Abstract
Background
Castor (Ricinus communis L.) seeds contain unusual fatty acid, hydroxy fatty acid (HFA) used as a chemical feedstock for numerous industrial products. Castor cultivation is limited by the potent toxin ricin in its seeds and other poor agronomic traits, so it is advantageous to develop a suitable HFA-producing crop. Significant research efforts have been made to produce HFA in model Arabidopsis, but the level of HFA produced in transgenic Arabidopsis is much less than the level found in castor seeds which produce 90% HFA in seed oil.
Results
We designed a transformation construct that allowed co-expression of five essential castor genes (named pCam5) involved in HFA biosynthesis, including an oleate \(\Delta\) 12-hydroxylase (FAH12), diacylglycerol (DAG) acyltransferase 2 (DGAT2), phospholipid: DAG acyltransferase 1–2 (PDAT1-2), phosphatidylcholine (PC): DAG cholinephosphotransferase (PDCT) and Lyso-PC acyltransferase (LPCAT). Transgenic Arabidopsis pCam5 lines produced HFA counting for 25% in seed oil. By knocking out Arabidopsis Fatty acid elongase 1 (AtFAE1) in pCam5 using CRISPR/Cas9 technology, the resulted pCam5-atfae1 lines produced over 31% of HFA. Astonishingly, the pCam5-atfae1 line increased seed size, weight, and total oil per seed exceeding wild type by 40%. Seed germination, seedling growth and seed mucilage content of pCam5-atfae1 lines were not affected by the genetic modification.
Conclusions
Our results provide not only insights for future research uncovering mechanisms of HFA synthesis in seed, but also metabolic engineering strategies for generating safe HFA-producing crops.
Similar content being viewed by others
Background
Castor seed oil contains 80–90% of ricinoleic acid (12-hydroxy-octadeca-9-enoic acid, 18:1OH), a typical hydroxy fatty acid (HFA) widely used as an industrial raw material for manufacturing high-grade lubricant, paint, coating, plastic, and pharmaceutical products [1, 2]. Castor production is hampered by the presence of deadly toxin ricin and potent allergenic 2S albumins [3,4,5]. Other limiting factors include narrow growth adaptation to tropical regions and labor-intensive hand-harvesting due to not simultaneously maturation of seeds [6]. As such, it is desirable to develop new HFA-producing crops that are safe and suitable for agronomic practices.
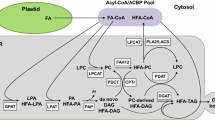
Pathways for seed oil (triacylglycerol, TAG) biosynthesis have been well studied. During seed development, fatty acids (FAs) are synthesized in plastids, exported to the cytosol, and activated to acyl-coenzyme As (acyl-CoAs). The acyl-CoAs are transferred into glycerol-3-phosphate (G3P) to synthesize TAG in the endoplasmic reticulum (ER) [7, 8] (Fig. 1). In the ER, TAGs are synthesized mainly through the de novo biosynthetic pathway or Kennedy pathway [7, 9,10,11], which consists of three sequential acylations of acyl-CoAs to a G3P backbone by glycerol-3-phosphate acyltransferase (GPAT) to produce lysophosphatidic acid (LPA), followed by LPA acyltransferase (LPAT) to generate phosphatidic acid (PA), and PA is then converted to 1,2-sn-diacylglycerol (DAG, or de novo DAG, DAG(1)) by PA phosphatase (PAP). Finally, the DAG is acylated by 1,2-sn-diacylglycerol acyltransferase (DGAT) to produce TAG (Fig. 1). In the cytosol, acyl-CoAs can also be directly incorporated into phosphatidylcholine (PC) through an acyl editing cycle [7, 8, 12,13,14,15]. Lyso-PC acyltransferase (LPCAT) is involved in the forward acylation of sn-2 lyso-PC using acyl-CoA and the reverse reactions of de-acylation sn-2 PC to yield acyl-CoA [7, 15,16,17,18]. The de-acylation of sn-2 PC can also occur with phospholipase A (PLA2)-type activity to yield a free FA, which is then activated to acyl-CoA [19]. Because the FA on the sn-2 PC is the substrate for FA-modifying enzymes, such as desaturases and hydroxylases, rapid de-acylation and re-acylation of PC cause the acyl-CoA pool to be enriched with modified FAs (mFAs), which can subsequently be used for TAG synthesis. Besides the Kennedy pathway, multiple routes utilizing PC lead to TAG formation. PC can be converted to DAG (PC-derived DAG, or DAG(2)) through the removal of the head group from the PC by PC:DAG cholinephosphotransferase (PDCT) [20,21,22]; therefore, acyl-CoAs on the PC are directed to DAG for TAG synthesis. PC-derived DAG can be produced by the reverse action of CDP-choline: DAG cholinephosphotransferase (CPT) [23], a lipase-based mechanism using phospholipase C (PLC), or phospholipase D plus PAP [24]. Because FAs in sn-2-PC can be modified, the conversion of PC into DAG also provides a means to increase the amount of mFAs in sn-2-TAG. Moreover, FA on the sn-2 PC can be transferred to the sn-3 position of DAG by phospholipid:DAG acyltransferase (PDAT) [25,26,27]. Recent metabolic research using Arabidopsis indicates that there is a third DAG pool, bulk-PC-derived DAG (or DAG(3)), a slowly turned over pool likely equilibrated by PDCT-mediated PC-DAG interconversion [28,29,30] (Fig. 1). AtDGAT1 produces TAG from a rapidly produced PC-derived DAG(2) pool, whereas AtPDAT1 and plant DGAT2 utilize bulk-PC-derived DAG(3) pool [28] (Fig. 1). Furthermore, there is growing evidence of existing membrane-associated complexes (or metabolons) made up of enzymes for some or all the reaction steps in a given pathway [28, 31]). The overall control of TAG biosynthesis has also been addressed for the importance of cellular, organellar, and sub-organellar localization of enzymes, structural proteins, and substrate pools [11, 32].
HFA-containing TAG synthesis pathways in Arabidopsis seeds. TAG synthesis is depicted by Kennedy pathways (green arrows) and PC-mediated pathways (red arrows). Arabidopsis genes are highlighted in yellow. Castor genes found to enhance HFA accumulation in Arabidopsis are highlighted in red. Numbers in circles are enzymatic steps targeted in this study. DAG(1) is de novo DAG. DAG(2) is initial rapid formation from PC-derived DAG [55]. DAG(3) is a bulk slowly turned over DAG pool. Dashed arrow indicates DAG(2) can be phase partitioned into DAG(3). FA numerical symbols: 18:1, oleic acid; 18:1OH, ricinoleic acid. Abbreviations: CoA, co-enzyme A; DAG, diacylglycerol; DGAT, diacylglycerol acyltransferase; FAE1, fatty acid elongase1; FAH12, oleate Δ12–hydroxylase; FAS, fatty acid synthesis; G3P, glycerol-3-phosphate; GPAT, glycerol-3-phosphate acyltransferase; LPA, lysophosphatidic acid; LPAT, lysophosphatidic acid acyltransferase; LPCAT, lysophosphatidylcholine acyltransferase; PA, phosphatidic acid; PAP, phosphatidic acid phosphatase; PC, phosphatidylcholine; PDAT, phospholipid:DAG acyltransferase; PDCT, PC:DAG cholinephosphotransferase; TAG, triacylglycerol
Unlike Arabidopsis and most commercial oilseeds, castor has evolved to co-ordinately synthesize and incorporate HFA (18:1OH) into the seed at 90% of TAG [33]. To explore the potential of HFA synthesis in a non-HFA producer, Arabidopsis has been used as a model for studying castor genes and pathways for HFA accumulation in seed [34] (Table 1). The first castor gene isolated and demonstrated to be responsible for 18:1OH synthesis is oleate \(\Delta\) 12-hydroxylase (RcFAH12) gene, which converts oleic acid (18:1) to 18:1OH at sn-2-PC [35]. Seed-specific expression of RcFAH12 in Arabidopsis reveals four non-native HFA in TAG, 18:1OH, 18:2OH, 20:1OH and 20:2OH at 7.8%, 6.6%, 2.5% and 0.4%, respectively (total HFA at 17.3%) [36]. The results suggest that Arabidopsis endogenous AtFAE1 is capable of elongating 18:1OH or 18:2OH to 20:1OH or 20:2OH, respectively, or AtFAD3 is responsible for desaturating 18:1OH or 20:1OH to 18:2OH or 20:2OH, respectively [36]. To simplify the FA profile and provide more 18:1 substrate for RcFAH12, RcFAH12 is expressed in the Arabidopsis fatty acid elongase1 (fae1) mutant, deficient in elongation of 18:1 to 20:1 in seeds [37], and a resulted stable transgenic line is named as CL37 that accumulates 18:1OH and 18:2OH at 13.6% and 3.5%, respectively, with an average of total HFA at 17% [38]. Similarly, expression of a FAH12 in various Arabidopsis mutant backgrounds results in the accumulation of total HFA at approximately 17% in seed oil [39, 40] (Table 1). Undesirably, overexpression of a FAH12 gene in Arabidopsis, including CL37 decreases seed oil content and seed weight [39, 41,42,43,44] (Table 1). The limited accumulation of HFA in Arabidopsis and negative impact on seed total oil content and weight are explained by constrains or bottlenecks of 18:1OH flux into TAGs [20, 21, 26], β-oxidation of unutilized 18:1OH [45], and/or feedback inhibition of FA synthesis [7, 46]. In CL37, inefficient utilization of 18:1OH induces posttranslational inhibition of plastid localized acetyl-CoA carboxylase activity, resulting in a decrease of de novo FA synthesis which ultimately leads to the decreased seed oil content [7, 46]. CL37 has been used to test additional castor genes for their ability to boost HFA content in seed oil. By co-expression of various acyltransferases in CL37, total HFA levels increase to 25–34% in seed TAG [21, 26, 27, 41, 42, 47, 48] (Table 1). Overexpression of additional castor genes in CL37 including castor DGAT2 (RcDGAT2) [41], RcPDAT1A [26] or RcPDAT1-2 [27], and RcPDCT [21], not only increase HFA to 25–27% but also recover seed oil content and weight (Table 1). Overexpression of RcLPAT2 [42, 48], as well as RcLPAT3B and RcLPATB [48], also increased HFA levels in CL37. In contrast, the expression of a castor phospholipase A2-alpha in the CL37 line decreases HFA levels [19] (Table 1). Another hypothesis of limiting HFA accumulation in transgenic Arabidopsis is that competition occurs between endogenous and transgenic isozymes for common FAs versus HFAs [49]. Indeed, when RcFAH12 and RcDGAT2 are co-expressed in a null Arabidopsis dgat1-2 mutant, the HFA level is further increased to 31% in seed oil [49]. Expression of all three castor acyltransferases, RcGPAT9, RcLPAT2 and RcPDAT1A, in CL37 enhances HFA up to 34% [47]. In addition, overexpression of Arabidopsis WRINKLED1 (AtWRI1) [44], a transcription factor for fatty acid biosynthesis, or SEIPIN1 (AtSEI1) [43, 50], a lipid droplet (LD) associated protein localize at the ER–LD junctions during de novo LD formation, also enhance HFA and oil contents in seeds (Table 1).
As described above, no more than four castor genes have been simultaneously co-expressed in Arabidopsis. To further enhance HFA production, we co-expressed five castor genes, RcFAH12 RcDGAT2, RcPDAT1-2, RcPDCT, and RcLPCAT, in Arabidopsis, which resulted in pCam5 lines. Through CRISPR/Cas9 genome editing, we deleted AtFAE1 in pCam5, generating pCam5-atfae1 lines. We found that pCam5-atfae1 lines increased not only HFA content in seeds but also seed size and weight dramatically. Therefore, pCam5-atfae1 lines contain the highest amount of HFA per Arabidopsis seed ever reported. The mechanisms underlying the enhancement of HFA production and seed development were discussed.
Results
Analysis of transgenic Arabidopsis pCam5 expressing five genes from castor
To maximize HFA production in transgenic plants, we included the most critical gene, RcFAH12, which is directly responsible for converting 18:1 to 18:1OH in developing seeds [35]. We also included RcDGAT2, RcPDAT1-2 and RcPDCT, because these genes have been demonstrated to facilitate the channeling of 18:1OH to TAG in transgenic Arabidopsis through various pathways [21, 26, 27, 41] (Fig. 1). Besides, we decided to include RcLPCAT, as biochemical evidence indicates that it is involved in a rapid acyl-editing between HFA–CoA and HFA–PC [7, 15,16,17,18] (Fig. 1).
We constructed these five genes into one transfer DNA (T-DNA) and designated it as pCam5 (Fig. 2a). Transgenic Arabidopsis carrying pCam5 T-DNA would allow the expression of these five genes simultaneously. Transgenic lines resistant to BASTA herbicide (indicating carrying pCam5 T-DNA) were analyzed for FA composition. As HFA is our targeted metabolite, we selected the lines with the highest amount of HFA in seeds. Four T1 pCam5 transformants were obtained. T2 seeds harvested from these four plants were analyzed for FA composition. Compared with WT which does not produce HFA, the transgenics produced three HFAs, 18:1-OH, 18:2-OH, and 20:1-OH, at 3.9–14.7%, 1.3–2.3%, and 0.4–2.2%, respectively (Additional file 1: Table S1). Line 1 (pCam5 1) and line 4 (pCam5 4) contained relatively higher total HFA at 8.3% and 18.3%, respectively (Additional file 1: Table S2). As T2 seeds are segregating populations, 13 T3 off-springs from pCam5 1 lines and 20 T3 off-springs from pCam5 4 lines were analyzed for FA compositions to identify homozygous individuals. As shown in Additional file 1: Table S2, total HFA contents in these T3 seeds ranged from 8.3% (pCam5 1–7) to 22.5–23% (pCam5 1–12, pCam5 1–16) among pCam5 1 lines, and from 12.0% (pCam5 4–11) to 22.8% (pCam5 4–5, pCam5 4–12) among pCam5 4 lines. Noticeably, the total HFA level in these lines can be roughly grouped into low levels (8.3–16.9%) or high levels (22.5–23%) (Additional file 1: Table S2). The results suggested that these five transgenes were either hemizygous or homozygous. The top T3 seeds from pCam5 1–12, pCam5 1–16, and pCam5 4–5 were grown to obtain T4 generation seeds, and eight T4 off-springs from each of these top lines were examined for their FA composition. As anticipated, total HFA contents were comparable among these off-springs, showing 24.4–26.5%, 22.9–26.6%, and 21.4–25.3%, respectively (Additional file 1: Table S3, Fig. 2b). The results indicated that pCam5 1–12-8, pCam5 1–16-7, pCam5 1–16-8 and pCam5 4–5-2 were homozygous lines. For each specific FA composition, HFAs accumulated 16.7–20.7% for 18:1OH, 1.4–4.1% for 18:2OH, and 2.3–3.5% for 20:1OH in transgenic lines, pCam5 1–12-n, pCam5 1–16-n, and pCam5 4–5-n, respectively (Fig. 3). Other FA levels were also changed: linoleic (18:2), α-linolenic (18:3), 11-eicosenoic (20:1), erucic (22:1) acids decreased from 29.7% to 17–20%, 17.8% to 5.4–7.5%, 18.1% to 9.2–13.9%, 1.9% to 0.5–0.8%, respectively (Fig. 3), palmitic (16:0) and stearic (18:0) acids increased from 9.8% to 10.9–17.5% and from 3.3% to 4.5–7.7%, respectively (Fig. 3). 18:1 level was comparable between WT (15.6%) and pCam5 4–5–n (15.4%), but the level increased in pCam5 1–12–n (17.9%) and pCam5 1–16–n (18.2%) (Fig. 3). Continued analysis on T5 seeds from pCam5 1–12-8-n, pCam5 1–16-7-n, pCam5 1–16-8-n and pCam5 4–5-2-n revealed no significant changes in FA composition compared with their corresponding T4 seeds (Additional file 1: Table S4, Fig. 2b) indicating that these five transgenes were stably inherited. Compared with the CL37 [38], which accumulated total HFA at 15.8–16.2% (Additional file 1: Table S4), a homozygote T5 line (pCam5 1–16-7–2) accumulated approximately 9% more HFA showing 25% (Additional file 1: Table S4). To verify the expression of these five transgenes, Reverse transcription PCR (RT–PCR) and quantitative RT–PCR (RT–qPCR) were performed for samples from developing seeds of pCam5 1–16-8 at various developmental stages. All five transgenes were expressed during seed development, where RcFAH12 and RcDGAT2 had a bell-shaped pattern and RcPDAT1-2, RcPDCT and RcLPCAT rose sharply at late stages (Additional file 2: Fig. S1a, b).
HFA content of pCam5 vector transformants. a T-DNA structure of pCam5 vector of five castor gene sets (promoter–gene–terminator). Set1, Phas-FAH12-Nos (2986 bp); Set2, FAE1-RcPDAT1-2-PinII (3324 bp); Set3, Napin-RcLPCAT-Nos (2794 bp); Set4, Phas-RcDGAT2-PinII (2989 bp); Set5, FAE1-RcPDCT-Nos (2050 bp). Each set was sequentially cloned into pCambia3300 vector. The restriction enzyme sites used for cloning are marked on the vector. The size of entire five expression sets is 14.3 kb, and the vector backbone of pCambia 3300 is 8.4 kb, resulting in pCam5 vector in size of 22.7 kb. Phas, phaseolin promoter (1500 bp); FAE1, Fatty acid elongase 1 promoter (946 bp); Napin, napin promoter (1152 bp); Nos, Nos terminator (255 bp); PinII, PinII terminator (406 bp); FAH12 (1164 bp), DGAT2 (1023 bp), RcPDAT1-2 (1,983 bp), LPCAT (1398 bp), PDCT (838 bp), LB, Left border; RB, Right Border. b HFA content in T2 and T4 seeds generations of pCam5 transformants
Fatty acid composition of pCam5 transgenic T4 seeds. Comparison of changes according to fatty acid composition of wild-type seeds and pCam5 transgenic seeds of three independent lines (pCam5 1–12-n, pCam5 1–16-n, and pCam5 4–5-n, Additional file 1: Table S3). Error bars represent SEM of the mean. Statistical analysis is one-way ANOVA with Dunnett’s multiple comparison test (**p < 0.01, ***p < 0.001). ND; not detected
Analysis of pCam5-atfae1 lines produced by knock-out Arabidopsis FAE1 in pCam5 transgenic background through genome editing
FAs 20:1 and 20:1OH are produced through elongation of 18:1 and 18:1OH, respectively, by FAE1 [51, 52] (Figs. 1, 4a). Knock-out (KO) FAE1 would block the formation of 20:1 and 20:1OH and consequently, increase 18:1 and 18:1OH accumulation. As 18:1 is the substrate for FAH12 to produce 18:1OH, the increased 18:1 could also contribute to 18:1OH accumulation (Fig. 1). To test our hypothesis, we applied CRISPR/Cas9 technology along with egg-specific Cas9 expression system [53] (Fig. 4b) and knocked out AtFAE1 in two independent homozygous lines, pCam5 1–12 and pCam5 1–16 (Additional file 1: Table S4). Thirty-five transformants showing resistance to hygromycin were selected. PCR was performed on the leaf genomic DNA using a primer containing the predicted mutation region of the FAE1 gene, and lines having a PCR band smaller than that of the wild-type FAE1 gene were selected. These DNA–PCR products were subjected to Sanger sequencing (Fig. 4c). As a result, we found four independent fae1 knock-out lines in which the FAE1 gene was deleted at 82 bp, or added 1 bp in A or T with a deletion of 82 bp, or added at 45 bp, and we designated these four lines as pCam5-atfae1 5, pCam5-atfae1 9, pCam5-atfae1 19, and pCam5-atfae1 28, respectively (Fig. 4d). Compared with the background seeds (pCam5 1–12-8 and pCam5 1–16-8) which produced 18:1 and 20:1 at averages of 19.7–20.2% and 13.4–12.7%, respectively, pCam5-atfae1 lines (T2 generation) increased 18:1 to 32.0–35.6% and decreased 20:1 to 0.3–0.5% (Fig. 5a, Additional file 1: Table S5). For HFAs, 18:1OH and 18:2OH increased slightly from averages of 18.4–20.4% and 3.4–3.8%, respectively, in pCam5 lines to averages of 22.2–23.4% and 4.2–6.0%, respectively, in pCam5-atfae1 lines (Fig. 5a, Additional file 1: Table S5); 20:1OH dropped from averages of 3.1–3.4% in background pCam5 lines to 0% in pCam5-atfae1 lines (Fig. 5a, Additional file 1: Table S5). We observed a small increase in total HFAs content from an average of 25.3–27.2% in pCam5 background to averages of 27.6–28.9% in T2 generation of pCam5-atfae1 lines (Fig. 5a, Additional file 1: Table S5). As described in the introduction, the CL37 line [38] is a stable transgenic Arabidopsis expressing RcFAH12 in fae1 mutant background that eliminates almost all 20:1 and 20:1OH. We compared FA compositions between CL37 and T3 pCam5-atfae1 lines. As shown in Fig. 5b and Table S6, CL37 accumulates 18:1OH and 18:2OH at averages of 14.1% (± 0.16% SD) and 2.9% (± 0.12% SD), respectively, while the pCam5-atfae1 lines accumulated 18:1OH and 18:2OH at averages of 24.1–26.2% and 3.7–3.9%, respectively (Additional file 1: Table S6). pCam5-atfae1 5–1 showed a highest level of HFA at 31.9% (Additional file 1: Table S6). Furthermore, the HFA content in T4 generation of pCam5-atfae1 lines was examined, and the results showed that 18:1OH and 18:2OH levels were accumulated at similar levels to that of T3 seeds (Fig. 5c, Additional file 1: Table S7). For common FAs, CL37 contains little higher contents in 16:0, 18:2, and 18:3 at 18.0%, 23.8% and 7.1%, respectively, than that of pCam5-atfae1 lines at averages of 12.1–12.4%, 18.5–21.2% and 5.2–5.8%, respectively (Additional file 1: Table S7). CL37 had 18:0 and 18:1 at 6.0% and 27.5%, respectively, comparable to that of pCam5-atfae1, showing averages at 6.3–6.4% and 23.9–26.9%, respectively (Additional file 1: Table S7). For non-HFA, the disappearance of 20:1 in pCam5-atfae1 coincided with the increases of 16:0 from 9.6–9.7% to 12.1–12.4%, 18:1 from 19.7–20.2% to 23.9–26.9%, and 18:2 from 17.1–18.5% to 18.5–21.2% (Additional file 1: Table S7). To verify that transgenic genes were expressed in pCam5-atfae1 5–9 line, RT–PCR and RT–qPCR were performed for samples from developing seeds at various stages. We found that all five genes were expressed during stage one to stage six of pCam5-atfae1 5–9 line (Additional file 2: Fig. S1c, d).
Generation of FAE1 knockout using CRISPR/Cas9 system in pCam5 transgenic line. a Fatty acid synthesis by FAE1 in transgenic Arabidopsis expressing RcFAH12. A solid arrow indicates strong flux, dashed arrow indicates weak flux. b CRISPR/Cas9 vector map for knockout of FAE1 gene in Arabidopsis. HygR, hygromycin resistance gene is driven by CaMV promoter; Cas9 gene is driven by Egg cell-specific promoter; sgRNA is driven by U6 promoter; LB/RB, left border, right border; CaMV P, Cauliflower mosaic virus gene promoter; EC2.1 P, Egg cell-specific promoter; U6 26P, U6 29P, U6 promoter. c Schematic diagram of design for sgRNAs in FAE1 gene. Two single guide RNA (sgRNA1, 2) positions for FAE1 gene editing. To identify the deletion pattern of the FAE1 gene, two primers are designed. d Sanger sequencing result of four independent pCam5-atfae1 lines with addition or deletion of FAE1 gene. Red color indicates PAM(NGG) sequence and blue color indicates mismatch sequence compared to wild type
Fatty acid analysis of transgenic seeds with knock-out of FAE1 in pCam5 lines. pCam5 1–12-8 and pCam5 1–16-8 are controls. Fatty acid composition in seeds of pCam5-atfae1 5, 9, 19, and 28 at T2 (a), T3 (b), and T4 (c) generations. Error bars represent SEM of the mean. Statistical significance is indicated by different letters and tested by one-way ANOVA with Tukey’s multiple comparison test (*p < 0.05). ND; not detected
Comparison of oil content, seed size, and seed germination among wild type, CL37, pCam5 1–16-7–2, and pCam5-atfae1 28–3-6
To determine the oil content of the seeds, the total fatty acid methyl ester (FAME) content was measured by gas chromatography (GC) for wild type (WT, 0% HFA), and three transgenics, CL37 (17% HFA), pCam5 1–16-7–2 (25.3% HFA), and pCam5-atfae1 28–3-6 (30.5% HFA). WT contained a little higher level of FAME per mg dry weight (DW) (254.72 µg/mg) than that of pCam5 1–16-7–2 (227.1 µg/mg DW), CL37 (216.48 µg/mg DW), and pCam5-atfae1 28–3-6 (214.22 µg/mg DW) (Fig. 6a). However, pCam5-atfae1 28–3-6 had significant higher amount of total FAME content per seed (6.54 µg/seed) than that of CL37 (3.68 µg/seed), and pCam5 1–16-7–2 (3.9 µg/seed) (background control of pCam5-atfae1 28–3-6) (Fig. 6b). WT had 5.5 µg/seed of FAME (Fig. 6b). To examine the effect of genes on HFA content, we calculated the non-HFA–FAME and HFA–FAME. WT contains the most non-HFA–FAME at 254.72 µg/mg DW followed by CL37 (180.36 µg/mg DW), pCam5 1–16-7–2 (167.98 µg/mg DW), and pCam5-atfae1 28–3-6 (146.26 µg/mg DW) (Fig. 6c). In term of non-HFA–FAME content per seed, WT also contains the most at 5.5 µg/seed, however, followed by pCam5-atfae1 28–3-6 (4.48 µg/seed), CL37 (3.06 µg/seed), and pCam5 1–16-7–2 (2.9 µg/seed) (Fig. 6d). Regarding HFA–FAME, pCam5-atfae1 28–3-6 contained the most at 67.94 µg/mg, or 2.06 µg/seed, followed by pCam5 1–16-7–2 (59.1 µg/mg, 1 µg/seed) and CL37 (36.12 µg/mg, 0.6 µg/seed) (Fig. 6e, f).
Comparison of oil (total FAs) contents among transgenic lines (pCam5 and pCam5-atfae1), wild type (WT), and CL37. a Total FAME per mg DW. b Total FAME per seed. c non-HFA FAME per mg DW. d non-HFA FAME per seed. e HFA FAME per mg DW. f HFA FAME per seed. The results are measured in five technical replicates. Error bars represent SEM of the mean (n = 5). Statistical analysis is one-way ANOVA with Turkey’s multiple comparison test (*p < 0.05, ***p < 0.001). ND; not detected
The size and weight of WT, CL37, pCam5 1–16-7–2 and pCam5-atfae1 28–3-6 were measured. The average width of WT was 0.29 mm, slightly wider than CL37 (0.23 mm) and pCam5 1–16-7–2 (0.27 mm). However, pCam5-atfae1 28–3-6 increased to 0.30 mm (Fig. 7a). Seed length did not differ between WT and pCam5 16–7-2, but seed length of pCam5-atfae1 28–3-6 was 0.59 mm, significantly larger than that of CL37 (Fig. 7b). The seed size (estimated by multiplying the length and width of the seed) of pCam5-atfae1 28–3-6 was significantly bigger than that of WT, CL37 and pCam5 1–16-7–2 (Fig. 7c). Seed sizes at descend order are pCam5-atfae1 28–3-6 (0.18 mm2) > WT (0.13 mm2) > pCam5 16–7-2 (0.12 mm2) > CL37 (0.09 mm2). The 100 seed weight of pCam5-atfae1 28–3-6 was 3.06 mg, significantly heavier than that of WT (2.16 mg/100 seeds), pCam5 1–16-7–2 (1.72 mg/100 seeds), and CL37 (1.7 mg/100 seeds) (Fig. 7c, d, e). Compared with CL37, pCam5-atfae1 28–3-6 increased by 100% in seed size and weighed 80% heavier. When compared with WT, pCam5-atfae1 28–3-6 exceeded by 40% both in seed size and weight.
Characterization of seeds in transgenic pCam5 and pCam5-atfae1 lines. a Seed width. b Seed length. c Seed size (width × length) of wild type, CL37, pCam5 1–16-7–2, and pCam5-atfae1 28–3-6. The results are measured in triplicate (n = 20). d Seed weight was measured in five technical replicates (n = 100). e Image of Arabidopsis seed. Scale bar = 1 mm. Error bars represent SEM of the mean. Statistical significance is indicated by different letters and tested by one-way ANOVA with Tukey’s multiple comparison test (*p < 0.05)
To test the effect of HFA on seed germination and seedling development, seed germination rate and percentage of healthy seedlings (seedlings with open cotyledons) are counted. The time to reach 50% of the maximum germination (T50) was observed in the following equal or ascend order: WT (36 h) = fae1 (36 h) = pCam5 1–12-8 (36 h) = pCam5-atfae1 5–10 (36 h) < pCam5-atfae1 28–3 (48 h) < pCam5 1–16-7 (60 h) (Fig. 8a). The time to reach 50% of the maximum number of healthy seedlings is in the following ascend order: WT (60 h) < fae1 (72 h) < pCam5-atfae1 5–10 (84 h) < pCam5 1–12-8 (96 h) = pCam5-atfae1 28–3 (96 h) = pCam5 1–16-7 (96 h) (Fig. 8b).
Germination rate and healthy seedlings in pCam5 and pCam5-atfae1 transgenic lines. a Germination rate. b Percentage of healthy seedlings with fully opened cotyledons in wild type, fae1, two pCam5 lines (1–16-7, 1–12-8), and two pCam5-atfae1 lines (28–3, 5–10). The emergence of the radicle was counted as germination. The appearance of roots and green fully opened cotyledons was scored as healthy seedlings (or established seedlings). Data represent means of three biological replicates ± SEM
Comparison of seedling growth and mucilage among WT, fae1 and pCam5-atfae1
To further examine the seedling growth, the leaf size and root length of the seedlings after 10 days of imbibement were measured for WT, fae1 and pCam5-atfae1. As shown in Fig. 9, fae1 had smaller leaf size and shorter root length than WT (Fig. 9). However, the leaf size of pCam5-atfae1 is between WT and fae1, and the root length of pCam5-atfae1 is statistically same as the of WT (Fig. 9). These results indicate that pCam5-atfae1 restored the growth inhibition by fae1 to a level close to WT. It is reported that transgenic Arabidopsis expressing a T-6b oncogene from A. tumefaciens increased seed size and oil content in mature seeds but decreased seed starch and seed coat mucilage content at the same time [54]. We compared mucilage content among WT, fae1 and pCam5-atfae1. As shown in Fig. 10, the mucilage content in fae1 was reduced compared to that of WT, but in pCam5-atfae1, the mucilage content was restored to WT level (Fig. 10). The results indicated that pCam5-atfae1 increased size but at the same time also increased seed coat mucilage content.
Seedling phenotype of pCam5-atfae1 28–3-6. a Seedling growth phenotype. The picture was taken for 10-day-old seedlings grown on ½ MS media (n = 3). Scale bars = 10 mm. The width of the true leave (b) and root length (c) were measured by Image J program (n = 3). Data represent means of three biological replicates ± SEM. Statistical analysis is one-way ANOVA with Turkey’s multiple comparison test (*p < 0.05, ***p < 0.001, ****p < 0.0001)
Analysis of mucilage by ruthenium red (RR) staining. Seeds of wild type (a), fae1 (b), and pCam5-atfae1 28–3-6 (c) were stained with RR (n = 50). Scale bars = 250 µm. d Mucilage area was calculated by HKBasic program using Nikon Eclipse Ci–L microscope (n = 10). The mucilage area was divided by the seed area for normalization (n = 10). Data represent means of three biological replicates ± SEM. Statistical analysis is one-way ANOVA with Turkey’s multiple comparison test (***p < 0.001)
Discussion
The current commercial source of HFA is castor oil and its production is limited by undesirable agronomic traits [6]. To investigate if high levels of HFA can be produced in a common oilseed crop, we targeted multiple genes through different approaches. We devised a five-gene stacker, pCam5, that contained RcFAH12, RcDGAT2, RcPDAT1-2, RcPDCT and RcLPCAT. These genes encode key enzymes in the synthesis and incorporation of HFA into TAG (Fig. 1). We introduced pCam5 into Arabidopsis which has been used as a model to study non-native HFA accumulation in seed oil. Homozygous transgenic Arabidopsis lines expressing pCam5 produced total HFA at averages of 24–25.3% (Additional file 1: Table S3), which is comparable to previously reported HFA levels at 25–27% through co-expressing of single RcDGAT2 [41], RcPDAT1-2/RcPDAT1A [21, 26, 27], or RcPDCT [21] in CL37. However, CL37 is generated in the fae1 background, so we assessed the effect of the elongase by knocking out AtFAE1 in pCam5 transgenics through gene editing. The pCam5-atfae1 lines increased total HFA content up to 29.1% (± 1.4% SD) (Additional file 1: Table S6; Fig. 5). Compared among CL37, pCam5 and pCam5-atfae1, 20:1OH and 20:1 were found only in pCam5 lines at averaging 3.1–3.4% and 12.7–13.4%, respectively (Additional file 1: Table S7; Fig. 5), indicating that the mutation of fae1 in CL37 or deletion of AtFAE1 in pCam5-atfae1 prevented the elongation of 18:1OH to 20:1OH and 18:1 to 20:1, respectively. Considering similar levels of 18:2OH (averages 3.4–4.2%) displayed in pCam5 and pCam5-atfae1 (Additional file 1: Table S7), the increased total HFA in pCam5-atfae1 was due to the increased 18:1OH content which rose from averages of 18.4–20.4% in pCam5 to averages 24.6–25.8% in pCam5-atfae1 (Additional file 1: Table S7). The results indicated that efficient incorporation of 18:1OH–CoA into TAG occurred in pCam5-atfae1 and the increased 18:1OH was at the expense of 20:1OH. For non-HFA, the blocked production of 20:1 in pCam5-atfae1 coincided with the increases of 16:0 from 9.6–9.7% to 12.1–12.4%, 18:1 from 19.7–20.2% to 23.9–26.9%, and 18:2 from 17.1–18.5% to 18.5–21.2% (Additional file 1: Table S7). It is expected that knocking out of AtFAE1 shifted 20:1 to 18:1. Since 18:1 was the substrate for FAD2 for synthesizing 18:2, an increase of 18:2 was also expected. However, the mechanisms of the increase of 16:0 were less clear. Based on 16:0 content in WT at 8.1% and in fae1 at 11.1% (Additional file 1: Table S4), similar levels were observed in pCam5 at 9.6–9.7% and in pCam5-atfae1 at 12.1–12.4% (Additional file 1: Table S7). The increase in 16:0 would not be directly associated with the expression of castor genes, it could be due to the changes that occurred in plastid during FA synthesis and/or in exporting of 16:0 to cytosol and ER for TAG assembly. We observed that pCam5-atfae1 lines dramatically increased seed size and weight. pCam5-atfae1 28–3-6 showed 1.8-fold bigger or heavier than CL37, 1.5-fold bigger and 1.8-fold heavier than pCam5 1–16-1–7, and 1.4-fold bigger or heavier than WT (Fig. 7). Total FAME per seed was measured 1.8-, 1.7- and 1.2-fold higher than that of CL37, pCam5 1–16-1–7 and WT, respectively (Fig. 6b). The dramatic increases in seed size and weight indicated that the five castor genes (RcFAH12, RcDGAT2, RcPDAT1-2, RcLPCAT, RcPDCT) over-expressed and AtFAE1 silenced in pCam5-atfae1 lines co-ordinately may enhance the seed development and reserve accumulation, surpassing WT.
Increases of de novo FA biosynthesis and input of FA flux from plastid to ER must occur to support seed growth of pCam5-atfae1. Overexpression of RcFAH12 alone in WT Arabidopsis or various mutant backgrounds including CL37 accumulates limited HFA at approximately 17% in the seed oil and concomitantly reduces seed oil and weight [21, 26, 38,39,40, 42, 44] (Table 1). Arabidopsis utilizes mainly PC-derived DAG(2) to produce TAG [20, 55] (Fig. 1). The PC-derived DAG(2) is synthesized through de novo DAG → PC → PC-derived DAG(2) [28] (Fig. 1). When RcFAH12 is expressed in Arabidopsis seeds, the de novo DAG containing 18:1OH is not efficiently converted to 18:1OH–PC by the Arabidopsis gene (e.g., AtPDCT), thus the converting step is a bottleneck to TAG synthesis. Over-expression of RcPDCT in CL37 increases HFAs from 17% to 25–27% and recovered seed oil content as well [21] (Table 1). Besides, the last step of TAG synthesis catalyzed by DGAT or PDAT is also bottlenecks in CL37, as overexpression of RcDGAT2 or RcPDAT1A increases HFA content and recovers seed oil content [26, 27, 41]. RcPDCT, RcDGAT2 and RcPDAT1A have been demonstrated to facilitate the utilization of 18:1OH to TAG through channeling 18:1OH–CoA, 18:1OH–DAG, and 18:1OH–PC to TAG [21, 26, 27, 41]. Metabolic analysis reveals that feedback inhibition of de novo FA synthesis, mainly through post-translational inhibition of acetyl-CoA carboxylase is triggered by unutilized HFA during TAG assembly, leads to reductions in HFA level and seed oil [46]. In fact, more efficient utilization of HFA for TAG synthesis by co-expression of RcDGAT2 or RcPDAT1A in CL37 alleviates the reduced acetyl-CoA carboxylase (ACCase) activity [46]. Compared with pCam5, pCam5-atfae1 accumulated more 18:1OH which is favorable substrate to castor genes. It is plausible that in pCam5-atfae1 lines, the multiple bottlenecks were simultaneously overcome because of more efficient channeling of 18:1OH into TAG by RcPDCT, RcDGAT2, and RcPDAT1-2, which enhanced de novo FA synthesis and led to the increased TAG assembly and seed development.
Genetic modifications in pCam5-atfae1 may provide additional metabolic enzyme complexes (metabolons) favorable in channeling C18 FA into TAG. In WT Arabidopsis, AtDGAT1 is the major enzyme that can rapidly utilize the PC-derived DAG(2) for TAG synthesis [28] (Fig. 1). Based on protein–protein interactions, AtDGAT1 interacts with AtPDCT and AtLPCAT2 [28], which forms a metabolon. The forward reaction of LPCAT transfers the acyl chain from CoA to LPC [15, 56], and the reverse LPCAT reaction can directly produce acyl-CoA [18] to be utilized for TAG biosynthesis (Fig. 1). PDCT carries out head group exchange between PC and DAG and is the key enzyme responsible to generated PC-derived DAG(2). In pCam5-atfae1, RcLPCAT and RcPDCT could also interact with AtDGAT1 forming additional metabolons. Besides, the absence of AtFAE1 would eliminate the competition with LPCATs for substrate C18 FA–CoA, allowing more C18 FA to be transferred to PC by LPCATs. Furthermore, RcLPCAT or RcPDCT has been shown strong activity in utilizing C18 FA [18, 22]. These features may collectively enable AtDGAT1 to assemble more PC-derived DAG(2) into TAG, resulting in increased seed oil biosynthesis. The PC-derived DAG(2) can turn to a larger and more slowly turned over bulk PC-derived DAG(3) pool, which is kept in equilibrium with PC by PDCT [28]. In the null mutant dgat1-1, AtPDAT1 becomes a major enzyme transferring FA from PC to form PC-derived DAG(3) [28] for TAG assembly [17, 57]. When RcDGAT2 is overexpressed in dgat1-1, RcDGAT2 competes with AtPDAT1 for PC-derived DAG(3) [28]. Since RcDGAT2 interacts with AtPDCT, AtPDAT1, and AtLPCAT2 [28], it was likely additional metabolons were formed in pCam5-AtFAE1 by interactions among enzymes from castor (RcDGAT2, RcPDAT1-2, RcPDCT, RcLPCAT) and Arabidopsis (AtPDAT1, AtPDCT, AtLPCAT2). These metabolons may efficiently utilize PC-derived DAG(3) for enhanced TAG accumulation in pCam5-atfae1. Although the exact mechanisms need to be investigated, pCam5 and pCam5-atfae1 provide important materials for metabolic studies to understand the metabolic complexes, substrate preference and pools, and pathways for TAG synthesis in seed oils.
We compared seed germination and seedling establishment among WT, fae1, pCam5, and pCam5-atfae1 (Fig. 8). Although seed germination and seedling establishment of pCam5 and pCam5-atfae1 lines were slower than that of WT and fae1, they all had a 92–100% germination rate in 4 days (Fig. 8), a reasonable time required for Arabidopsis seed germination [58]. After 6 days of seed imbibition, seedling establishment in pCam5 and pCam5-atfae1 reached 77–83% and 81–91%, respectively, slightly lower than that of WT or fae1 at 97% (Fig. 8). The results indicate that seeds from pCam5 and pCam5-atfae1 are viable. We also compared seedling growth (Fig. 9) and seed coat mucilage content (Fig. 10) among WT, fae1 and pCam5-atfae1. Our results indicated that pCam5-atfae1 recovered the adverse effect of fae1 on seedling growth and seed coat mucilage content. Therefore, pCam5-atfae1 increased seed size and oil content but displayed normal seedling growth and mucilage content. We are currently investigating if other carbohydrates such as sugar or starch are also proportionally increased in pCam5-atfae1 seeds.
A pCam5-atfae1 line increased seed biomass and seed size by 40% compared to WT (Fig. 7). Before this research, the highest amount of HFA per seed is 1.81 µg HFA/seed [41] (Table 1), a pCam5-atfae1 line produced 1.99 µg HFA/seed, the highest amount of HFA in a seed ever achieved. Ectopic expression of an Agrobacterium tumefaciens T-DNA oncogene, T-6b, also increases Arabidopsis seed biomass, oil content and seed size by 10% [54]. This is due to, in part, the expression of T-6b resulting in increased expression of WRI1 and DGAT1 and ultimately increasing the seed TAG [54]. Until now, only Camelina has been transformed with castor FAH12, the resulted plants produce 15% HFA [59]. By co-expression of RcFAH12 and a lesquerella KCS18 (or PfKCS3 and PfFAE1) [52], the resulted transgenics produce up to about 20.9% of HFA [60]. The similarity between Arabidopsis and Camelina as hosts to express castor gene for HFA production would allow to apply knowledge from Arabidopsis to other crops, including Camelina, rapeseed, and soybeans. Besides, the design of pCam5 allows one transformation process to express five different key genes essential for HFA synthesis, which avoids two or more separated procedures and thus shorten the time to generate HFA-producing plants. Our strategies demonstrated in this study can help design future HFA-producing crops through genetic engineering.
Methods
Plant materials, Arabidopsis transformation and growth
Arabidopsis thaliana wild type, Columbia-0 (Col-0), was used for the transformation of recombinant vectors. CL37 transgenic plant producing 17% HFA was used as the control [38]. Wild type and transgenic plants were germinated in MS medium containing 1% sucrose, the seedlings were transferred to soil or were directly germinated, and then grown in soil. Arabidopsis was grown on a control growth chamber under the conditions of 22 °C, 16 h light/8 h dark photoperiod. The light intensity was 100 µmol m–2 s–1. Agrobacterium strain, GV3101, was used to transform pCam5 (pCambia-set1-set2-set3-set4-set5) vector into Arabidopsis Col-0 by floral dipping. The harvested seeds were selected on MS medium supplemented with 1% (w/v) sucrose, 10 mg/L DL–phosphinothricin (PPT), 100 mg/L of carbenicillin and 0.8% (w/v) plant agar. Selected T1 individuals showing resistance to PPT were transferred to soil for further experiments. T2 seeds and their decedents T3 and T4 were continued for PPT selection. As T2 seeds are segregating population, homozygous lines were identified by selecting T3 seeds with resistant to PPT and higher concentration of HFA than their segregants. Stable homozygous lines were further confirmed at T4 generation.
Plant expression vector construction for co-expression of five genes
A vector containing five castor genes was constructed as the following method. Five castor genes involved in HFA synthesis and accumulated in TAG were contained in a vector; each gene has each promoter and terminator. Each of the cDNA RcFAH12, RcPDAT1-2, RcDGAT2, RcPDCT and RcLPCAT were synthesized from mRNA isolated from castor developing seeds and cloned into pENTR/D-TOPO vector (Invitrogen, USA) to obtain pENTR–RcFAH12, pENTR–RcPDAT1-2, pENTR–RcDGAT2, pENTR–RcLPCAT and pENTR–RcPDCT, respectively. To minimize the gene silencing, three kinds of seed-specific promoters including phaseolin promoter from soybean, Arabidopsis FAE1 promoter from Arabidopsis and Brassica Napin promoter from Brassica napus were used [61,62,63]. These promoters were amplified by PCR with the promoter cloned using vectors pGEM–phaseolin, pMDC32–AtFAE1, and pMDC–Napin as templates, respectively. Nos and Pin II were used as two types of terminators. These sequences were obtained by PCR amplification of the PinII terminator from the pBI221 vector and Nos terminator from the pPZP–3'PINII-Bar vector. Primers were designed to ligate the promoter, gene, and terminator at once (Additional file 1: Table S8). Each of the PCR products was inserted into the pUC19 vector by the infusion cloning method. Five subcloning vectors were completed after sequencing. Each gene expression set (promoter–gene–terminator) in five subcloning vectors was PCR-amplified using primers with unique restriction enzymes and cloned into the restriction site of the multi cloning site of the pCambia3300 plant expression vector stepwise. First, the Phas-FAH12-Nos set was inserted into EcoRI site of pCambia3300 and cloned to complete pCambia3300–Set1. Second, pCambia3300–set1–set2 was constructed by inserting the AtFAE1–RcPDAT1-2–PinII set into the SmaI site of pCambia3300–set1. Next, the Napin–RcLPCAT–Nos set was inserted into XbaI site to construct pCambia3300–set1–set2–set3. In addition, the pCambia3300–set4 was prepared by inserting the Phase-RcDGAT2–PinII set into the EcoRI/BamHI site in pCambia3300, and the AtFAE1–RcPDCT–Nos set was further inserted into the BamHI/HindIII site to prepare pCambia3300–set4–set5. To insert 5 gene expression sets into one vector, we cut the ApaI/HindIII in pCambia3300–set4 with ApaI/EcoRI in the vector of pCambia3300–set1–set2–set3 using ApaI site on one side and blunt-end ligation to construct pCambia3300–set2–set3–set4. In this vector, Phas-FAH12–Nos set1 was inserted into the EcoRI site again to construct pCambia2200–set1–set4–set2–set3. Finally, we constructed the pCambia3300–set1–set4–set2–set3–set5 (pCam5) by inserting the AtFAE1–RcPDCT–Nos set into the AscI site of pCambia3300–set1–set4–set2–set3.
CRISPR/Cas9 vector cloning for Arabidopsis FAE1 knockout
To knockout the Arabidopsis fatty acid elongase 1 (FAE1) gene, two guide RNAs (5′-GCTGCAAAAGTCTTCCGCGG-3′ and 5′-AAGTTAACCCTAGAGAGATC-3′) were designed using the CRISPR–RGEN tool available online (http://www.rgenome.net). The BsaI enzyme site was attached to the front of each primer and PCR was performed using the pCBC–DT1T2 vector as a template. The PCR product was purified by a PCR Purification kit (QIAGEN, Germany). This product was put into pHEE401E-egg vector and treated with BsaI enzyme to proceed with the golden gate reaction [53]. After cloning, Sanger sequencing was performed to confirm whether the guide RNA sequence was in the vector. This vector was transformed into pCam5 lines producing 25% HFA in seed oil.
Fatty acid analysis
The fatty acid composition and total oil content of Arabidopsis seeds was analyzed using gas chromatography (GC). First, 500 μl toluene and 500 μl of 5% H2SO4 dissolved in methanol were added and methylation was performed in 85 °C water bath. Pentadecanoic acid (15:0) was used as the internal standard. After methylation at 85 °C for 2 h, the sample was cooled and 1 ml of 0.9% NaCl was added. Then 1 ml of n-hexane was added and violently mixed. The sample tube was centrifuged at 3000 rpm for 2 min. The supernatant was transferred to a new 6ml round tube. The step from adding 1 ml of n-hexane to transferring supernatant was repeated three times. After total of 3 ml supernatant was purged with nitrogen gas, the 200 μl of n-hexane was added thereto. The FAMEs dissolved were analyzed by GC-2010 plus (Shimadzu, Japan) with flame ionization detector and DB-23 column (30 m × 0.25 mm, 0.25 \(\mathrm{\mu m}\) film, Agilent, USA). The range of oven temperature is from 190 °C to 230 °C at 3 °C/min.
Measure of seed length, width, size, and weight
Arabidopsis seed weight was measured using an electronic scale (OHAUS, USA) capable of measuring up to the fourth decimal point. The seed weight (n = 100) was measured, followed by 3 repetitions, and then the mean values of the three lines were measured. To check the seed size, the length and width of 30 seeds were measured using an optical microscope (NIKON, Japan) in triplicate. Seed sizes were calculated by multiplying the width by length using the software, Image J (http://imagej.nih.gov/ij/).
Measure of seed germination rate, cotyledon opening, seedling growth and mucilage
To measure the germination rate and seedling growth, the seed was sterilized in 70% ethanol for 1 min and 0.5% (w/v) NaOCl for 5 min and then washed in distilled water three times. The seeds were subjected to stratification at 4 °C for 3 days. Next, three repetitions of 30 seeds for germination test or 100 seeds for seedling growth observation were made on half strength of MS media containing 1% (w/v) sucrose and 0.8% (w/v) plant agar, and cultured in a culture chamber of 23 °C at 16 h light/8 h dark condition. The germination rate was measured at intervals of 12 h and confirmed up to 144 h. Germination rates were counted when a radicle emerged from the seed. Seedlings with two fully opened cotyledons at a 180-degree angle were considered healthy seedlings. The seedling growth was checked by measuring the leaf width in triplicate using the Image J program (n = 3). The three repetitions of 50 seeds were stained with 0.01% ruthenium red to check the mucilage of the seed coat. All experiment steps for mucilage analysis were followed by a previously published paper [64]. To quantify the mucilage area, mucilage area was calculated except for seed area by HKBasic program using Eclipse Ci-L microscope (NIKON, Japan) and divided by seed area (n = 10).
Reverse transcriptase PCR and Quantitative RT-PCR
Total RNA was isolated from the developing siliques having different stages from 1 to 18 days after flowering. RNA extraction has followed the method mentioned in Onate-Sanchez et al. [65]. RNA concentration and quality were determined by measuring the A260/A280 ratio using a DS-11 spectrophotometer (Denovix, USA). RNA samples were treated with DNase I (Thermo Fisher Scientific, USA). 1 μg of cDNA was synthesized using a PrimeScript 1st strand cDNA synthesis kit (Takara, Japan) following the recommendation of the manufacturer. Total RNA was extracted in triplicate and RT–qPCR was performed in triplicate on each sample in StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, USA) using SYBR green master mix (TOYOBO, Japan). The expression level was determined using the ΔΔCт method. The ΔCт value was obtained by subtracting the Cт value of the target gene and the Cт value of the endogenous control. In addition, we obtained ΔCт by subtracting the Cт value of the treated group and the Cт value of the control group. The relative expression levels were obtained by subtracting the two ΔCт values, which was a ΔΔCт. After that, the value of \({2}^{(-\Delta \Delta {\mathrm{C}}_{\mathrm{T}})}\) was obtained for a comparison of relative expression. eIF4a was used as a control gene to normalize relative expression level. For the expression of quantitative RT–PCR, the expression of each gene of DAF1 was set to 1. The primers of RT–PCR and RT–qPCR are described in Additional file 1: Table S9. The stages were divided into 6 stages for 18 days after flowering, indicating the seed development stage. For example, stage 1 indicates 1–3 days after flowering.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional files.
Abbreviations
- TAG:
-
Triacylglycerol
- acyl-CoA:
-
Acyl-coenzyme A
- FA:
-
Fatty acid
- HFA:
-
Hydroxy fatty acid
- DAG:
-
Diacylglycerol
- PC:
-
Phosphatidylcholine
- FAH12:
-
Oleate \(\Delta\) 12-hydroxylase
- GPAT:
-
Glycerol-3-phosphate acyltransferase
- LPAT:
-
Lysophosphatidic acid acyltransferase
- DGAT:
-
1,2-sn-Diacylglycerol acyltransferase
- PDAT:
-
Phospholipid:DAG acyltransferase
- LPCAT:
-
Lyso-PC acyltransferase
- PDCT:
-
PC:DAG cholinephosphotransferase
- FAE1:
-
Fatty acid elongase 1
- FAME:
-
Fatty acid methyl ester
- WT:
-
Wild type
- DW:
-
Dry weight
- ER:
-
Endoplasmic reticulum
- RT–PCR:
-
Reverse transcription PCR
- RT–qPCR:
-
Quantitative reverse transcription PCR
References
Ogunniyi DS. Castor oil: a vital industrial raw material. Bioresour Technol. 2006;97(9):1086–91.
Patel VR, Dumancas GG, Kasi Viswanath LC, Maples R, Subong BJ. Castor oil: properties, uses, and optimization of processing parameters in commercial production. Lipid Insights. 2016;9:1–12.
Chen GQ, He X, McKeon TA. A simple and sensitive assay for distinguishing the expression of ricin and Ricinus communis agglutinin genes in developing castor seed (R. communis L.). J Agric Food Chem. 2005;53(6):2358–61.
Balint GA. Ricin: the toxic protein of castor oil seeds. Toxicology. 1974;2(1):77–102.
Bashir ME, Hubatsch I, Leinenbach HP, Zeppezauer M, Panzani RC, Hussein IH. Ric c 1 and Ric c 3, the allergenic 2S albumin storage proteins of Ricinus communis: complete primary structures and phylogenetic relationships. Int Arch Allergy Immunol. 1998;115(1):73–82.
Severino LS, Auld DL, Baldanzi M, Candido MJD, Chen G, Crosby W, Tan D, He XH, Lakshmamma P, Lavanya C, et al. A review on the challenges for increased production of castor. Agron J. 2012;104(4):853–80.
Bates PD. Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim Biophys Acta. 2016;1861(9 Pt B):1214–25.
Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. Acyl-lipid metabolism. Arabidopsis Book. 2013;11: e0161.
Barron EJ, Stumpf PK. Fat metabolism in higher plants XIX. The biosynthesis of triglycerides by avocado-mesocarp enzymes. Biochim Biophys Acta. 1962;60(2):329–37.
Weiss SB, Kennedy EP, Kiyasu JY. The enzymatic synthesis of triglycerides. J Biol Chem. 1960;235:40–4.
Chapman KD, Ohlrogge JB. Compartmentation of triacylglycerol accumulation in plants. J Biol Chem. 2012;287(4):2288–94.
Lands WE. Lipid metabolism. Annu Rev Biochem. 1965;34:313–46.
Lager I, Glab B, Eriksson L, Chen G, Banas A, Stymne S. Novel reactions in acyl editing of phosphatidylcholine by lysophosphatidylcholine transacylase (LPCT) and acyl-CoA: glycerophosphocholine acyltransferase (GPCAT) activities in microsomal preparations of plant tissues. Planta. 2015;241(2):347–58.
Głąb B, Beganovic M, Anaokar S, Hao M-S, Rasmusson AG, Patton-Vogt J, Banaś A, Stymne S, Lager I. Cloning of glycerophosphocholine acyltransferase (GPCAT) from fungi and plants: a novel enzyme in phosphatidylcholine synthesis. J Biol Chem. 2016;291(48):25066–76.
Bates PD, Fatihi A, Snapp AR, Carlsson AS, Browse J, Lu C. Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol. 2012;160(3):1530–9.
Wang L, Shen W, Kazachkov M, Chen G, Chen Q, Carlsson AS, Stymne S, Weselake RJ, Zou J. Metabolic interactions between the lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell. 2012;24(11):4652–69.
Xu J, Carlsson AS, Francis T, Zhang M, Hoffman T, Giblin ME, Taylor DC. Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biol. 2012;12:4.
Lager I, Yilmaz JL, Zhou XR, Jasieniecka K, Kazachkov M, Wang P, Zou J, Weselake R, Smith MA, Bayon S, et al. Plant acyl-CoA: lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J Biol Chem. 2013;288(52):36902–14.
Bayon S, Chen G, Weselake RJ, Browse J. A small phospholipase A2-alpha from castor catalyzes the removal of hydroxy fatty acids from phosphatidylcholine in transgenic Arabidopsis seeds. Plant Physiol. 2015;167(4):1259–70.
Bates PD, Browse J. The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J. 2011;68(3):387–99.
Hu Z, Ren Z, Lu C. The phosphatidylcholine diacylglycerol cholinephosphotransferase is required for efficient hydroxy fatty acid accumulation in transgenic Arabidopsis. Plant Physiol. 2012;158(4):1944–54.
Lu C, Xin Z, Ren Z, Miquel M, Browse J. An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA. 2009;106(44):18837–42.
Slack CR, Campbell LC, Browse JA, Roughan PG. Some evidence for the reversibility of the cholinephosphotransferasecatalysed reaction in developing linseed cotyledons in vivo. Biochim Biophys Acta. 1983;754(1):10–20.
Bates PD, Stymne S, Ohlrogge J. Biochemical pathways in seed oil synthesis. Curr Opin Plant Biol. 2013;16(3):358–64.
Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA. 2000;97(12):6487–92.
van Erp H, Bates PD, Burgal J, Shockey J, Browse J. Castor phospholipid: diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol. 2011;155(2):683–93.
Kim HU, Lee KR, Go YS, Jung JH, Suh MC, Kim JB. Endoplasmic reticulum-located PDAT1-2 from castor bean enhances hydroxy fatty acid accumulation in transgenic plants. Plant Cell Physiol. 2011;52(6):983–93.
Regmi A, Shockey J, Kotapati HK, Bates PD. Oil-producing metabolons containing DGAT1 use separate substrate pools from those containing DGAT2 or PDAT. Plant Physiol. 2020;184(2):720–37.
Bates PD, Durrett TP, Ohlrogge JB, Pollard M. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 2009;150(1):55–72.
Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9(3):338–52.
Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11(6):1151–65.
Chapman KD, Aziz M, Dyer JM, Mullen RT. Mechanisms of lipid droplet biogenesis. Biochem J. 2019;476(13):1929–42.
Binder RG, Kohler GO, Goldblatt LA, Applewhite TH. Chromatographic analysis of seed oils—fatty acid composition of castor oil. J Am Oil Chem Soc. 1962;39(12):513–7.
Lee KR, Chen GQ, Kim HU. Current progress towards the metabolic engineering of plant seed oil for hydroxy fatty acids production. Plant Cell Rep. 2015;34(4):603–15.
Van De Loo FJ, Broun P, Turner S, Somerville C. An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci. 1995;92(15):6743–7.
Broun P, Somerville C. Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol. 1997;113(3):933–42.
Kunst L, Taylor DC, Underhill EW. Fatty-acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem. 1992;30(4):425–34.
Lu C, Fulda M, Wallis JG, Browse J. A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J. 2006;45(5):847–56.
Dauk M, Lam P, Kunst L, Smith MA. A FAD2 homologue from Lesquerella lindheimeri has predominantly fatty acid hydroxylase activity. Plant Sci. 2007;173(1):43–9.
Smith MA, Moon H, Chowrira G, Kunst L. Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta. 2003;217(3):507–16.
Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J. 2008;6(8):819–31.
Shockey J, Lager I, Stymne S, Kotapati HK, Sheffield J, Mason C, Bates PD. Specialized lysophosphatidic acid acyltransferases contribute to unusual fatty acid accumulation in exotic Euphorbiaceae seed oils. Planta. 2019;249(5):1285–99.
Lunn D, Wallis JG, Browse J. Overexpression of seipin1 increases oil in hydroxy fatty acid-accumulating Seeds. Plant Cell Physiol. 2018;59(1):205–14.
Adhikari ND, Bates PD, Browse J. WRINKLED1 rescues feedback inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physiol. 2016;171(1):179–91.
Moire L, Rezzonico E, Goepfert S, Poirier Y. Impact of unusual fatty acid synthesis on futile cycling through beta-oxidation and on gene expression in transgenic plants. Plant Physiol. 2004;134(1):432–42.
Bates PD, Johnson SR, Cao X, Li J, Nam JW, Jaworski JG, Ohlrogge JB, Browse J. Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc Natl Acad Sci USA. 2014;111(3):1204–9.
Lunn D, Wallis JG, Browse J. Tri-hydroxy-triacylglycerol is efficiently produced by position-specific castor acyltransferases. Plant Physiol. 2019;179(3):1050–63.
Kim HU, Park M-E, Lee KR, Suh MC, Chen GQ. Variant castor lysophosphatidic acid acyltransferases acylate ricinoleic acid in seed oil. Ind Crop Prod. 2020;150:112245.
van Erp H, Shockey J, Zhang M, Adhikari ND. Reducing isozyme competition increases target fatty acid accumulation in seed triacylglycerols of transgenic Arabidopsis. Plant Physiol. 2015;168(1):36–46.
Cai Y, Goodman JM, Pyc M, Mullen RT, Dyer JM, Chapman KD. Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell. 2015;27(9):2616–36.
Lemieux B, Miquel M, Somerville C, Browse J. Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor Appl Genet. 1990;80(2):234–40.
Moon H, Smith MA, Kunst L. A condensing enzyme from the seeds of Lesquerella fendleri that specifically elongates hydroxy fatty acids. Plant Physiol. 2001;127(4):1635–43.
Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015. https://doi.org/10.1186/s13059-015-0715-0.
Jin Y, Hu J, Liu X, Ruan Y, Sun C, Liu C. T-6b allocates more assimilation product for oil synthesis and less for polysaccharide synthesis during the seed development of Arabidopsis thaliana. Biotechnol Biofuels. 2017;10:19.
Bates PD, Browse J. The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front Plant Sci. 2012. https://doi.org/10.3389/fpls.2012.00147.
Karki N, Johnson BS, Bates PD. Metabolically distinct pools of phosphatidylcholine are involved in trafficking of fatty acids out of and into the chloroplast for membrane production. Plant Cell. 2019;31(11):2768–88.
Zhang M, Fan J, Taylor DC, Ohlrogge JB. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 2009;21(12):3885–901.
Rivero L, Scholl R, Holomuzki N, Crist D, Grotewold E, Brkljacic J. Handling Arabidopsis plants: growth, preservation of seeds, transformation, and genetic crosses. Methods Mol Biol. 2014;1062:3–25.
Lu C, Kang J. Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 2008;27(2):273–8.
Snapp AR, Kang J, Qi X, Lu C. A fatty acid condensing enzyme from Physaria fendleri increases hydroxy fatty acid accumulation in transgenic oilseeds of Camelina sativa. Planta. 2014;240(3):599–610.
Slightom JL, Sun SM, Hall TC. Complete nucleotide sequence of a French bean storage protein gene: phaseolin. Proc Natl Acad Sci USA. 1983;80(7):1897–901.
Rossak M, Smith M, Kunst L. Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol Biol. 2001;46(6):717–25.
Kridl JCMD, Rose RE, Scherer DE, Knutzon DS, Radke SE, Knauf VC. Isolation and characterization of an expressed napin gene from Brassica rapa. Seed Sci Res. 1991;1:209–19.
Voiniciuc C, Schmidt MH, Berger A, Yang B, Ebert B, Scheller HV, North HM, Usadel B, Gunl M. MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol. 2015;169(1):403–20.
Onate-Sanchez L, Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes. 2008;1:93.
Funding
This work was supported by grants from the Mid-Career Researcher Program of the National Research Foundation of Korea (NRF-2020R1A2C2008175), Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1A6A3A13038532), the New Breeding Technologies Development Program (Project No. PJ016533), and the Next Generation BioGreen21 program (Project No. PJ015714), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
HUK designed the research; MEP and HUK did experiments; MEP, KRL, and HUK analyzed the data and MEP, GQC and HUK wrote a manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Fatty acid composition of T2 seeds from pCam5 T1 plants. Table S2. Fatty acid composition of T3 seeds from pCam5 1 and pCam5 4 independent T2 plants (n = T3 line number). Table S3. Fatty acid composition of T4 seeds from pCam5 1–12, pCam5 1–16 and pCam5 4–5 independent T3 plants (n = T4 line number). Table S4. Fatty acid composition of T5 seeds from pCam5 1–12-8, pCam5 1–16-7, pCam5 1–16-8 and pCam5 4–5-2 independent T4 plants (n = T5 line number). Table S5. Fatty acid composition of T2 seeds pCam5-atfae1 1–12 and pCam5-atfae1 1–16 independent T3 plants (n = T2 pCam5-atfae1 line number). Table S6. Fatty acid composition of T3 seeds from pCam5-atfae1 5, pCam5-atfae1 9 and pCam5-atfae1 28 independent T2 plants (n = T3 pCam5-atfae1 line number). Table S7. Fatty acid composition of T4 seeds from pCam5-atfae1 5–9, pCam5-atfae1 28–3 independent T3 plants (n = T4 pCam5-atfae1 line number). Table S8. Promoter–gene–terminator was linked by infusion method and primer list used for pUC19 subcloning. Table S9. Primers were used in this study.
Additional file 2: Fig. S1.
Expression pattern of five transgenes (RcFAH12, RcPDAT1-2, RcPDCT, RcLPCAT and RcDGAT2) in the developing seed in pCam5 1–16-8 (a, b) and pCam5-atfae1 5–9 line (c, d). AtACT2 and eIF4a were used as controls for RT–PCR and RT–qPCR, respectively. RcFAH12, RcPDAT1-2, RcPDCT, RcLPCAT and RcDGAT2 expression analyses were performed using their specific primers (Table S9). The stages were divided into 6 stages for 18 days after flowering (DAF). Stage 1: 1 ~ 3 DAF, stage 2: 4 ~ 6DAF, stage 3: 7 ~ 9 DAF, stage 4: 10 ~ 12 DAF, stage 5: 13 ~ 15 DAF, stage 6: 16 ~ 18 DAF.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Park, ME., Lee, KR., Chen, G.Q. et al. Enhanced production of hydroxy fatty acids in Arabidopsis seed through modification of multiple gene expression. Biotechnol Biofuels 15, 66 (2022). https://doi.org/10.1186/s13068-022-02167-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-022-02167-1