. 2000;18(10):76–9.
Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE. 2009;4(7): e6441.
Van Bogaert INA, Zhang J, Soetaert W. Microbial synthesis of sophorolipids. Process Biochem. 2011;46(4):821–33.
Huang FC, Peter A, Schwab W. Expression and characterization of CYP52 genes involved in the biosynthesis of sophorolipid and alkane metabolism from Starmerella bombicola. Appl Environ Microbiol. 2014;80(2):766–76.
Saerens KMJ, Van Bogaert INA, Soetaert W. Characterization of sophorolipid biosynthetic enzymes from Starmerella bombicola. FEMS Yeast Res. 2015;15(7): fvo075.
Jezierska S, Claus S, Van Bogaert I. Yeast glycolipid biosurfactants. FEBS Lett. 2018;592(8):1312–29.
De Clercq V, Roelants SLKW, Castelein MG, De Maeseneire SL, Soetaert WK. Elucidation of the natural function of sophorolipids produced by Starmerella bombicola. J Fungi. 2021;7(11):917.
Kobayashi Y, Li Q, Ushimaru K, Hirota M, Morita T, Fukuoka T. Isolation and characterization of novel naturally occurring sophorolipid glycerides. Bioresour Technol Rep. 2023;1(22):101399.
Acknowledgements
The authors wish to thank Sven Dierickx for the creation of the full knockout strains.
Funding
ZD is indebted to the Chinese Scholarship Council for a scholarship (Grant Number 201706150093) and to Ghent University special research funds for extension of this Grant (BOF21/CDV/064).
Author information
Authors and Affiliations
Contributions
SR, SDM and BD were involved in the experimental study design and described the new function in a patent application. KM and GL performed the data analysis and interpretation. SB, EB, MC, NDF, GL, ZD, TD and MDM wrote the manuscript. SDM and SR edited the manuscript. WS and BD supervised this work, were involved in the data interpretation and edited the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
This work is the subject of a patent application (WO 20232/13677A2) filed by UGent. A license agreement has been signed with AmphiStar, a UGent spin-off created by Wim Soetaert and Sophie Roelants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
13068_2024_2557_MOESM1_ESM.docx
Supplementary Material 1. Supplementary Figure 1: Schematic overview of the SL biosynthetic pathway as previously described in literature; Supplementary Figure 2: Deletion cassette employed for the full knockout of the SBLE gene using the URA3 selectable marker; Supplementary Figure 3: Deletion cassette employed for the full knockout of the AT gene using the URA3 selectable marker. Supplementary Figure 4: Deletion cassette employed for the full knockout of the AT gene using the hygromycin resistance marker (hph). Supplementary Figure 5: UPLC-ELSD chromatograms of supernatant of wild type S. bombicola and the Δsble_full knockout strain and the sophorose standard.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Roelants, S.L.K.W., Bovijn, S., Bytyqi, E. et al. Bubbling insights: unveiling the true sophorolipid biosynthetic pathway by Starmerella bombicola. Biotechnol Biofuels 17, 113 (2024). https://doi.org/10.1186/s13068-024-02557-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-024-02557-7