Abstract
Background
Previous work aimed at understanding the gene regulatory networks (GRNs) governing caudal hindbrain formation identified morphogens such as Retinoic Acid (RA) and Fibroblast growth factors (FGFs), as well as transcription factors like hoxb1b, hoxb1a, hnf1ba, and valentino as being required for rhombomere (r) r4-r6 formation in zebrafish. Considering that the caudal hindbrain is relatively complex – for instance, unique sets of neurons are formed in each rhombomere segment – it is likely that additional essential genes remain to be identified and integrated into the caudal hindbrain GRN.
Methods
By taking advantage of gene expression data available in the Zebrafish Information Network (ZFIN), we identified 84 uncharacterized genes that are expressed in r4-r6. We selected a representative set of 22 genes and assayed their expression patterns in hoxb1b, hoxb1a, hnf1b, and valentino mutants with the goal of positioning them in the caudal hindbrain GRN. We also investigated the effects of RA and FGF on the expression of this gene set. To examine whether these genes are necessary for r4-r6 development, we analyzed germline mutants for six of the genes (gas6, gbx1, sall4, eglf6, celf2, and greb1l) for defects in hindbrain development.
Results
Our results reveal that r4 gene expression is unaffected by the individual loss of hoxb1b, hoxb1a or RA, but is under the combinatorial regulation of RA together with hoxb1b. In contrast, r5/r6 gene expression is dependent on RA, FGF, hnf1ba and valentino – as individual loss of these factors abolishes r5/r6 gene expression. Our analysis of six mutant lines did not reveal rhombomere or neuronal defects, but transcriptome analysis of one line (gas6 mutant) identified expression changes for genes involved in several developmental processes – suggesting that these genes may have subtle roles in hindbrain development.
Conclusion
We conclude that r4-r6 formation is relatively robust, such that very few genes are absolutely required for this process. However, there are mechanistic differences in r4 versus r5/r6, such that no single factor is required for r4 development while several genes are individually required for r5/r6 formation.
Similar content being viewed by others
Background
During vertebrate embryogenesis, cells in the presumptive hindbrain are sorted into seven molecularly and neuroanatomically unique segments called rhombomeres (r1-r7). Segmentation creates regional diversity along the anterior-posterior (A-P) axis of the hindbrain and sets the stage for proper neuronal development and cranial neural crest migration [1,2,3]. The vertebrate hindbrain is responsible for regulating complex processes such as breathing, heartbeat, circulation, wakefulness and cranio-facial development [4, 5]. Precise hindbrain patterning is critical for the development of a fully functional central nervous system (CNS) and defects in this process have been linked to neurological disorders like autism [6, 7]. Thus, it is crucial to understand the regulatory mechanisms underlying hindbrain formation.
In the zebrafish, the earliest factors to be expressed in the hindbrain include the posteriorizing morphogens Retinoic acid (RA) and Fibroblast growth factors (FGFs) [8,9,10,11,12,13]. Around 6 h post fertilization (hpf), RA is detected in the posterior paraxial mesoderm from where it diffuses throughout the neural tube creating a spatial gradient with the caudal hindbrain being exposed to higher concentrations of RA [14,15,16,17,18,19]. FGF signaling (fgf3 and fgf8a) is detected as early as 8hpf in the presumptive r4 [19, 20]. Expression of these morphogens initiates the division of the hindbrain primordium into rostral and caudal regions. The subsequent expression of transcription factors (TFs) like hoxb1b, hoxb1a, hnf1ba, valentino and krox20 further subdivides the caudal hindbrain into r4, r5 and r6. Expression levels of these factors have been manipulated to gain insight into how they interact with one another. For example, loss of RA signaling results in posterior expansion of r4 gene expression domains (e.g. hoxb1a), and reduced expression of r5/r6 genes like hnf1ba and valentino [16, 17, 21]. Similarly, combined knockdown of Fgf8 and Fgf3 leads to the loss of krox20 and valentino expression in r5 [19, 20]. Accordingly, mutations in the hnf1ba and valentino genes cause mis-patterning of r5/r6, with the posterior expansion of hoxb1a and efnb2a, loss of krox20 expression in r5, and loss of abducens motor neurons [22, 23]. Germline mutants for hoxb1b have a smaller r4, with mis-patterned cranial motor neurons, and partial loss of Mauthner neuron formation [24,25,26]. Similar neuronal defects are also seen in hoxb1a mutants [24, 26]. These results have led to a model for the gene regulatory network (GRN) underlying caudal hindbrain formation. This model posits that RA triggers caudal hindbrain patterning by initiating the expression of hoxb1b (in r4-r7) and hnf1ba (in r5/r6) [16]. hoxb1b turns on the expression of hoxb1a, which sustains its own expression through an autoregulatory loop. Thus, the expression of the paralog group 1 (PG1) hox genes leads to the formation and specification of r4 [24,25,26,27,28,29]. hnf1ba together with FGF signaling activates valentino expression in r5/r6 [23, 30], which in turn regulates krox20 expression in r5. [20, 31]. Thus, within the caudal hindbrain GRN, there seems to be two relatively linear pathways that regulate r4 and r5/r6 formation, while cross-talk between these pathways maintains the integrity of each rhombomere.
Despite numerous genetic approaches – initially using chemical (ENU; [32,33,34] [22]) and retroviral [35,36,37] mutagens, but more recently also applying TILLING [38], Zinc Finger Nucleases [24] and TALENs [24] – RA, FGF, hoxb1b, hoxb1a, hnf1ba, and valentino remain the key factors required for caudal hindbrain formation in zebrafish. This is a surprisingly small number considering that other GRNs associated with developmental processes (e.g. germ layer differentiation in sea urchin [39], embryonic development in C. elegans [40], pancreas formation [41], mouse neural tube specification [42] and zebrafish endoderm formation [43]) are more complex [41] — implying that there may be additional genes acting within the caudal hindbrain GRN. It is possible that shortcomings of genetic screens (such as the necessary bias towards readily detectable phenotypes [32, 33, 37, 44]) may have overlooked other genes acting in the caudal hindbrain GRN. Indeed, induction of Hoxb1 and Hoxa1 in murine embryonic stem (ES) cells [45,46,47,48], MO-knockdown of hoxb1b and hoxb1a [27, 49, 50], and overexpression of hoxb1b and hoxb1a [51, 52] has identified additional genes expressed in the caudal hindbrain. While a few of these genes may have roles in the hindbrain (e.g. migration of neural crest cells and neuronal patterning and differentiation; [48, 50]), most have not been assayed functionally. Hence, the goal of our study was to identify novel regulators required for caudal hindbrain development and position them within the GRN governing caudal hindbrain formation.
We reasoned that potential regulators should be expressed in r4-r6 at early stages of development—similar to the TFs hoxb1b, hoxb1a, hnf1ba, valentino and krox20. To find such genes, we analyzed the gene expression data deposited in the Zebrafish Information Network (ZFIN) and identified 107 genes that are expressed in r4, r5 and r6 during the first 24 h of zebrafish development. The majority (n = 84) of these 107 genes have not been extensively characterized previously, suggesting that they may represent novel regulators of caudal hindbrain formation. To test this, we selected 22 representative genes and assayed their expression patterns in zebrafish mutants for hoxb1b, hoxb1a, hnf1ba, and valentino. We also investigated the effects of the morphogens RA and FGF on the expression of these genes. Lastly, we assayed germline mutants for six of the genes (gas6, gbx1, sall4, eglf6, celf2, and greb1l) for defects in hindbrain and neuronal patterning. Strikingly, our results show that genes expressed in r4 are not affected by the loss of hoxb1b or hoxb1a. Loss of RA and FGF signaling also does not affect r4 gene expression (except for dusp2, dusp6, spry1, fgf3 and fgf8 - which are components of the FGF signaling pathway itself). Instead, we find that all tested r4 genes are under the combinatorial regulation of RA and hoxb1b. Furthermore, we observe that hoxb1a (either directly or indirectly) represses the expression of gbx1 in r4, revealing a novel relationship between hoxb1a and gbx1. In contrast to the situation in r4, r5/r6 gene expression requires each of RA, FGF, hnf1ba and valentino – whereby loss of any one of those four factors blocks r5/r6 gene expression. Lastly, analysis of hindbrain and neuronal markers revealed that mutations in gas6, gbx1, sall4, eglf6, celf2, and greb1l are not sufficient to cause detectable developmental defects in the caudal hindbrain. Nevertheless, transcriptome analysis of gas6 mutants identified expression changes in many genes involved in a variety of developmental processes, indicating that these mutants may have very subtle phenotypes. In summary, by positioning 22 novel genes into the caudal hindbrain GRN, we demonstrate that gene regulation in r4 is robust with no single gene being essential, whereas r5/r6 gene expression is susceptible to disruption of either RA, FGF, hnf1ba or valentino function. We also identify novel interactions between r4 and r5/r6 genes – highlighting the importance of cross-talk between the two gene-sets in maintaining the specific molecular identity of each rhombomere.
Methods
Zebrafish care
Wildtype (WT) and mutant zebrafish embryos were collected through natural matings. All embryos were staged according to previously described morphological criteria [53]. All zebrafish lines were raised in the University of Massachusetts Medical School Zebrafish Facility.
In situ hybridization
Embryos were collected at various timepoints between 11hpf and 24hpf and were fixed in 4% paraformaldehyde and stored in 100% methanol at − 20°C. In situ hybridization (ISH) was performed as previously described and was followed by a color reaction using NBT/BCIP or INT/BCIP in 10% polyvinyl alcohol [54]. Synthesis of RNA probes for the genes dusp6, dusp2, krox20, hoxb1a, fgf3, fgf8 and valentino has been previously described [55]. 800-1000 bp of coding sequence for the genes pax2, spry1, hoxd4a, dm20, efnb2a, sall4, greb1l, egfl6, hoxb2a, engrailed1b, irx7, meis1a, tox3, sema3fb, mpz, gas6, hoxb3a, hoxa3, isl1/2, neurod6b, atoh1b, olig4 and nr2f2 were cloned and used for probe synthesis. The ccnjl, cefl2, col15a1b and gbx1 probes were purchased from the Zebrafish International Resource Center (ZIRC). Each ISH experiment included a positive control of wildtype embryos stained with the same probe as the experimental samples. Control and experimental ISH reactions were stopped when control embryos reached optimal staining. Panels shown in each figure were not necessarily processed on the same day.
Immunostaining
For whole-mount immunostaining, embryos were fixed in 4% paraformaldehyde/8% sucrose/1× PBS. Fluorescent antibody staining was performed as described previously [56]. Primary antibodies were used to detect Mauthner neurons (3A10; 1:100; Developmental Studies Hybridoma Bank [DSHB]), and Abducens motor neurons (mouse anti-Zn8; 1:1000; DSHB). The secondary antibody used was goat anti-mouse Alexa Fluor 488 (1:200; Molecular Probes A11001).
Imaging
Embryos between 11hpf and 19hpf were suspended in 3% methyl cellulose for imaging. Images were captured using a Leica M165 FC microscope equipped with a Leica DFC310 FX camera. 24 hpf, 48 hpf and 4 days post fertilization (dpf) old embryos were de-yolked and flat-mounted in 70% glycerol for imaging on bridged coverslips. Whole-mount embryos were imaged with a Nikon Eclipse E600 microscope equipped with a Nikon 20× Plan Fluor objective and flat-mounted embryos were imaged with a Zeiss Axiocam 503 color camera. Captured images were cropped and adjusted (limited to contrast and levels) in Adobe Photoshop.
Pharmacological treatments
A 250 mM stock solution of SU5402 (a competitive inhibitor of the Fgf receptor tyrosine kinase; Calbiochem) and a 1 M stock solution of 4(Diethylamino)-benzaldehyde (DEAB – small molecule inhibitor of RALDH enzyme involved in RA synthesis; Aldrich) was diluted in DMSO and stored in − 20 °C. To block RA signaling, embryos were soaked in 10uM DEAB starting at 4 hpf. The drug was never washed off and embryos were collected and fixed for ISH at 12hpf, 14hpf 16hpf, 19hpf and 24hpf. Similarly, to block FGF signaling, embryos were soaked in 50uM of SU5402 from 7hpf to 12hpf. After which, embryos were thoroughly rinsed in aquarium water [12] and allowed to develop till 12hpf, 14hpf, 16hpf, 19hpf and 24hpf when they were collected and fixed for ISH.
Design and injection of single-strand guide RNAs for CRISPR/Cas9 mediated genome editing
gas6 (alleles um296, um297, um298 and um299) and gbx1 (alleles um300 and um301) mutants were generated using the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat)/Cas9 genome editing system. Target sites for gas6 (5’-ATGAGGGAGCTGGTGTGGAGC-3′) and gbx1 (5’-CCAGATAGT- TTCTACCCCCC-3′) were determined using the CHOPCHOP web tool for genome editing [57]. Oligos containing a T7 promoter sequence, the target sequence, and an additional constant region were created and annealed according to previously described methods [55, 58]. These templates were transcribed in vitro using T7 RNA polymerase (Promega) to generate single-stranded guide RNAs (sgRNAs) for microinjection. A linearized plasmid encoding cas9 was also transcribed in vitro using the SP6 mMessage mMachine Kit (Ambion). 200 ng each of the sgRNA and cas9 mRNA were combined and 2-4 ng of this mixture was injected into early 1-cell stage embryos.
Genotyping zebrafish lines
The zebrafish mutants valentinob337 [22], hnf1bahi2169 [35], hoxb1bum197 and hoxb1aum191 [24] were genotyped as previously described. The greb1lsa17608, egfl6sa21615, sall4sa14110 and celf2sa33469 lines were identified via TILLING [59] and mutant alleles were ordered from ZIRC. Mutant alleles were genotyped by sequencing PCR products amplified from genomic DNA using primers.
5’-TGTGAAAATTTCCTTGCTGTGT-3′ and 5’-CTGAAGGGCAGAATACGG-3′ for greb1lsa17608, 5’-ATCACAGATCCTGGGACAGC-3′ and 5’-AAAAGCATTGGATGCA- GCTC-3′ for egfl6sa21615, 5’-GGGCATGAGGAGAGTATGGA-3′ and 5’-TCTTTCAG- CCCACTGTCACTC-3′ for sall4sa14110, and 5’-CTTTGTTGGCGACCATTGA-3′ and 5’-AAAGCGACAAAAACAGATTCG-3′ for celf2sa33469. gbx1 mutants (alleles um300 and um301) were genotyped by Hpy188III restriction digest of PCR products amplified from genomic DNA using primers 5’-TGTCTCATTCGTCATTACCGTC-3′ and 5’-AAGTTTCCGTGAAATTGAGGAG-3′. gas6 mutants (alleles um296, um297, um298 and um299) were genotyped by XcmI restriction digest of PCR products amplified from genomic DNA using primers 5’-GCGAACACATTGAGCAAGAA-3′ and 5’-CATCG- CTAATGCTTCATCCA-3′.
Genotyping embryos post ISH and immunostaining
hoxb1a, hnf1ba and valentino homozygous mutants are not viable as adults. As a result, all embryos assayed in this study were collected from crosses of heterozygous parents. After ISH and immunostaining, embryos were thoroughly rinsed in 1xPBS solution and individually genotyped. Representative genotyping data for hoxb1a mutants are shown in Additional file 1: Figure S1. Embryos lacking r5 krox20 staining represent valentino and hnf1ba homozygous mutants. Mutants ordered from ZIRC were genotyped as described above.
mRNA injections
All mRNAs for microinjection were synthesized in vitro using the Sp6 mMessage mMachine Kit (Ambion). 100 ng/ul each of GFP [51] and hoxb1a [28] mRNA were combined and 1-2 ng of this mixture was injected into early 1-cell stage embryos.
olig2 reporter line in gas6 mutant background
The transgenic line Tg (olig2:EGFP)vu12 was crossed into the gas6 mutant background and subsequently a gas6 homozygous mutant line was generated carrying the olig2:eGFP transgene. This line was used in preparing the RNA-seq library as well as studying the status of Olig2+ oligodendrocyte progenitor cells in mutant background.
RNA-seq library preparation
gas6 mutant embryos carrying the olig2:EGFP transgene were raised to 48hpf. Using the GFP signal as a guide, hindbrains were dissected from homozygous gas6 transgenic mutants. Hindbrains were also dissected from Tg (olig2:EGFP)vu12 embryos as control samples. Pools of dissected tissues were deyolked and total RNA was extracted using the RNeasy Mini Kit (Qiagen). Similarly, WT and hoxb1bum197 mutants were collected at 18hpf and total RNA was extracted from pools of dechorionated, deyolked, whole embryos. For each RNA-seq experiment, three libraries were synthesized from 3μg RNA for each WT and mutant sample using the TruSeq Stranded mRNA Library Prep Kit (Illumina). All libraries were analyzed for quality on a bioanalyzer prior to sequencing (Agilent 2100 BioAnalyzer).
Processing and analysis of RNA-seq data
Fastq files were analyzed as previously described [55] using the University of Massachusetts Medical School Dolphin web interface [60]. Differentially-expressed (DE) genes were identified as those with a greater than 2-fold change in expression between the WT and mutant samples. RNA-seq data is available at GEO under accession number GSE113437.
Quantitative PCR
Total RNA was extracted from whole embryos (WT and hoxb1b−/− at 18hpf), or from dissected hindbrain tissue (gas6−/− and WT at 48hpf) using the RNeasy Mini Kit (Qiagen). Approximately 100 ng of RNA was used to reverse transcribe cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The qPCR reaction was carried out using SYBR Green qPCR Master Mix (BioTool) on an Applied Biosystems 7300 PCR System.
Results
Derivation of gene-sets expressed in r4 and r5/r6 of the zebrafish hindbrain
To generate a list of candidate genes for function in the formation of r4-r6 of the vertebrate hindbrain, we turned to the gene expression database hosted at ZFIN [61]. We downloaded the “Expression data for wildtype fish” file and searched for genes whose annotation include the terms “hindbrain”, “rhombomere 4”, “rhombomere 5” or “rhombomere 6”. This produced a list of 1820 entries (Additional file 2: Table S1). We eliminated 146 records representing expressed sequence tags (ESTs), as these entries are not fully annotated, resulting in 1674 genes. To further characterize these genes, we next manually reviewed the expression patterns deposited in ZFIN. Since we were particularly interested in genes controlling rhombomere formation, we excluded 480 genes that are only expressed later than 24hpf – when rhombomere formation is already completed – leaving 1194 genes. We also expect genes controlling rhombomere formation to be expressed throughout the corresponding rhombomere. For instance, hoxb1a and valentino, which are respectively active in r4 and r5/r6 formation [5, 22, 23], are expressed throughout the entire corresponding rhombomere, while islet1, which is required for the differentiation of specific neurons, is expressed only in a subset of cells in each rhombomere. After restricting ourselves to genes expressed throughout one, or more, rhombomeres, we were left with 107 genes expressed in r4, r5 or r6 prior to 24hpf. Specifically, 68 of these genes are expressed in r5 and/or r6 (r5/r6 gene-set), while 39 are expressed in r4 (r4 gene-set). Notably, expression of these genes is not necessarily exclusive to r4 or r5/r6, but many of them are also expressed in additional rhombomeres – particularly r3. Our literature review revealed that a relatively small fraction of these 107 genes has previously reported roles in hindbrain formation. Specifically, eleven members of the r4 gene-set (28%) and twelve of the r5/r6 gene-set (18%) have been previously implicated in hindbrain formation, indicating that a large number of uncharacterized genes are expressed in zebrafish r4-r6.
PG1 hox function is not required for expression of many r4 genes
We next set out to position the r4 and r5/r6 gene-sets within the GRN controlling caudal hindbrain formation. Previous work demonstrated that mutations in the PG1 hox TFs hoxb1a and hoxb1b disrupt proper hindbrain formation in zebrafish. In particular, hoxb1b mutants possess smaller r4 and r6, while hoxb1a mutants display a mis-specified r4 [24,25,26], suggesting that PG1 hox TFs may regulate the r4 GRN.
To directly test if genes from the r4 gene-set are key components of a PG1 hox-regulated r4 GRN, we analyzed expression of the r4 gene-set by ISH in hoxb1a and hoxb1b mutant zebrafish (Fig. 1). Specifically, since mutants for two known key r5 regulators (hnf1ba and valentino) show near-complete loss of r5 gene expression [22, 23], we examined if PG1 hox mutants display a similarly profound effect on expression of the r4 gene-set. Zebrafish hoxb1b is required for expansion of the r4 domain, but not for r4 formation [24, 26]. Accordingly, homozygous hoxb1b um197/um197 mutants (hereafter referred to as hoxb1b mutants) possess a narrow r4 domain that nevertheless expresses hoxb1a and is capable of generating both Mauthner cells and nVII facial neurons [24, 26], albeit at a lower rate than wildtype r4. We generated ISH probes for 14 genes from the r4 gene-set and find that all 14 remain expressed in hoxb1b mutant fish, although their expression domains are reduced in size due to the smaller r4 (Fig. 1, column ii). Although there may be subtle changes in expression of some of the r4 genes tested, these are much less pronounced than what is observed in hnf1ba and valentino mutants ([22, 23]; such differences may instead result from slight variations in the ISH processing), suggesting that hoxb1b is not a key regulator of r4 formation. In contrast to hoxb1b, hoxb1a is required for r4 formation [24]. In particular, homozygous hoxb1aum191/um191 mutant embryos (hereafter referred to as hoxb1a mutants), have reduced hoxb1a expression, lack r4-specific Mauthner cells, and the nVII facial neurons fail to migrate out of r4. While this disruption of r4 formation suggests that r4 gene expression might be generally reduced in hoxb1a mutants, we instead find that expression of the r4 gene set persists in hoxb1a mutants (Fig. 1, column iii), at least until 24hpf (Additional file 3: Figure S2). Previous work [24, 26] showed that expression of several r4 genes is only subtly affected in hoxb1a/hoxb1b double mutants relative to single mutants. While this suggests that the PG1 hox genes do not display significant functional redundancy, it remains possible that a subset of r4 genes may be redundantly regulated by the two PG1 hox genes.
PG1 hox function is not required for expression of most r4 genes. Expression of r4 genes was assayed via ISH in (i) WT, (ii) hoxb1b mutant, (iii) hoxb1a mutant, (iv) hnf1ba mutant and (v) valentino mutant zebrafish. The genes assayed include a hoxb1a, b dusp2, c dusp6, d spry1, e fgf3, f fgf8, g efnb2a, h meis1a, i ccnjl, j irx7, k sall4, l greb1l, m egfl6, n hoxb2a and o engrailed1b. krox20 (red) which is expressed in r3 and r5, was used to assign the expression domains of several genes, as indicated. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging. Black arrows point to the r4 domain in 12hpf and 24hpf embryos. White brackets mark the normal and expanded r4 domains in embryos staged at 14hpf, 16hpf and 19 hpf
Our finding that PG1 hox function is not required for expression of the 14 tested r4 genes led us to assess more broadly if hindbrain gene expression is hox-dependent. To address this, we took advantage of the viability of homozygous hoxb1b mutants and used RNA-seq to identify hoxb1b-dependent genes during zebrafish embryogenesis. Comparing the hoxb1b mutant transcriptome to that of wildtype embryos revealed 866 differentially expressed genes at 18hpf (Additional file 4: Figure S3) (Additional file 5: Table S2). Comparison to the hindbrain-expressed genes identified in our database search demonstrated that only 85 of these genes are affected in hoxb1b mutants (seven up-regulated and 78 down-regulated; Additional file 4: Figure S3B). Thus, by this measure, ~ 5% (85/1674) of zebrafish hindbrain genes are hoxb1b regulated (although the fraction may be lower, since some of these genes are also expressed in non-hindbrain tissue). Furthermore, of the 85 genes, only four (mpz, fgf8a, cyp26b1 and desma) have rhombomere-restricted expression patterns (Additional file 5: Table S2). Also, while fgf8 and mpz were identified as upregulated in hoxb1b mutants by our RNA-seq analysis, we did not detect obvious changes in expression of these genes by ISH (Figs. 1Fii and 3Cii). Since RT-qPCR on independently collected samples validated our RNA-seq analysis (Additional file 4: Figure S3D), it is possible that gene expression changes identified by RNA-seq are too subtle for detection by ISH. Indeed, the change in expression of fgf8 and mpz is less than 2.5-fold and we find that the majority of hoxb1b-regulated genes identified by RNA-seq show relatively subtle changes in expression, such that ~ 93% of the down-regulated genes are reduced by less than 4-fold and only three genes are down-regulated by more than 10-fold (Additional file 5: Table S2). We conclude that, while many genes may be hoxb1b-regulated in the zebrafish embryo, only a few of these genes are expressed in the hindbrain and the observed changes in expression levels are relatively subtle. Hence, our ISH analysis in hoxb1a and hoxb1b mutants, together with our RNA-seq analysis of hoxb1b mutants, suggests that PG1 hox genes may not be absolutely required for r4 gene expression. Further, a recent RNA-seq analysis of Hoxa1 mutant mouse embryos (murine Hoxa1 is functionally analogous to zebrafish hoxb1b) identified 1537 Hoxa1-dependent genes [48], but only 31 genes are shared between the zebrafish and mouse data sets (Additional file 6: Figure S4), suggesting that PG1 hox genes may in fact regulate distinct sets of genes in different species.
A subset of r4 genes is regulated by FGF, but not RA, signaling
The possibility that PG1 hox genes are not required for r4 gene expression suggests that other factors may be involved. In particular, the RA and FGF signaling pathways are known to function in hindbrain development [8,9,10,11,12,13]. To determine if expression of the r4 gene-set is dependent on RA or FGF signaling, we treated wildtype embryos with 50uM SU5402 (a competitive inhibitor of the FGF receptor tyrosine kinase; [19]) or 10uM DEAB (a competitive inhibitor of RALDH, the enzyme required for conversion of retinaldehyde to retinoic acid; [10, 24]). We find that inhibition of FGF signaling blocks expression of dusp2 (Fig. 2Bii), dusp6 (Fig. 2Cii), spry1 (Fig. 2Dii), fgf3 (Fig. 2Eii), and fgf8 (Fig. 2Fii), in r4, although the effect on spry1 is difficult to assess since the anterior spry1 expression domain appears to have expanded. In contrast, inhibition of Fgf signaling does not block expression of hoxb1a or the remaining members of the r4 gene-set, with the exception of engrailed1b (Fig. 2). Notably, all genes affected by loss of FGF signaling are themselves involved in the FGF signaling pathway, confirming the extensive use of feedback loops in this pathway [62,63,64]. Furthermore, inhibiting RA signaling does not block expression of the r4 genes tested, again with the exception of engrailed1b (Fig. 2Miii). The fact that expression of the r4 gene-set is not lost upon disrupting PG1 Hox TFs, FGF signaling or RA signaling suggests that it is either regulated independently of these signaling pathways or is under combinatorial control.
Simultaneous loss of hoxb1b and RA function disrupts expression of r4 genes. Expression of r4 genes was assayed via ISH in (i) WT embryos treated with 50uM DMSO, (ii) WT embryos treated with 50uM SU5402 (iii) WT embryos treated with 10uM DEAB, (iv) hoxb1b mutant embryos treated with 10uM DEAB and (v) hoxb1b mutant embryos treated with 10uM DMSO. The genes assayed include a hoxb1a, b dusp2, c dusp6, d spry1, e fgf3, f fgf8, g efnb2a, h meis1a, i irx7, j greb1l, k egfl6, l hoxb2a and m engrailed1b. krox20 (red), which is expressed in r3 and r5, was used to position the expression domains of several genes, as indicated. In panels (Jiv), (Jv), (Kiv) and (Kv), pax2 is the second blue marker which labels the MHB. Black arrows point to r4, red arrows to r3 and white arrows to the MHB. White and black brackets indicate r4 and r2 size, respectively. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19 hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging. This figure also shows that a subset of r4 genes is regulated by FGF signaling (Bii, Cii, Dii, Eii and Fii) and that r4 genes are not affected by the loss of RA signaling (embryos in row iii)
Simultaneous loss of hoxb1b and RA function disrupts expression of r4 genes
We previously found that hoxb1a expression is unaffected when RA and hoxb1b function is disrupted independently, but is lost when these signals are disrupted simultaneously [24]. To determine if the r4 gene-set is similarly regulated, we treated hoxb1b mutant embryos with 10uM DEAB and assayed gene expression by ISH. We find that expression of ten members of the r4 gene-set is completely lost when hoxb1b and RA signaling are simultaneously disrupted (Fig. 2, column iv). Two genes, fgf8 (Fig. 2Fiv), and egfl6 (Fig. 2Kiv), show residual expression in the hindbrain, but these two genes are normally expressed also in the anterior hindbrain and previously published mouse data showed an expansion of the r2/r3 domains upon disruption of RA signaling [65]. Hence, the residual fgf8 and egfl6 expression detected in DEAB-treated hoxb1b mutants may be derived from r2/r3, not from r4. However, expression of greb1l (Fig. 2Jiv) – which is also expressed in the anterior hindbrain – is completely lost, indicating that not all genes are regulated in the same manner. A recent study reported subtle changes in fgf3/8a expression patterns in hoxb1b mutants and suggested that hoxb1b may regulate FGF signaling [26]. In line with this observation, we show that expression of FGF pathway components (fgf3, fgf8, dusp2, dusp6 and spry1) is lost upon simultaneous disruption of hoxb1b and RA function, indicating that FGF signaling is downstream of hoxb1b and RA activity in the hindbrain. Notably, simultaneous loss of hoxb1b and RA function has no effect on pax2 (Fig. 2Jiv and Kiv) and fgf8 (Fig. 2Fiv) expression at the mid-hindbrain boundary (MHB), indicating that co-regulation is specific to the region where hoxb1b is expressed. We carried out an analogous experiment to test if hoxb1b and FGF also cooperate to control r4 gene expression but find that even brief treatment of hoxb1b mutants with SU5402 dramatically disrupts embryogenesis (Additional file 7: Figure S5), precluding us from assaying hindbrain gene expression. We conclude that expression of the r4 gene-set (including FGF pathway components) requires both hoxb1b and RA function.
Expression of the r5/r6 gene-set is dependent on hnf1ba and valentino function
hnf1ba is the earliest-acting TF in zebrafish r5/r6 where it controls expression of the mafB gene valentino. Indeed, previous work demonstrated that both hnf1ba and valentino function is required for the expression of several r5/r6 genes [23]. In order to determine if expression of our r5/r6 gene-set is also dependent on hnf1ba and valentino, we generated ISH probes for eight genes (gbx1, tox3, sema3fb, mpz, gas6, celf2, nr2f2 and col15a1b) and assessed their expression in homozygous hnf1bahi2169/hi2169 (referred to as hnf1ba mutant) and homozygous valentinob337/ b337 (referred to as valentino mutant) embryos. For each of the eight genes, we find that expression is dramatically reduced in r5 and r6 of both mutant lines (Fig. 3, columns iv and v). Expression of tox3, sema3fb, gas6, nr2f2 and col15a1b appears to be completely lost in r5/r6 of both mutant lines, whereas residual expression of celf2 (Fig. 3Eiv) and gbx1 (Fig. 3Hiv) is detected in hnf1ba mutants and weak mpz (Fig. 3Civ, v) expression is observed in both hnf1ba and valentino mutants. This is in agreement with previous reports [66] demonstrating that slight expression of some r5 genes may persist in these mutants.
Expression of the r5/r6 gene-set is dependent on hnf1ba and valentino. Expression of r5/r6 candidate genes was assayed via ISH in (i) WT, (ii) hoxb1b mutant, (iii) hoxb1a mutant, (iv) hnf1ba mutant and (v) valentino mutant zebrafish lines. The genes assayed include a tox3, b sema3fb, c mpz, d gas6, e celf2, f nr2f2, g col15a1b and h gbx1. krox20 (red), which is expressed in r3 and r5, was used to position the expression domains of several genes as indicated. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging. Black brackets mark the smaller r5 domain. Red bracket in (Hiii) indicates expression of gbx1 throughout the hindbrain, as a result of reappearance of gbx1 expression in r4 of hoxb1a mutants
hoxb1b is initially broadly expressed in the caudal hindbrain [10, 16] and previous work demonstrated that several r5/r6 genes (including seven genes from the r5/r6 gene set; Additional file 1: Table S1) are up-regulated following hoxb1b overexpression [51], suggesting that hoxb1b may regulate gene expression in r5/r6. We therefore assayed expression of the r5/r6 gene-set also in hoxb1b and hoxb1a mutant embryos (Fig. 3, column ii and iii), but find that r5/r6 gene expression persists in PG1 hox mutants. We conclude that hnf1ba and valentino, but not hoxb1b or hoxb1a, are required for gene expression in r5 and r6.
Expression of the r5/r6 gene-set requires FGF and RA signaling
Previous work demonstrated that FGF signaling is required for r5/r6 formation [19, 20]. Since hnf1ba expression is independent of FGF [20], FGF must control r5/r6 formation downstream of this transcription factor. Indeed, FGF reportedly acts together with hnf1ba to regulate valentino and krox20 expression [23, 30]. Hence, by determining if the r5/r6 gene-set is FGF independent (like hnf1ba) or FGF dependent (like valentino and krox20), we can better understand the GRN controlling r5/r6 formation. Additionally, RA is required for formation of r5/r6 and for the expression of r5/r6-restricted genes such as hnf1ba, valentino, and krox20 [16]. Strikingly, our analyses revealed that inhibition of either FGF or RA signaling in wildtype embryos blocks expression of all genes in the r5/r6 gene-set (Fig. 4), with the one exception of a narrow domain of residual nr2f2 expression in r6 (Fig. 4Fii) of SU5402 treated embryos. Hence, r5/r6 gene expression is dependent both on the activity of the hnf1ba and valentino TFs, as well as on RA and FGF signaling.
Expression of the r5/r6 gene-set requires FGF and RA signaling. Expression of r5/r6 genes was assayed via ISH in (i) WT embryos treated with 50uM DMSO, (ii) WT embryos treated with 50uM SU5402 and (iii) WT embryos treated with 10uM DEAB. The genes assayed include a tox3, b sema3fb, c mpz, d gas6, e celf2, f nr2f2, g col15a1b and h gbx1. krox20 (red), which is expressed in r3 and r5, was used to position the expression domain of some genes as indicated. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 12hpf, 14hpf, 16hpf and 19hpf were imaged as whole-mounts. 24hpf embryos were flat-mounted for imaging
hnf1ba establishes the posterior boundary of r4 gene expression
Gene expression boundaries in the developing hindbrain are initially established via repressive interactions at the level of transcription. For instance, hoxb1a, efnb2a and fgf3 expression expands caudally from r4 into presumptive r5 in hnf1ba mutants [23, 36], indicating that hnf1ba represses r4 gene expression (directly or indirectly) to establish the r4/r5 border. In order to determine if the expression domains of genes in the r4 and r5/r6 gene-sets are similarly established by repressive interactions, we examined r4 gene expression in hnf1ba and valentino mutants, as well as r5/r6 gene expression in the PG1 hox mutants.
For the r4 gene-set, we find that dusp6 (Fig. 1Civ), spry1 (Fig. 1Div), and egfl6 (Fig. 1Miv), show expansion of the r4 expression domain into r5 of hnf1ba mutants, while fgf8, irx7, greb1l and eng1b expression is not affected. In contrast, expression of the r4 gene-set is not affected in valentino mutants, with the exception of efnb2a (Fig. 1Gv), which may show a slight expansion into r5 (previously shown in [23]). For the r5/r6 gene-set, we do not observe expansion into r4 in either of the PG1 hox mutants (Fig. 3, columns ii and iii). We conclude that hnf1ba restricts expression of many, but not all, genes in the r4 gene-set to presumptive r4, but that valentino and the PG1 hox genes are not required to establish gene expression boundaries for the r4 and r5/r6 gene-sets.
gbx1 expression requires hnf1ba and valentino in r5/r6 and is repressed by hoxb1a in r4
The gbx1 gene displays an interesting expression pattern in that it is expressed throughout the hindbrain at 24hpf, except in r4 (Fig. 3Hi). To better understand the regulation of gbx1 expression, we analyzed its expression pattern in hoxb1b, hoxb1a, hnf1ba and valentino mutants. We find that gbx1 expression is lost in r5/r6 of valentino mutants and that only a narrow expression domain persists in r5 of hnf1ba mutants (Fig. 3Hiv, v), indicating that hnf1ba and valentino are required for gbx1 expression in r5/r6. Strikingly, gbx1 expression is restored to the r4 domain of 24hpf hoxb1a mutant embryos, but not of hoxb1b mutant embryos (Figs. 3Hiii and 5Ai), suggesting that gbx1 might be repressed by hoxb1a in r4. gbx1 is actually expressed in wildtype r4 at ~10hpf, but this expression disappears coincident with the onset of hoxb1a expression (Fig. 5b), again suggesting that hoxb1a may repress gbx1 expression. To test this directly, we overexpressed hoxb1a by injection of synthetic mRNA into 1–2 cell stage embryos and assayed gbx1 expression at 24hpf by ISH. We find that ~ 80% (27/34) injected embryos display a clear decrease in gbx1 hindbrain expression (Fig. 5c). We conclude that hoxb1a, either directly or indirectly, represses gbx1 expression in r4 (Fig. 5d) and that its expression in r5/r6 is regulated similarly to the genes in the r5/r6 gene-set.
gbx1 expression is repressed by hoxb1a in r4. a gbx1 expression is restored in hoxb1a mutant embryos. Embryos from a cross of hoxb1a heterozygous parents were assayed by ISH for gbx1 expression, producing two phenotypes (i, ii). Subsequent genotyping revealed that homozygous hoxb1a mutants express gbx1 in r4 (ii, iv). b gbx1 is initially expressed in r4 (iv), but disappears (v, vi) when hoxb1a expression is activated (i, ii, iii). c A mixture of hoxb1a and GFP mRNA was injected into 1-cell stage embryos and successfully injected embryos (identified by GFP expression) were stained for gbx1 expression at 24hpf. The observed reduction in gbx1 expression demonstrates that hoxb1a is capable of repressing gbx1 expression. d Hypothetical model depicting the potential relationship between gbx1 and hoxb1a. hoxb1a could either repress gbx1 directly (solid red T bar) or indirectly by activating a repressor (X; blue arrow) or repressing an activator (Y; orange T bar)
gas6, gbx1, sall4, egfl6, celf2 and greb1l function is not required for r4-r6 formation
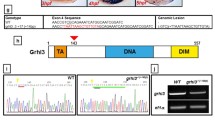
Considering the number of genes assigned to the r4 and r5/r6 gene-sets by our database search, it is surprising that previous large-scale mutagenic screens identified only a few genes required for r4-r6 formation [22, 33, 37]. While this finding may indicate redundancy in the r4 and r5/r6 GRNs, the fact that targeted mutagenesis studies have identified additional genes required for r4-r6 formation (e.g. PG1 hox genes [24,25,26, 67,68,69] and krox20 [38, 70]) may instead suggest that the original screens did not reached saturation. To directly test if genes in the r4 and r5/r6 gene-sets are required for rhombomere formation, we selected six genes (gas6, sall4, egfl6, celf2, greb1l and gbx1) for further analysis. Germline mutations for four of these genes (sall4, egfl6, celf2 and greb1l) have been generated by community-based mutagenesis projects [59, 71] and are available from the zebrafish resource center (ZIRC; Table 1). These four mutations were procured from ZIRC in the form of fertilized embryos and raised in our laboratory. Genotyping and sequencing confirmed the presence of the expected mutations (Table 1) (Additional file 8: Figure S6). Since gas6 and gbx1 mutants were not available from the resource center, we generated these by CRISPR/Cas9-mediated mutagenesis. We designed a sgRNA targeting the gas6 start codon in exon 1 and identified two gas6 mutant founders from five fish screened (Fig. 6a-c). One founder did not transmit mutation to its offspring (n = 0/92), but the other transmitted mutations to 20% (n = 12/60; Table 2) of its F1 offspring. Sequencing revealed that this founder transmitted four different mutant alleles (Additional file 9: Data S1) where each allele carried a different four nucleotide deletion, but translation of each mutant allele is nevertheless predicted to produce an out of frame product that terminates at the same premature stop codon (residue 99; Fig. 6d) (Additional file 9: Data S1). ISH and RT-qPCR analyses further revealed that gas6 transcripts are lacking in gas6 mutants, possibly as a result of nonsense-mediated mRNA decay (Fig. 6e). In an analogous fashion, a sgRNA was designed to exon 1 of gbx1 (Additional file 10: Figure S7A) and we identified two founders (Additional file 10: Figure S7B) (Table 2). Of the two founders, only one produced offspring and this founder transmitted two different mutations. Both of these alleles produce premature STOP codons N-terminal to the homeodomain (Additional file 10: Figure S7D) (Additional file 11: Data S2).
Scheme for generating gas6 mutant line. a Schematic showing the 20 nucleotide (orange text) target site in exon 1 of gas6. CAA represents the PAM sequence (blue box) and ATG (green box) is the start codon. XcmI target sequence is indicated by the dotted red line, the red arrow denotes the cut site. b sgRNA and Cas9 mRNA was injected into 1-cell stage embryos. Injected embryos were raised to 24hpf and genomic DNA was extracted from a pool of embryos. XcmI digest of PCR products amplified from genomic DNA (extracted from injected embryos) reveal the presence of a mutation (red box in gel). c Injected embryos were raised to give rise to F0 adults. These fish were crossed with WT adults to raise the F1 generation. At 3 months age, genomic DNA was extracted from fin-clips from individual F1 fish and genotyped as in panel B. d Sequencing of F1 genomic DNA revealed transmission of four different mutant alleles (um296, um297, um298, um299), each with a different 4 nucleotide deletions (orange dashes). Each mutant allele codes for 96 out of frame amino acids (gray boxes) followed by a premature stop codon. e One quarter of the embryos collected from a cross of two heterozygous parents lack gas6 expression in r5/r6 (i). XcmI digest of PCR products amplified from genomic DNA extracted from embryos lacking gas6 expression were homozygous for mutant gas6 allele (iii, lane 5). cDNA was synthesized from total RNA extracted from WT and homozygous gas6 mutant fish. Quantitative RT-PCR using two different primer pairs (targeting the N and C termini, respectively) shows that homozygous gas6 mutants have significantly lower levels of gas6 mRNA (iv)
In order to assess whether gas6, gbx1, sall4, egfl6, celf2 and greb1l are required for rhombomere formation, we assayed expression of hoxb1a in r4, krox20 in r3 and r5, pax2 at the MHB boundary and in the otic vesicle, as well as of hoxd4a in r7 and anterior spinal cord, by ISH in each of the mutant lines. Upon breeding the mutant lines, we noted that the gas6, sall4, greb1l and egfl6 lines produced homozygous mutants at close to the expected ratio, while the celf2 and gbx1 lines produced few, or no, homozygous mutant offspring (Table 2). This is in agreement with previously published information were gbx1 mutants obtained via ENU mutagenesis, were also not viable as homozygotes [72]. We also find that homozygous gas6, sall4, and greb1l mutants are fertile, but homozygous egfl6 mutants are not. Therefore, our functional analyses made use of offspring from crosses of homozygous mutant parents for gas6, sall4 and greb1l, but offspring of heterozygous carriers for the other lines. Strikingly, we do not detect gene expression changes in any of the mutants (Fig. 7). We also do not detect any defects in rhombomere size, in the spacing of the rhombomere expression domains, or in the integrity of rhombomere boundaries. As a further test of rhombomere development, we examined the differentiation of rhombomere-specific neurons. Specifically, we used immunochemistry to visualize reticulospinal Mauthner neurons in r4 (Fig. 7, column iv) and nVI abducens neurons in r5/r6 (Fig. 7, column iii). Results from this analysis revealed the presence of normal and properly patterned neurons in each of the mutants. Hence, our results suggest that the gas6, gbx1, greb1l, celf2, egfl6, and sall4 genes may not be required for rhombomere-restricted gene expression or neuronal differentiation in the zebrafish hindbrain, although – since we did not assay all aspects of hindbrain development – we cannot exclude the possibility that these genes have other roles in hindbrain development.
gas6, gbx1, sall4, egfl6, celf2 and greb1l function is not required for r4-r6 formation. ISH for hindbrain markers (i) hoxb1a (blue, r4) and krox20 (red r3/5), (ii) pax2 (MHB), krox20 (r3/5) and hoxd4a (r7-anterior spinal cord), and immunostaining for neuronal markers detecting (iii) abducens motor neurons (four green dots in white boxes) in r5/r6 and (iv) Mauthner neurons (white arrows) in r4 was carried out on embryos collected from an a cross of WT fish, b cross of hoxb1a heterozygous mutants, c cross of gas6 homozygous mutants, d cross of gbx1 heterozygous mutants, e cross of greb1l homozygous mutants, f cross of a celf2 heterozygous and a homozygous mutant, g cross of egfl6 heterozygous mutants and h cross of sall4 homozygous mutants. All embryos are oriented in dorsal view with anterior to the top. Embryos collected at 14hpf and 18hpf were imaged as whole-mounts. 48hpf embryos were flat-mounted for imaging
A detailed analysis of gas6 mutants does not reveal hindbrain defects
To examine the possibility that the mutant lines may have subtle phenotypes that went undetected by our initial screening, we selected the gas6 mutant for in-depth analysis. Since krox20 and valentino expression is unaffected in gas6 mutants (Fig. 7Ci, ii and Fig. 8Ai), we reasoned that gas6 might act downstream of these TFs and therefore examined expression of two later-acting r5/r6 genes (hoxb3a and hoxa3). However, we find that expression of both genes persists in gas6 mutants (Fig. 8Ai and ii). We also examined the migration of nVII facial motor neurons from r4 into r5/r6 (Fig. 8Aiv, blue bracket), but do not detect any disruptions of this process in gas6 mutants. Lastly, r5 and r6 are the source of the initial wave of oligodendrocyte precursor cells (OPCs) (Fig. 8Av) in the hindbrain and we therefore examined expression of olig2 (a gene required for OPC formation) and dm20 (a marker of differentiated, myelin-producing oligodendrocytes) (Fig. 8Avi) in gas6 mutants, but do not find oligodendrocyte formation to be affected in gas6 mutants.
gas6 may only have subtle roles in caudal hindbrain development. a WT and gas6 mutant embryos were assayed for expression of valentino (r5/r6) (i), hoxb3a (r5-spinal cord) (ii), hoxa3 (r5/r6) (iii), islet1 (cranial nerves)(iv) and dm20 (oligodendrocyte marker) (vi) by ISH, as well as for the presence of OPCs and abducens neurons by crossing to the Tg (olig2:EGFP)vu12 line (v). In column (iv), yellow brackets mark cranial nerve V, blue brackets mark cranial nerve VII and red brackets mark cranial nerve X. White brackets indicate the presence of abducens (cranial nerve VI) in column (v). b Schemes showing RNA-seq library synthesis. Hindbrain tissue was dissected from 48 hpf gas6 mutant embryos in the olig2:eGFP background. Total RNA was collected from pools of hindbrain tissue and was used in library synthesis following the TruSeq Stranded mRNA Library Prep Kit (Illumina) protocol. c 1590 differentially expressed genes were identified from RNA-Seq where 41 out of the 928 up-regulated genes and 78 out of the 662 down-regulated genes were expressed in the hindbrain. GO terms related to Biological Processes were identified in both up-regulated and down-regulated genes using DAVID. d A subset of differentially expressed genes was validated via qPCR from independently collected hindbrain tissue samples. e ISH analysis of representative differentially expressed hindbrain genes (i) neurod6b, (ii) atoh1b and (iii) olig4 show no detectable change in expression pattern in gas6 mutant embryos
For a more global view of potential defects in gas6 mutants, we used RNA-seq to compare gene expression between homozygous gas6 mutants and wildtype embryos. Since gas6 is expressed exclusively in the hindbrain, we made use of dissected hindbrains from wildtype and gas6 mutants at 48 hpf (Fig. 8b). Our analysis identified 1590 genes with a 2-fold or greater change in expression between wildtype and mutant hindbrains (928 up-regulated and 662 down-regulated in gas6 mutants) (Additional file 12: Table S3) (Fig. 8c). Subsequent RT-qPCR analysis on ten differentially expressed genes (olig1, neurod6b, crabp2a, hoxb1a, krox20, atoh1a, atoh1b, ptf1a, olig4 and ncam1b) confirmed the gene expression changes observed by RNA-seq (Fig. 8d). Using the DAVID functional annotation tool [73], we find enrichment for genes associated with developmental processes like “nervous system development”, “forebrain development”, and “neural crest development”, but only a few genes are associated with each GO term. Furthermore, comparison to the hindbrain-expressed genes identified in our database search (Additional file 2: Table S1) revealed that only ~ 7.5% of the genes differentially expressed in gas6 mutants (41 up-regulated and 78 down-regulated; Fig. 8c) (Additional file 12: Table S3) are expressed in the hindbrain. However, when we use ISH to assess expression one upregulated (neurod6b) and two downregulated (atoh1b and olig4) genes, we do not detect any differences between wildtype and gas6 mutant embryos (Fig. 8e). We conclude that disruption of gas6 leads to changes in hindbrain gene expression, but these changes are too subtle to be detected by ISH and do not seem to affect rhombomere formation or neuronal patterning.
Discussion
Our goal for this study was to identify novel genes required for caudal hindbrain development and to position them within the corresponding GRN (Fig. 9). We used the ZFIN database to identify 84 genes that are expressed in r4-r6, but that are relatively uncharacterized. We focused on 22 representative genes and find important differences between r4 and r5/r6 gene expression. In particular, we find that r4 genes are under the combinatorial regulation of RA and hoxb1b while r5/r6 genes are under control of RA, FGF, hnf1ba and valentino in a regulatory arrangement where the loss of any one of these factors disrupts r5/r6 gene expression. Additionally, we identified several novel interactions between the r4 and r5/r6 gene-sets. This includes the repression of dusp6, spry1 and egfl6 by hnf1ba and repression of gbx1 by hoxb1a (Fig. 9). We also analyzed germline mutants for six genes (gas6, gbx1, sall4, eglf6, celf2, and greb1l), but we do not detect hindbrain defects in any of the mutants. However, transcriptome profiling of gas6 mutants identified differentially expressed genes involved in a variety of hindbrain related developmental processes – leading us to speculate that gas6 may play subtle roles in hindbrain development. Thus, our study suggests that the regulatory logic differs in r4 versus r5/r6, but that both GRNs are relatively robust with a limited number of genes being absolutely required for their integrity.
Proposed model depicting the GRN in the caudal hindbrain. Black arrows and bars represent regulatory relationships known prior to this study. Green arrows and red T bars represent relationships uncovered in this study. In this model, arrows (activating) and T bars (repressive) indicate interactions that have been observed, they do not indicate whether the interactions are direct or indirect. All r4 genes regulated by hoxb1b + RA are grouped in the purple box. Within r4, dusp2, dusp6 and spry1 are regulated by FGF signaling (yellow box). hnf1ba represses dusp6, spry1 and egfl6 expression while gbx1 expression is repressed in r4 by hoxb1a. All r5/r6 genes (light blue box) are regulated by RA, FGF, hnf1ba and valentino. Red star next to RA, FGFs, hoxb1b, hoxb1a, hnf1ba and valentino represent the key regulators of the caudal hindbrain – without these factors r4-r6 does not form properly
The r4 and r5/r6 gene regulatory networks operate by different mechanisms
Previous studies demonstrated that loss of PG1 hox function results in a mis-specified r4 [16, 24,25,26, 28, 29, 74, 75], leading us to hypothesize that all r4 genes are regulated by PG1 hox genes. Surprisingly, our ISH analysis of the r4 gene-set revealed continued expression in PG1 hox mutants. We note that, since PG1 hox mutants show defects in neuronal differentiation [24,25,26], hoxb1a and hoxb1b must regulate genes involved in neural differentiation. However, such genes are likely to be expressed in only a subset of cells in r4 and would not have been included in the r4 gene-set used for our analysis (which was restricted to genes expressed throughout r4). While a recent report indicates that expression of fgf pathway components is affected in hoxb1b mutants [26], we do not observe this, possibly due to differences in sensitivity of the ISH protocols used. Additionally, while our transcriptome analysis of hoxb1b mutants identified differentially expressed genes present in the hindbrain, the expression changes were relatively subtle. These results suggest that r4 gene regulation may require other factors in addition to PG1 hox genes. Indeed, we observe complete loss of expression of all tested r4 genes when hoxb1b and RA function is simultaneously disrupted, demonstrating that both factors are required to control r4 gene expression. Since RA signaling is unaffected by loss of PG1 hox function [26], these factors likely act in parallel (Fig. 9) – although it is unclear whether the combinatorial regulation by RA and hoxb1b acts directly at each r4 gene, or at an intermediary factor required to drive r4 gene expression. It is also unknown how RA signaling is initiated. A recent report concluded that RA signaling in the hindbrain is under control of pbx genes [26], but this effect is somewhat subtle, indicating that other as yet unknown factors may also control the RA pathway.
In r5/r6, the available data predict a relatively linear pathway where RA, FGF, hnf1ba and valentino control r5/r6 identity and disruption of any one of these factors causes r5/r6 defects. Additionally, at least in mice, r5 cells adopt an r6 fate in the absence of krox20 [76, 77] and combined mutations in the mouse PG3 hox genes Hoxa3 and Hoxb3 result in loss of r5/r6 specific abducens motor neurons [78], suggesting that krox20 and PG3 hox genes are also required for r5/r6 formation. In accordance with the prevailing model, we find that expression of all tested genes from the r5/r6 gene-set is abolished in hnf1ba and valentino mutants, as well as upon disruption of RA or FGF signaling. Notably, there are combinatorial interactions also in r5/r6 – for instance, hnf1ba and FGF act together to drive valentino expression [23, 30] – but the mechanism of combinatorial regulation differs between r4 and r5/r6. In r4, hoxb1b and RA function together to drive gene expression and either factor is sufficient to support expression. However, in r5/r6, neither hnf1ba nor FGF is sufficient to support r5/r6 gene expression. Hence, the r4 GRN appears less susceptible to disruptions than the r5/r6 GRN (Fig. 9). It is not clear why this would be the case, except that r4 is the earliest rhombomere to form and it acts as a key signaling center during hindbrain development, raising the possibility that there may have been greater evolutionary pressure to ensure that r4 forms properly.
Repressive interactions may represent a key function of the hindbrain GRNs
Cross-talk between r4 and r5/r6 genes is a crucial part of establishing rhombomere boundaries and maintaining the uniqueness of each rhombomere. An example of this is seen in hnf1ba mutants where there is posterior expansion of the r4 genes hoxb1a, fgf3, and efnb2a into the mispatterned r5/r6 domain [23]. In this study, we identified dusp6 (Fig. 1Civ), spry1 (Fig. 1Div), and eglf6 (Fig. 1Miv) as additional r4 genes whose expression domains are defined by hnf1ba-mediated repression. Importantly, hnf1ba is thought to act primarily as a transcriptional activator [79], raising the possibility that hnf1ba controls expression of a transcriptional repressor in r5/r6. Such an indirect effect may be mediated by krox20, which represses Hoxb1 (murine ortholog of zebrafish hoxb1a) expression in r4 [80,81,82,83,84]. In particular, krox20 activates the expression of Nab proteins, which are known negative regulators of transcription – making them possible candidates for mediating the effect of hnf1ba in repressing r4 gene expression [85]. Furthermore, the fact that only a subset of r4 genes is repressed by hnf1ba, suggests that additional factor (s) might be responsible for repressing the remaining r4 genes in an hnf1ba-independent manner.
We did not detect a reciprocal role for PG1 hox genes in repression of r5/r6 gene expression, but our experiments did demonstrate hoxb1a-mediated repression of gbx1 in r4 (Fig. 3Hiii and Fig. 5Ai). We do not know the mechanism for this repression, but it may be indirectly mediated by Nlz proteins – members of a subfamily of zinc-finger proteins. Previous work demonstrated that Nlz proteins, which are found in r4, act as transcriptional repressors and nlz loss of function leads to gene expression from adjacent rhombomeres expanding into r4 [86,87,88,89,90,91]. Since nlz1 expression is regulated by PG1 hox genes [87], gbx1 repression may be indirectly mediated by hoxb1a via Nlz proteins.
Members of the r4 and r5/r6 gene sets are not essential for hindbrain development
To test if members of the r4 and r5/r6 gene sets regulate caudal hindbrain formation, we analyzed germline mutants for six genes (gas6, gbx1, sall4, eglf6, celf2, and greb1l). Our results reveal that in all of these mutant lines, hindbrain patterning and the subsequent development of the r4-specific Mauthner neurons, and the r5/r6-specific abducens neurons are normal. Indeed, detailed transcriptome analysis of gas6 mutants identified differentially expressed genes involved in neuronal development, but the expression changes are subtle and cannot be detected by ISH. We cannot fully exclude the possibility that some residual gene activity persists in the specific mutants assayed. For instance, the egfl6 mutation affects a splice junction and some mutants may also harbor maternal transcripts or proteins. However, in three cases (gas6, sall4 and greb1l), we were able to assay the offspring of homozygous mutant parents, which eliminates the concern with maternal products. Furthermore, the viability of homozygous gbx1 and celf2 mutants was reduced, while egfl6 homozygous mutants were infertile, demonstrating that these genes are important, just not for hindbrain development. Based on these analyses, it appears that most members of the r4 and r5/r6 gene sets may not be individually essential for hindbrain development. Accordingly, we recently found that dusp6 and dusp2 homozygous mutants also have normal hindbrain and neuronal pattering [55]. Hence, our data suggest that caudal hindbrain development is robust, and genes involved in this process most likely have redundant roles such that the loss of a single gene will not cause gross developmental defects. However, we note that these analyses are not exhaustive. In particular, previous genetic screens were not performed to saturation and most r4-r6 genes identified herein have not yet been tested by deletion in the germline. It therefore remains possible that additional key genes acting in r4-r6 will be identified.
Conclusion
Gene regulatory networks are inherently complex, and this has been demonstrated in a variety of developmental processes in several model organisms. In this study we successfully positioned 22 previously uncharacterized genes into the existing GRN governing caudal hindbrain formation in the zebrafish (Fig. 9). Analysis of six mutant lines indicated that these genes are not absolutely required for r4-r6 formation but may have subtle roles. This leaves the previously reported factors RA, FGF, hoxb1a, hoxb1b, hnf1ba and valentino as key regulators of r4-r6 formation in the zebrafish. By extrapolation from work in the mouse [76,77,78], it is likely that krox20 and PG3 hox genes also play a role in r4-r6 development in the zebrafish. While this may seem to be a small number of essential genes, there are other GRNs that have a limited number of core regulatory factors, like that of the transcriptional network regulating ES cells. Biochemical and bioinformatic studies done in both mice and humans show that Oct4, Sox2 and Nanog are the master regulators controlling the pluripotency and self-renewal of embryonic stem cells. While there are other TFs are involved in the larger embryonic stem cell GRN, they all feed into the core Oct4-Sox2-Nanong circuit [92, 93]. In support of the complex nature of GRNs, we demonstrate that regulation of r4 and r5/r6 is achieved via different mechanism. Specifically, our results support a novel model wherein r4 genes are under the combinatorial regulation of RA and hoxb1b, whereas r5/r6 genes are downstream of the previously described RA, FGF hnf1ba and valentino factors. We also identify novel interactions between the two gene-sets where the most striking observation is the repression of gbx1 by hoxb1a in r4 (Fig. 9). In conclusion, our study demonstrates the distinct mechanisms of gene regulation in r4 and r5/r6 which stands as evidence to the complex nature of the GRN governing caudal hindbrain development in the zebrafish.
Abbreviations
- A-P:
-
Anterior-posterior
- CNS:
-
Central nervous system
- CRISPR:
-
Clustered Regularly Interspaced Short Palindromic Repeats
- DE:
-
Differentially expressed
- DEAB:
-
Diethylamino-benzaldehyde
- Dpf:
-
days post fertilization
- ENU:
-
N-ethyl-N-nitrosourea
- ES:
-
Embryonic stem
- EST:
-
Expressed sequence tag
- FGF:
-
Fibroblast growth factor
- GRN:
-
Gene regulatory network
- hpf:
-
Hours post fertilization
- ISH:
-
In situ hybridization
- MHB:
-
Mid-hindbrain boundary
- MO:
-
Morpholino
- OPC:
-
Oligodendrocyte progenitor cell
- PG1:
-
Paralog group 1
- r:
-
rhombomere
- RA:
-
Retinoic Acid
- sgRNA:
-
single-stranded guide RNA
- TALEN:
-
Transcription activator-like effector nuclease
- TF:
-
Transcription factor
- TILLING:
-
Targeted Induced Local Lesions
- WT:
-
Wildtype
- ZFIN:
-
The Zebrafish Information Network
- ZFN:
-
Zing finger nuclease
- ZIRC:
-
The Zebrafish International Resource Center
References
Chandrasekhar A. Turning heads: development of vertebrate branchiomotor neurons. Dev Dyn. 2004;229:143–61.
Chandrasekhar A, Moens C, Warren J, Kimmel C, Kuwada J. Development of branchiomotor neurons in zebrafish. Development. 1997;124:2633–44.
Trainor PA, Krumlauf R. Hox genes, neural crest cells and branchial arch patterning. Curr Opin Cell Biol. 2001;13:698–705. https://doi.org/10.1016/S0955-0674(00)00273-8.
Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–56. https://doi.org/10.1146/annurev.cellbio.042308.113423.
Moens CB, Prince VE. Constructing the hindbrain: insights from the zebrafish. Dev Dyn. 2002;224:1–17. https://doi.org/10.1002/dvdy.10086.
Rodier PM, Ingram JL, Tisdale B, Nelson S, Romano J. Embryological origin for autism: developmental anomalies of the cranial nerve motor nuclei. J Comp Neurol 1996;370:247–61. https://doi.org/10.1002/(SICI)1096-9861(19960624)370:2<247::AID-CNE8>3.0.CO;2-2.
Bingham SM, Sittaramane V, Mapp O, Patil S, Prince VE, Chandrasekhar A. Multiple mechanisms mediate motor neuron migration in the zebrafish hindbrain. Dev Neurobiol. 2010;70:87–99. https://doi.org/10.1002/dneu.20761.
Gavalas A, Krumlauf R. Retinoid signalling and hindbrain patterning. Curr Opin Genet Dev. 2000;10:380–6.
White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:2522–33.
Dupé V, Lumsden A. Hindbrain patterning involves graded responses to retinoic acid signalling. Development. 2001;128
Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–46.
Roy NM, Sagerström CG. An early Fgf signal required for gene expression in the zebrafish hindbrain primordium. Dev Brain Res. 2004;148:27–42. https://doi.org/10.1016/j.devbrainres.2003.10.005.
Marín F, Charnay P. Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development. 2000;127:4925–35.
Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–94.
Serpente P. Direct crossregulation between retinoic acid receptor and Hox genes during hindbrain segmentation. Development. 2005;132:503–13. https://doi.org/10.1242/dev.01593.
Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. https://doi.org/10.1016/j.ydbio.2005.07.015.
Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–87. https://doi.org/10.1242/dev.02706.
Linville A, Gumusaneli E, Chandraratna RAS, Schilling TF. Independent roles for retinoic acid in segmentation and neuronal differentiation in the zebrafish hindbrain; 2004. https://doi.org/10.1016/j.ydbio.2004.02.022.
Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol. 2002;12:1117–23.
Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–37.
Begemann G, Marx M, Mebus K, Meyer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev Biol. 2004;271:119–29. https://doi.org/10.1016/j.ydbio.2004.03.033.
Moens CB, Yan YL, Appel B, Force AG. Kimmel CB. Valentino: a zebrafish gene required for normal hindbrain segmentation. Development. 1996;122:3981–90.
Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–20. https://doi.org/10.1242/dev.01297.
Weicksel SE, Gupta A, Zannino DA, Wolfe SA, Sagerstrom CG. Targeted germ line disruptions reveal general and species-specific roles for paralog group 1 hox genes in zebrafish. BMC Dev Biol. 2014;14:25. https://doi.org/10.1186/1471-213X-14-25.
Zigman M, Laumann-Lipp N, Titus T, Postlethwait J, Moens CB. Hoxb1b controls oriented cell division, cell shape and microtubule dynamics in neural tube morphogenesis. Development. 2014;141:639–49. https://doi.org/10.1242/dev.098731.
Selland LG, Koch S, Laraque M, Waskiewicz AJ. Coordinate regulation of retinoic acid synthesis by pbx genes and fibroblast growth factor signaling by hoxb1b is required for hindbrain patterning and development. Mech Dev. 2018;150:28–41. https://doi.org/10.1016/j.mod.2018.02.005.
McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129:2339–54.
McClintock JM, Carlson R, Mann DM, Prince VE. Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001;128:2471–84.
Vlachakis N, Choe SK, Sagerström CG. Meis3 synergizes with Pbx4 and Hoxb1b in promoting hindbrain fates in the zebrafish. Development. 2001;128:1299–312.
Wiellette EL, Sive H. vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development. 2003;130
Labalette C, Bouchoucha YX, Wassef MA, Gongal PA, Le Men J, Becker T, et al. Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4. Development. 2011;138:317–26. https://doi.org/10.1242/dev.057299.
Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46.
Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, et al. The identification of genes with unique and essential functions in the development of the zebrafish. Danio rerio Development. 1996;123:1–36.
Pöpperl H, Rikhof H, Cheng H, Haffter P, Kimmel CB, Moens CB. Lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6:255–67. https://doi.org/10.1016/S1097-2765(00)00027-7.
Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–29. https://doi.org/10.1101/gad946701.
Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–40. https://doi.org/10.1038/ng896.
Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci. 2004;101:12792–7. https://doi.org/10.1073/pnas.0403929101.
Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, Dominguez C, et al. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325:1402–5. https://doi.org/10.1126/science.1173474.
Li E, Davidson EH. Building developmental gene regulatory networks. Birth Defects Res C Embryo Today. 2009;87:123–30. https://doi.org/10.1002/bdrc.20152.
Murray JI. Systems biology of embryonic development: prospects for a complete understanding of the Caenorhabditis elegans embryo. Wiley Interdiscip Rev Dev Biol. 2018:e314. https://doi.org/10.1002/wdev.314.
Peter I, Davidson E. Genomic control process: development and evolution.1st edition. 2015.
Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, et al. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–89. https://doi.org/10.1242/dev.001966.
Chan T-M, Longabaugh W, Bolouri H, Chen H-L, Tseng W-F, Chao C-H, et al. Developmental gene regulatory networks in the zebrafish embryo. Biochim Biophys Acta - Gene Regul Mech. 2009;1789:279–98. https://doi.org/10.1016/J.BBAGRM.2008.09.005.
Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–24.
Gouti M, Gavalas A. Hoxb1 controls cell fate specification and proliferative capacity of neural stem and progenitor cells. Stem Cells. 2008;26:1985–97. https://doi.org/10.1634/stemcells.2008-0182.
Bami M, Episkopou V, Gavalas A, Gouti M. Directed neural differentiation of mouse embryonic stem cells is a sensitive system for the identification of novel Hox gene effectors. PLoS One. 2011;6:e20197. https://doi.org/10.1371/journal.pone.0020197.
De Kumar B, Parker HJ, Paulson A, Parrish ME, Pushel I, Singh NP, et al. HOXA1 and TALE proteins display cross-regulatory interactions and form a combinatorial binding code on HOXA1 targets. Genome Res. 2017;27:1501–12. https://doi.org/10.1101/gr.219386.116.
De Kumar B, Parker HJ, Paulson A, Parrish ME, Zeitlinger J, Krumlauf R. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Dev Biol. 2017;432:151–64. https://doi.org/10.1016/J.YDBIO.2017.09.033.
Choe S-K, Sagerström CG. Paralog group 1 hox genes regulate rhombomere 5/6 expression of vhnf1, a repressor of rostral hindbrain fates, in a Meis-dependent manner. Dev Biol. 2004;271:350–61. https://doi.org/10.1016/j.ydbio.2004.04.007.
Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–72. https://doi.org/10.1016/j.ydbio.2007.06.012.
Choe S-K, Zhang X, Hirsch N, Straubhaar J, Sagerstrom CG. A screen for hoxb1-regulated genes identifies ppp1r14al as a regulator of the rhombomere 4 Fgf-signaling center. Dev Biol. 2011;358:356–67. https://doi.org/10.1016/j.ydbio.2011.05.676.
van den Akker WMR, Durston AJ, Spaink HP. Identification of hoxb1b downstream genes: hoxb1b as a regulatory factor controlling transcriptional networks and cell movement during zebrafish gastrulation. Int J Dev Biol. 2010;54:55–62. https://doi.org/10.1387/ijdb.082678wv.
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. https://doi.org/10.1002/aja.1002030302.
Hauptmann G, Gerster T. Multicolor whole-mount in situ hybridization. In: Developmental biology protocols. New Jersey: Humana Press; 2000. p. 139–48.
Maurer JM, Sagerström CG. A parental requirement for dual-specificity phosphatase 6 in zebrafish. BMC Dev Biol. 2018;18:6. https://doi.org/10.1186/s12861-018-0164-6.
Zannino DA, Sagerström CG, Appel B. olig2-expressing hindbrain cells are required for migrating facial motor neurons. Dev Dyn. 2012;241:315–26. https://doi.org/10.1002/dvdy.23718.
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–7. https://doi.org/10.1093/nar/gku410.
Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. https://doi.org/10.1371/journal.pone.0098186.
Kettleborough RNW, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–7. https://doi.org/10.1038/nature11992.
Biocore-Dolphin. https://www.umassmed.edu/biocore/introducing-dolphin/. Accessed 19 Feb 2016.
The Zebrafish Information Network http://zfin.org/downloads. Accessed 24 July 2016.
Aragon F, Pujades C, Appel B, Force A, Kimmel C, Owens D, et al. FGF signaling controls caudal hindbrain specification through Ras-ERK1/2 pathway. BMC Dev Biol. 2009;9:61. https://doi.org/10.1186/1471-213X-9-61.
Smith TG, Karlsson M, Lunn JS, Eblaghie MC, Keenan ID, Farrell ER, et al. Negative feedback predominates over cross-regulation to control ERK MAPK activity in response to FGF signalling in embryos. FEBS Lett. 2006;580:4242–5. https://doi.org/10.1016/j.febslet.2006.06.081.
Bermudez O, Pages G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. AJP Cell Physiol. 2010;299:C189–202. https://doi.org/10.1152/ajpcell.00347.2009.
Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dollé P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85.
Choe S-K, Hirsch N, Zhang X, Sagerström CG. hnf1b genes in zebrafish hindbrain development. Zebrafish. 2008;5:179–87. https://doi.org/10.1089/zeb.2008.0534.
Carpenter EM, Goddard JM, Chisaka O, Manley NR, Capecchi MR. Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development. 1993;118:1063–75.
Chisaka O, Musci TS, Capecchi MR. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox geneHox-#150;1.6. Nature. 1992;355:516–20. https://doi.org/10.1038/355516a0.
Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–19.
Taillebourg E, Buart S, Charnay P. Conditional, floxed allele of the Krox20 gene. Genesis. 2002;32:112–3.
Varshney GK, Lu J, Gildea DE, Huang H, Pei W, Yang Z, et al. A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res. 2013;23:727–35. https://doi.org/10.1101/gr.151464.112.
Su C-Y, Kemp HA, Moens CB. Cerebellar development in the absence of Gbx function in zebrafish. Dev Biol. 2014;386:181–90. https://doi.org/10.1016/j.ydbio.2013.10.026.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. https://doi.org/10.1093/nar/gkn923.
Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–51.
Parker HJ, Krumlauf R. Segmental arithmetic: summing up the Hox gene regulatory network for hindbrain development in chordates. Wiley Interdiscip Rev Dev Biol. 2017;6:e286. https://doi.org/10.1002/wdev.286.
Voiculescu O, Taillebourg E, Pujades C, Kress C, Buart S, Charnay P, et al. Hindbrain patterning: Krox20 couples segmentation and specification of regional identity. Development. 2001;128:4967–78.
Garel S, Garcia-Dominguez M, Charnay P. Control of the migratory pathway of facial branchiomotor neurones. Development. 2000;127:5297–307.
Gaufo GO, Thomas KR, Capecchi MR. Hox3 genes coordinate mechanisms of genetic suppression and activation in the generation of branchial and somatic motoneurons. Development. 2003;130:5191–201. https://doi.org/10.1242/dev.00730.
Rey-Campos J, Chouard T, Yaniv M, Cereghini S. vHNF1 is a homeoprotein that activates transcription and forms heterodimers with HNF1. EMBO J. 1991;10:1445–57.
Desmazières A, Charnay P, Gilardi-Hebenstreit P. Krox20 controls the transcription of its various targets in the developing hindbrain according to multiple modes. J Biol Chem. 2009;284:10831–40. https://doi.org/10.1074/jbc.M808683200.
Giudicelli F, Taillebourg E, Charnay P, Gilardi-Hebenstreit P. Krox-20 patterns the hindbrain through both cell-autonomous and non cell-autonomous mechanisms. Genes Dev. 2001;15:567–80. https://doi.org/10.1101/gad.189801.
Theil T, Frain M, Gilardi-Hebenstreit P, Flenniken A, Charnay P, Wilkinson DG. Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development. 1998;125:443–52.
Seitanidou T, Schneider-Maunoury S, Desmarquet C, Wilkinson DG, Charnay P. Krox-20 is a key regulator of rhombomere-specific gene expression in the developing hindbrain. Mech Dev. 1997;65:31–42. https://doi.org/10.1016/S0925-4773(97)00051-8.
Garcia-Dominguez M, Gilardi-Hebenstreit P, Charnay P. PIASxbeta acts as an activator of Hoxb1 and is antagonized by Krox20 during hindbrain segmentation. EMBO J. 2006;25:2432–42. https://doi.org/10.1038/sj.emboj.7601122.
Mechta-Grigoriou F, Garel S, Charnay P. Nab proteins mediate a negative feedback loop controlling Krox-20 activity in the developing hindbrain. Development. 2000;127:119–28.
Nakamura M, Choe S-K, Runko AP, Gardner PD, Sagerström CG. Nlz1/Znf703 acts as a repressor of transcription. BMC Dev Biol. 2008;8:108. https://doi.org/10.1186/1471-213X-8-108.
Labalette C, Wassef MA, Desmarquet-Trin Dinh C, Bouchoucha YX, Le Men J, Charnay P, et al. Molecular dissection of segment formation in the developing hindbrain. Development. 2015;142:185–95. https://doi.org/10.1242/dev.109652.
Andreazzoli M, Broccoli V, Dawid IB. Cloning and expression of noz1, a zebrafish zinc finger gene related to Drosophila nocA. Mech Dev. 2001;104:117–20. https://doi.org/10.1016/S0925-4773(01)00359-8.
Runko AP, Sagerström CG. Nlz belongs to a family of zinc-finger-containing repressors and controls segmental gene expression in the zebrafish hindbrain. Dev Biol. 2003;262:254–67. https://doi.org/10.1016/S0012-1606(03)00388-9.
Runko AP, Sagerström CG. Isolation of nlz2 and characterization of essential domains in Nlz family proteins. J Biol Chem. 2004;279:11917–25. https://doi.org/10.1074/jbc.M310076200.
Hoyle J, Tang YP, Wiellette EL, Wardle FC, Sive H. Nlz gene family is required for hindbrain patterning in the zebrafish. Dev Dyn. 2004;229:835–46. https://doi.org/10.1002/dvdy.20001.
Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci. 2007;104:16438–43. https://doi.org/10.1073/pnas.0701014104.
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. https://doi.org/10.1016/j.cell.2005.08.020.
Acknowledgements
We are grateful to Dr. Scot Wolfe for advice regarding CRISPR/Cas9 mutagenesis, to Dr. Nathan Lawson for providing the Cas9 plasmid, to Dr. Zhaoxia Sun for providing the hnf1ba mutant line, to Dr. Cecilia Moens for providing the valentino mutant line and to the Sagerström lab for helpful discussions and ideas. We acknowledge ZFIN for hosting the zebrafish gene expression database and ZIRC for providing the celf2, egfl6, sall4 and greb1l mutant lines. The following monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242: Isl1/2 (39.4D5) antibody developed by T. M. Jessell and S. Brenner-Morton; 3A10 antibody developed by T. M. Jessell, J. Dodd, and S. Brenner-Morton; and Zn8 antibody developed by B. Trevarrow. This work was supported by NIH grant NS038183 to CGS.
Funding
This work was supported by NIH grant NS038183 to CGS. The NIH had no role in the design of the study, in the collection, analysis, and interpretation of data or in writing the manuscript.
Availability of data and materials
The RNA-seq datasets generated and analyzed in the present study is available in the GEO repository at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113437.
Author information
Authors and Affiliations
Contributions
PG participated in the design of the study, mined the ZFIN database, carried out in situ hybridization in hoxb1a, hoxb1b, valentino and hnf1ba mutant animals, generated the gas6 and gbx1 germ line mutants, performed all phenotype analysis, and drafted the manuscript. JMM carried out in situ hybridizations for the fgf3, fgf8, dusp2, dusp6 and spry1 genes. CGS conceived the study, secured funding, participated in study design, and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was submitted to and approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee and the University of Massachusetts Medical School Institutional Review Board.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Genotyping of embryos collected from cross of hoxb1a heterozygous parents. Several mutant lines used in this study are not viable as adults. As a result, many embryos used in assays were collected from crosses of heterozygous mutants. To ensure the presence of homozygous mutants in an assayed clutch, embryos were individually genotyped following ISH as outlined in the Methods section. Representative genotyping data for hoxb1a mutant embryos stained with (A) spry1, (B) dusp6, (C) egfl6 and (D) greb1l demonstrate that approximately one quarter of the embryos assayed are homozygous mutant (indicated with asterisks), while 100% of the clutch showed normal staining for the assayed gene. (PDF 967 kb)
Additional file 2:
Table S1. Identification of additional r4, r5 and r6 genes. This file represents the data downloaded from ZFIN and how it was parsed to generate a list of 107 rhombomere-restricted genes expressed in r4, r5 and r6. Sheet 1 lists 1820 genes that are expressed in the hindbrain, rhombomeres 4, 5 and 6. Sheet 2 represents 1194 genes that are expressed in the hindbrain, rhombomeres 4, 5 and 6 during the first 24 h development. Sheet 4 has the list of 107 genes that re restricted to rhombomeres 4, 5 and 6. Sheets 5, 6 and 7 represent the genes sorted according to their expression location and additional information associated to these genes are also listed in the last three sheets. (XLSX 112 kb)
Additional file 3:
Figure S2. Expression of the r4 gene set is unaffected in hoxb1a mutants at least until 24hpf. Expression of hoxb1a (A), meis1a (B), fgf3 (C) and egfl6 (D) was assessed in wildtype (i) and hoxb1a mutant (ii) zebrafish at 24hpf. The black brackets mark r4 and dotted circles represent the otic vesicles (OV). (PDF 1051 kb)
Additional file 4:
Figure S3. Generation and analysis of RNA-seq data from 18 hpf WT and hoxb1b mutant embryos. (A) Total RNA was collected from WT and hoxb1b mutant whole embryos and used for RNA-seq. (B) 866 differentially expressed genes were identified from RNA-Seq where seven of the 175 up-regulated genes and 78 of the 691 down-regulated genes are expressed in the hindbrain. (C) Top 20 GO terms for up-regulated and down-regulated genes. (D) A subset of genes was validated by RT-qPCR from independently collected samples. (PDF 589 kb)
Additional file 5:
Table S2. Detailed analysis of WT and hoxb1b mutant RNA-seq data. 866 differentially expressed genes identified from the RNA-Seq experiment are listed in sheet 2. All GO terms associated with Biological Processes for both up-regulated and down-regulated genes are shown in sheets 3 and 4. Sheet 5 shows the subset of differentially expressed genes that is expressed in the hindbrain. In sheet 5, the genes in column A were derived from the ZFIN database (refer to sheet 3 in Additional file 2: Table S1). (XLSX 215 kb)
Additional file 6:
Figure S4. Comparison between RNA-seq analyses of Hoxa1 mutant mouse embryos and hoxb1b mutant zebrafish embryos. RNA-seq analysis of Hoxa1 mutant mouse embryos was recently published in [48]. Comparing the mouse data set (A) with the 866 differentially expressed genes identified by our RNA-seq (B) revealed an overlap of 31 genes (C). Notably, none of these 31 genes has a rhombomere restricted expression pattern. (PDF 211 kb)
Additional file 7:
Figure S5. SU5402 disrupts embryogenesis in hoxb1b mutants. Wildtype (i) and hoxb1b mutant (ii) zebrafish embryos were treated with SU5402 and assayed at various developmental stages by brightfield microscopy (A, B, F, H), or ISH to detect expression of efnb2a/krox20 (C), meis1a/krox20 (D), irx7/krox20 (E) or greb1l/krox20 (G). Note that defects in development are readily detectable in hoxb1b mutants treated with 50uM SU5402 (Aii, Bii), but not in WT embryos treated with SU5402 (Ai, Bi), nor in hoxb1b mutants treated with DEAB (Biii). As a result of these severe developmental defects, hoxb1b mutant embryos treated with SU5402 showed no specific staining for the r4 genes tested. (PDF 853 kb)
Additional file 8:
Figure S6. Genotyping data for sall4, egfl6, celf2 and greb1l mutants. sall4, egfl6, celf2 and greb1l mutants generated by TILLING were procured from ZIRC. In each case, the mutation introduces a single nucleotide change (A; red text) causing a premature stop codon, except for egfl6 where the point mutation disrupts an essential splice site in exon 8. (B) Sequencing traces showing expected single nucleotide changes in each mutant line. (PDF 758 kb)
Additional file 9:
Data S1. Amino acid sequences of wildtype and mutant gas6 alleles. Amino acid sequences of four mutant gas6 alleles (um296, um297, um298, um299) aligned to the wildtype sequence shows that all four mutant alleles code for a premature stop codon after 96 amino acids. (DOCX 18 kb)
Additional file 10:
Figure S7. Scheme for generating gbx1 mutant line. (A) Schematic showing the 20 nucleotide (orange text) target site in exon 1 of gbx1. CCT represents the PAM sequence (blue box) and ATG (green box) is the start codon. Hpy188III target sequence is represented by the dotted red line, the red arrow denotes the cut site. (B) sgRNA and Cas9 mRNA was injected into 1-cell stage embryos. Injected embryos were raised to 24hpf and genomic DNA extracted from a pool of embryos. Hpy188III digest of PCR products amplified from genomic DNA (extracted from injected embryos) reveal the presence of a mutation (red boxes in gel). (C) Injected embryos were raised to give rise to F0 adults. These fish were crossed with WT adults to raise the F1 generation. At 3 months age, genomic DNA was extracted from fin-clips of individual F1 fish and genotyped as described in panel B. (D) Sequencing of F1 genomic DNA revealed transmission of two different mutant alleles; one allele (um300) has a 26-nucleotide insertion (green text) and the second allele (um301) has a two-nucleotide deletion (orange dashes). The resulting amino acid sequence is shown in the form of grey (amino acid sequence identical to wildtype) and yellow (out of frame amino acid sequence) boxes. Both mutant alleles result in premature stop codons upstream of the homeodomain. (E) Hpy188III digest of PCR products amplified from genomic DNA (extracted from individual F2 embryos) reveal the absence of homozygous mutants. (PDF 772 kb)
Additional file 11:
Data S2. Amino acid sequences of wildtype and mutant gbx1 alleles. Amino acid sequences of two mutant gbx1 alleles (um300 and um301) aligned to the wildtype sequence shows that the mutant alleles introduce premature stop codons. (DOCX 14 kb)
Additional file 12:
Table S3. Detailed analysis of WT and gas6 mutants RNA-seq data. 1590 differentially expressed genes were identified from the RNA-Seq experiment and is shown in sheet 2. All the GO terms associated with Biological Processes for both up-regulated and down-regulated genes are listed in sheets 3 and 4. Sheet 5 shows the subset of differentially expressed genes that are expressed in the hindbrain. (XLSX 889 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ghosh, P., Maurer, J.M. & Sagerström, C.G. Analysis of novel caudal hindbrain genes reveals different regulatory logic for gene expression in rhombomere 4 versus 5/6 in embryonic zebrafish. Neural Dev 13, 13 (2018). https://doi.org/10.1186/s13064-018-0112-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13064-018-0112-y