Abstract
Background
The effect of extracellular microenvironment (hypoxia and pH) has been regarded as a key hallmark in cancer progression. The study aims to investigate the effects of carbonic anhydrase IX (CAIX), a key hypoxia-inducible marker, in triple-negative breast cancer (TNBC) in correlation with clinicopathological parameters and predicting survival outcomes.
Methods
A total of 323 TNBC cases diagnosed at the Department of Anatomical Pathology, Singapore General Hospital from 2003 to 2013 were used. Immunohistochemical staining (IHC) was performed using CAIX antibody and digital mRNA quantification was performed using NanoString assays. CAIX membranous expression was correlated with clinicopathological parameters using Chi-squared test or Fisher’s exact tests. Disease-free survival (DFS) and overall-survival (OS) were estimated using Kaplan–Meier analysis and compared between groups with the log-rank test.
Results
Forty percent of TNBCs were observed to express CAIX protein and demonstrated significant association with larger tumour size (P = 0.002), higher histological grade (P < 0.001), and significantly worse disease-free survival (DFS) and overall survival (OS) (after adjustment: HR = 2.99, 95% CI = 1.78–5.02, P < 0.001 and HR = 2.56, 95% CI = 1.41–4.65, P = 0.002, respectively). Gene ontology enrichment analysis revealed six significantly enriched cellular functions (secretion, cellular component disassembly, regulation of protein complex assembly, glycolytic process, cellular macromolecular complex assembly, positive regulation of cellular component biogenesis) associated with genes differentially expressed (CAIX, SETX, WAS, HK2, DDIT4, TUBA4α, ARL1). Three genes (WAS, SETX and DDIT4) were related to DNA repair, indicating that DNA stability may be influenced by hypoxia in TNBC.
Conclusions
Our results demonstrate that CAIX appears to be a significant hypoxia-inducible molecular marker and increased CAIX protein levels are independently associated with poor survival in TNBC. Identification of CAIX-linked seven gene-signature and its relationship with enriched cellular functions further support the implication and influence of hypoxia-mediated CAIX expression in TNBC tumour microenvironment.
Similar content being viewed by others
Background
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer with high five year mortality which is partly due to the lack of therapeutic target specificity on common breast cancer receptors such as oestrogen receptor (ER), progesterone receptor (PR) or the human epidermal growth factor receptor 2 (HER2) [1]. Further classification of TNBCs can be grouped into four molecular subgroups, driving many studies focusing on immunotherapy and new development in endocrine targeted treatments to identify potential targeted therapies [2]. Hypoxic microenvironment in tumour cells occurs in most solid malignancies, evolving tumours into an aggressive oncogenic metabolism, increasing metastasis and enhancing resistance to clinical therapies [3,4,5]. Studies have also shown that hypoxia markers such as hypoxia inducible factor 1 (HIF-1) and hypoxia-driving factors are associated poorly in TNBC outcomes [6,7,8].
HIF-1 is a heterodimeric protein composed of a constitutively expressed HIF-1ß subunit and an O2- regulated HIF-1α subunit [9, 10]. Increased HIFα activates target genes involved in tumour proliferation, angiogenesis, metabolism, apoptosis and metastasis [4]. Additionally, HIFα and its regulated proteins including carbonic anhydrase nine (CAIX) and glucose transporter 1 (GLUT1) are highly expressed in several type of cancers and are associated with dismal prognosis [11,12,13,14]. HIF-1 regulates key aspects of cancer biology, including pH regulation in glycolysis, through CAIX [15]. Over-expression of CAIX was observed in several solid tumours, and its link with invasiveness has given rise to the hypothesis that CAIX expression may contribute to advanced disease and tumour progression [11, 15]. Increased CAIX expression has been shown to be more common in TNBC compared to other subtypes of breast cancer and a marker of poor prognosis [11, 16]. Therefore, we investigated the impact of hypoxia-dependent CAIX in both protein and transcriptional expression on TNBC biology and outcome in order to elucidate its potential role as a therapeutic target in a subset of TNBC patients.
Methods
Study design and clinicopathological parameters
A total of 323 archival formalin-fixed paraffin-embedded (FFPE) TNBC specimens from patients diagnosed between 2003 and 2013 at the Department of Anatomical Pathology, Singapore General Hospital were analysed. 17 cases were excluded due to depleted tumour regions and/or IHC staining artefacts. Only IHC-proven invasive TNBC immunophenotype in female patients was included in the study while those with history of neoadjuvant chemotherapy, radiotherapy, and concomitant cancers were excluded. Clinicopathological parameters were reviewed (Tables 1, 2). The Centralized Institutional Review Board of SingHealth provided ethical approval for the retrospective study.
Tissue microarray (TMA) construction
Tissue Microarray (TMA) was constructed as previously described [17], using tumour regions which was selected based on pathological assessment of > 50% of the sample being tumour area.
Immunohistochemistry and immunoscoring
Tissue microarray sections of 4 µm thickness were cut onto Bond Plus slides (Leica Biosystems Richmond) and heated at 60 °C for 20 min. The slides were then incubated with primary antibodies specific for HIF-1α (rabbit monoclonal, Abcam, Cambridge, MA, US, diluted 1:200) and CAIX (rabbit monoclonal, Cell Marque, Rocklin, CA, USA, diluted 1:100) using Leica Bond Max autostainer (Leica Biosystems Melbourne) and Roche Ventana Benchmark Ultra (Ventana Medical Systems Arizona), respectively. Details of antibodies, labelling patterns and dilution factors can be found in Additional file 1: Table S1. Positive controls used for HIF-1α include glioblastoma and tonsil tissue, while renal cell carcinoma tissue was used as a positive control for CAIX. Antibodies were detected with diaminobenzidine substrate (DAB) as the chromogen, and counterstained with hematoxylin.
Immunoscoring was done by two trained pathologists to determine the staining intensity and percentage of tumour cells stained in each TMA core. Semi-quantitative H-score was used and calculated using intensity and percentage expressed, respectively. The H-score was calculated as follows: (3 × % strong staining) + (2 × % moderate staining) + (1 × % weak staining). To analyse HIF-1α expression, only homogenously and darkly stained nuclei were considered, and a median H-score of ≥ 1 was considered positive. The staining of CAIX was scored as positive using a median H-score of ≥ 1 for membrane staining. Tumours were then categorized into “CAIX-negative” and “CAIX-positive” subsets based on the median H-score of ≥ 1.
RNA extraction and NanoString gene expression measurement
RNA was extracted from four FFPE sections of 10 µm thickness using the RNeasy FFPE kit (Qiagen, Hilden, Germany) on a QIAcube automated sample preparation system (Qiagen, Hilden, Germany), and was quantified by an Agilent 2100 Bioanalyzer system (Agilent, Santa Clara, CA, USA). A total of 100 ng of functional RNA (> 300 nucleotides) was assayed on the nCounter MAX Analysis System (NanoString Technologies, Seattle, WA, USA). The NanoString counts were normalized using the positive control probes as well as the housekeeping genes, as previously reported [18]. The count data were then logarithmically transformed prior to further analysis. A total of 386 genes in the NanoString panel were tested for significant differences between CAIX positive and CAIX negative groups.
Gene ontology (GO) enrichment analysis
Seven genes that were significantly differentially expressed were analysed for gene ontology (GO) enrichment using an R package (topGO) and stringent selection criteria to avoid false positive results to effectively cluster functional genes into different biological processes. Significant ontology terms were determined by a P value < 0.05 in this study.
Follow-up and statistical analysis
Follow-up data were obtained from electronic medical records. Disease-free survival (DFS) and overall-survival (OS) were defined as the time from diagnosis to recurrence or death/date of last follow-up, respectively.
Statistical analysis was performed using SPSS for Windows, Version 15. The relationship between the association the clinicopathological parameters and hypoxia-related protein biomarkers was tested using Chi-square test or Fisher’s exact test. Survival outcomes were estimated with the Kaplan–Meier analysis and compared between subgroups with the log-rank statistics. Multivariate Cox Regression was carried out to evaluate the effect of CAIX tumour cell expression level with survival adjusted to the effects of age, grade, tumour size, lymph node stage, lymph node positivity and/or HIF-1α H score; multivariate analysis was also carried out on combinatorial CAIX/HIF1α tumour cell expression level with survival adjusted to the effects of age, grade, tumour size and lymph node stage.
Genes that were significantly differentially expressed between the two sample groups (positive-CAIX, negative-CAIX) were identified using Student t-tests with Welch’s correction and was used to determine differentially expressed genes (DEGs). Multiple testing corrections were applied using the method of Benjamini and Hochberg. The selection of seven significantly differentially expressed genes was based on statistical significance (P < 0.05) using t-tests (on the expression values) and multiple testing corrections (method of Benjamini and Hochberg), as seen in Additional file 1: Figure S1. Hierarchical clustering using complete linkage on Euclidean distances for both samples and genes generated a heat map, and is coloured by the gene expression levels (log2 counts) which has been mean centred and scaled by standard deviation on a per gene basis with the highest expression in red and the lowest expression in blue (Fig. 4).
All gene expression and survival data for the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) and The Cancer Genome Atlas (TCGA) were obtained from cBioPortal (http://www.cbioportal.org/) [19,20,21]. Statistical significance was defined by P value < 0.05.
Results
Positive CAIX membrane staining is associated with larger tumour size, higher histological grade and poorer survival rates
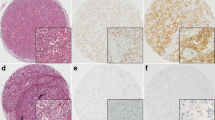
Positive CAIX membranous staining in tumour cells was present in approximately 39.5% of the TNBC cohort (121/306) (Fig. 1). Approximately 45.9% of the tumour showed HIF-1α expression (141/307). However, the expression was variable throughout the tumour with some accentuation near areas of necrosis.
Significant associations were found between CAIX positivity in tumour cells and clinicopathological features such as larger tumour size (P = 0.002) and higher histological grade (P < 0.001) in Table 1. However, positive HIF-1α expression did not show any significant association with any clinicopathological parameters (Additional file 1: Table S2).
Furthermore, TNBC patients with CAIX-positive expression had significantly worse disease-free survival (DFS) and poorer overall-survival (OS) ([DFS: HR = 2.77, 95% CI 1.78 to 4.31, P < 0.001], and [OS: HR = 2.48, 95% CI = 1.50–4.09, P < 0.001]) (Fig. 2). Moreover, after adjustment by age, grade, tumour size and lymph node positivity, there is a significant difference between positive CAIX expression and negative CAIX expression in TNBC patients on their survival outcomes ([OS: HR = 2.99, 95% CI = 1.78–5.02, P < 0.001], and [DFS: HR = 2.56, 95% CI = 1.41–4.65, P = 0.002]).
However, survival analysis for HIF-1α expression in TNBC patients found no statistical differences in DFS and OS ([DFS: P = 0.137], and [OS: P = 0.807]). Although significant correlation between CAIX and HIF-1α protein expression in tumours was observed (P = 0.013) (Table 2); further adjustments by age, grade, tumour size, lymph node stage and HIF-1α H score in survival outcomes of CAIX tumour expression, HIF-1α did not affect CAIX risks on poorer survival and prognostic outcomes ([DFS: HR = 2.95, 95% CI 1.75 to 5.00, P < 0.001], and [OS: HR = 2.43, 95% CI 1.34 to 4.41, P = 0.004]).
Co-expression of HIF-1α and CAIX protein in TNBC patients is linked with poorer survival rates
In addition, patients with both HIF-1α and CAIX protein co-expression were more likely to have shorter DFS (HR = 3.07, 95% CI 1.72 to 5.49, P < 0.001) and poorer OS (HR = 2.30, 95% CI 1.20 to 4.39, P = 0.012) (Table 3). After accounting for age, grade, tumour size and lymph node stage, there is a statistically significant association in patients with both HIF-1α and CAIX protein co-expression and survival outcomes ([DFS: HR = 4.46, 95% CI 2.26 to 8.81, P < 0.001], and [OS: HR = 3.30, 95% CI 1.57 to 6.94, P = 0.002]) (Table 3).
Expression level of hypoxia CAIX-linked genes (CAIX, DDIT4, TUBA4α, HK2 and ARL1, WAS , SETX) is significantly higher in CAIX-positive and CAIX-negative TNBCs, respectively
Out of the 306 viable CAIX TNBC tumours identified for immunoscoring, 105 “positive” and 152 “negative” tumour samples had NanoString RNA data. Samples from four benign breast tumours were also included in this analysis. Student t-tests with Welch’s correction revealed seven genes (CAIX, Carbonic Anhydrase IX; HK2, Hexokinase 2; TUBA4α, Tubulin Alpha 4α; DDIT4, DNA damage inducible transcript 4; SETX, Senataxin; WAS, WASP Actin Nucleation Promoting Factor; ARL1, ADP Ribosylation Factor Like GTPase 1) that showed differential expression (P < 0.05).
Amongst the differentially expressed genes (DEGs), four genes (CAIX, DDIT4, TUBA4α, HK2) reported significant upregulated expression level in our CAIX-positive TNBC cohort (Fig. 3A–D and Additional file 1: Table S3). On the contrary, the remaining three DEGs (ARL1, WAS, SETX) reported significant downregulated expression level in our CAIX-positive TNBC cohort (Fig. 3E–G and Additional file 1: Table S3). Within the seven genes, CAIX have been reported to have a similar gene expression profile with DDIT4 and HK2 in our TNBC cohort in the heat map (Fig. 4).
Six differentially expressed genes (CAIX, HK2, TUBA4α, DDIT4, SETX , WAS) are associated with key cellular pathways modulating tumorigenesis
Gene ontology enrichment analysis identified significant functional enrichment in expression of genes related to secretion (CAIX, HK2, and TUBA4α), cellular component disassembly (DDIT4, HK2, and SETX), regulation of protein complex assembly (SETX and WAS), glycolytic process (DDIT4 and HK2), cellular macromolecular complex assembly (SETX and WAS) and positive regulation of cellular component biogenesis (SETX and WAS) between the positive and negative CAIX groups in TNBCs. Taken together, these six pathways share six genes which are CAIX, HK2, TUBA4α, DDIT4, SETX and WAS (Table 4).
Low (WAS , SETX) and high (ARL1, DDIT4, TUBA4α, CAIX, HK2) mRNA expression is associated with poorer overall-survival rates in TNBC
Comparison of the prognosis of seven DEGs in TNBC observed that low (SETX and WAS) and high (ARL1, DDIT4, TUBA4α, CAIX, HK2) mRNA expression is associated with poorer overall-survival in our SGH TNBC database (SETX, P < 0.05; WAS, P < 0.001; ARL1, P = 0.07934; DDIT4, P < 0.01; TUBA4α, P = 0.1503; CAIX, P = 0.2001; HK2, P = 0.2224) (Table 5, and Additional file 1: Figure S2; Table S4).
Comparative survival analysis on DEG expression between SGH, METABRIC and TCGA patient database
Low WAS gene expression had poorer OS in all three databases (SGH, P < 0.001; METABRIC, P < 0.05; TCGA, P = 0.3709) (Table 5, and Additional file 1: Figure S2a; Table S4); however, high WAS gene expression reported poorer OS post-290 months in the METABRIC database. Moreover, high ARL1 gene expression also demonstrated poorer overall-survival (OS) in all three databases (SGH, P = 0.07934; METABRIC, P = 0.09737; TCGA, P < 0.05) (Table 5, and Additional file 1: Figure S2b, Table S4). Similarly, high DDIT4, high TUBA4α, high CAIX, and high HK2 gene expression showed poorer OS in SGH and METABRIC databases, respectively (SGH, P < 0.01, P = 0.1503, P = 0.2001, and P = 0.2224; METABRIC, P = 0.07328, P = 0.4021, P < 0.001, and P = 0.2111) (Table 5, and Additional file 1: Figure S2c, e–g; Table S4).
However, low DDIT4, TUBA4α, CAIX, and HK2 gene expression reported poorer OS in TCGA database, respectively (P = 0.08319, P < 0.05, P = 0.09129, and P < 0.01) (Table 5, and Additional file 1: Figure S2c, e–g; Table S4).
Furthermore, low SETX gene expression had poorer overall-survival (OS) in SGH and METABRIC databases (SGH, P < 0.05; METABRIC, P = 0.1404) (Table 5, and Additional file 1: Figure S2d; Table S4); however, high SETX gene expression reported poorer OS post-290 months in METABRIC database. High SETX gene expression had poorer OS in TCGA database (TCGA, P = 0.2142) (Table 5, and Additional file 1: Figure S2d; Table S4).
Discussion
In the present study, we investigated the role of two important hypoxia-regulated markers (HIF-1α and CAIX) and found that increased expression in both CAIX protein and mRNA transcriptional levels are indicators of poorer survival in TNBC. However, HIF-1α protein expression failed to demonstrate any such association with either survival or clinicopathological factors. Interestingly, our results showed that HIF-1α protein expression is not a confounding factor in prognosis of patients expressing CAIX protein. However, co-expression of CAIX and HIF-1α protein in TNBC patients had the poorest prognosis. Furthermore, our study also identified seven CAIX-linked hypoxia genes with prognostic value in our TNBC cohort: DDIT4, ARL1, WAS, SETX, HK2, TUBA4α and CAIX which have been known to be hypoxia-regulated in vitro.
Our results were in agreement with CAIX protein in breast cancer studies, where 50% of basal-like breast cancers usually have high grade tumours expressing CAIX [22, 23]. Previous clinical studies in invasive breast cancer have also demonstrated the association of CAIX with poor outcome, suggesting that CAIX expression is linked to an aggressive phenotype [11, 16, 24, 25]. Over-expression of CAIX and carbonic anhydrase XII (CAXII) has also been associated with poor DFS in invasive breast cancer. However, the role of CAXII remains unclear and there have been conflicting reports about its role in TNBC. Chen et al. have shown that CAIX correlated with CAXII (R = 0.376, P = 0.0001) in a cohort of invasive breast cancer [26]. However, our study did not include CAXII and thus, unable show any correlation findings.
Furthermore, our study did not manage to find any prognostic value in HIF-1α protein expression, suggesting that HIF-1α may not be a reliable marker for hypoxia in TNBC. Although there are many markers to assess hypoxia in tumours, such as HIF-1α, X-Box Binding Protein 1 (XBP1), GLUT1 and Vascular endothelial growth factor (VEGF) [7, 8], the results however have been conflicting in various studies. Drawbacks associated with the modification of these hypoxia-responsive protein markers are their potential regulation by non-hypoxia-related factors such as stress, growth factor application, oncogene activation, cell culture densities, local pH, and metabolite concentrations [27]. Therefore, generating hypoxia signatures from in vivo tissue, despite the presence of contaminating stromal tissue, seem to be more robust than those generated from in vitro experiments [28]. Yehia et al. assessed the relative expression of HIF-1α among three breast cancer groups (TNBC, HER2+, ER+/PR+), with TNBC expression results differed only slightly and with little to no statistical significance from the other subgroups, and that HER2 positive tumours showed the highest levels of expression for all studied parameters [29]. This further supports that HIF-1α may not be an exclusive candidate marker for TNBC. Previous findings have demonstrated that HIF-1α was undetectable within minutes after re-oxygenation [30], suggesting that CAIX possibly activates hypoxic condition independently of HIF-1α, as CAIX protein persists longer than HIF-1α. Thus, CAIX as a biomarker for hypoxia could be more suitable as it is more stable and persists longer than HIF-1α.
Moreover, previous findings show that CAIX in high density cultures is induced via the phosphatidylinositol-3-kinase (PI3K) pathway [31] and by the mitogen-activated protein kinase (MAPK) pathway during both normoxia and hypoxia conditions [32]. Taken together, these observations suggest that CAIX expression may also be driven by other HIF-1α-independent signalling pathways to induce hypoxic conditions in the cells. Therefore, CAIX may be a better biomarker for cancer hypoxia.
The seven CAIX-linked hypoxia genes identified in our study have been linked to modulate key functions in tumourigenesis such as DNA repair, metastasis, innate immunity and metabolism in Additional file 1: Table S5. Notably, three of the genes (DDIT4, WAS, SETX) are linked to DNA repair functions. DNA damage inducible transcript 4 (DDIT4) acts as an independent prognostic factor for TNBC resistant to neoadjuvant chemotherapy [33]. DDIT4 activity supposedly enhances cancer cell resistance to mTOR inhibitors, thereby increasing cancer cells chemoresistance. Our results further support the notion of significant association between high DDIT4 mRNA level with poor survival, and reported upregulation in DDIT4 expression in our CAIX-positive TNBC cohort. Induced DDIT4 expression under cellular stressors and other chemical molecules (e.g. glucocorticoids, endoplasmic reticulum stress inducers, etc.) suggests its role in DNA repair under hypoxic conditions [34].
In the other two genes (WAS, SETX) linked to DNA repair functions, both downregulated WAS and SETX mRNA expression is associated with poorer overall-survival. Similarly, a subset of TNBC with increased expression of WAS and SETX mRNA showed better survival in other studies [35, 36]. Gene SETX role in tumourigenesis has been linked to its function in maintaining genome integrity via the coordination of transcription, DNA replication and DNA damage response [35], whereas gene WAS encodes for the cytoskeletal regulator, Wiskott-Aldrich syndrome protein (WASP), which plays a key role in tumourigenesis via binding to double strand breaks, regulating RNA Polymerase II activity and facilitating actin polymerization [37]. Its influence on actin filament dynamics and facilitation of actin reorganization, such as branching and crosslinking, are inherent in metastasis and invasion [37, 38]. Moreover, WASP and Arp2/3 complex have been reported to be recruited to damaged DNA double-strand breaks sites to promote double-strand breaks clustering and homology-directed repair [38, 39].
Thus, these further supports that the integrity of DNA-repair mechanism may be essential for protection against hypoxia-mediated DNA damage [36, 40, 41]. These biological categories have known functional relationships on breast cancer development and the aforementioned genes’ value as diagnostic markers and therapeutic targets deserves further investigation.
Within our seven gene DEG signature, TUBA4α is linked to metastasis, HK2 and CAIX is linked to promoting tumourigenesis, while the remaining ARL1 is linked to innate immunity [42]. Our results showed that these four genes were upregulated within the CAIX-positive group and associated with poorer survival outcomes in this subset of TNBC patients. Upregulation of TUBA4α disrupts the optimal tubulin isotype compositions in cell [43] and the dynamics of microtubule polymerisation and depolymerisation are of key importance in spindle formation during mitosis [44]. Moreover, upregulation of HK2 drives glucose metabolism and promotes sufficient number of metabolic intermediates to support anabolic processes (such as nucleic acid, lipid and protein synthesis), which is characteristic of rapidly dividing cancer cells [45]. While upregulation of CAIX disrupts pH balance [46], resulting in a hypoxic environment, which is also regulated under hypoxic condition through the hypoxia inducible factor (HIF1) cascade, promoting tumorigenesis. Thus, these genes are associated with aggressive cancer features and proliferation within the tumour microenvironment, reflecting the poorer survival outcome in our study.
Our study has several limitations. Since the FFPE blocks used in TMA construction were dated from 2003 to 2013, the tissue quality may be considered a limitation of this study. Tissue quality may contribute to the reduction of antigenicity and decrease in the sensitivity of the IHC reaction, leading to reduced protein detection. Furthermore, the FFPE tissue quality may also affect the amount of viable RNA for NanoString extraction and experiments. Although this study was conducted on a limited number of patient samples, the data indicates that quantification of hypoxia-related genes in TNBC can have potential prognostic value regardless of treatment type. Moreover, it is imperative that the clinical relevance of the seven hypoxia-linked gene signatures to be validated in independent studies with larger patient cohorts. Protein expression of the aforementioned genes showing significant association with survival is being studied in ongoing follow-up studies.
Conclusion
In conclusion, our study demonstrated that CAIX expression is independently associated with a poorer clinical and survival outcome in TNBC. Since hypoxia is increasingly being studied for being responsible for resistance against radiotherapy and emerging immunotherapy [47], the identification of the seven-genes associated with CAIX could be a step forward to test for hypoxia in TNBCs and possibly improve patients’ treatment regimen and prognosis. Thus, further studies on the seven-gene hypoxia panel are warranted.
Availability of data and materials
The data that support the findings of this study are available from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) and The Cancer Genome Atlas (TCGA) were obtained from cBioPortal (http://www.cbioportal.org/). The datasets generated and analysed during the current study are not publicly available due restrictions from institutional policy on human tissue data but are available from the corresponding author on reasonable request.
Abbreviations
- TNBC:
-
Triple-negative breast cancer
- ER:
-
Oestrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
- HIF-1:
-
Hypoxia inducible factor 1
- CAIX:
-
Carbonic anhydrase IX
- GLUT1:
-
Glucose transporter 1
- FFPE:
-
Formalin-fixed paraffin-embedded
- TMA:
-
Tissue Microarray
- IHC:
-
Immunohistochemical staining
- DAB:
-
Diaminobenzidine substrate
- GO:
-
Gene ontology
- DFS:
-
Disease-free survival
- OS:
-
Overall survival
- DEG:
-
Differentially expressed genes
- HK2:
-
Hexokinase 2
- TUBA4α:
-
Tubulin Alpha 4α
- DDIT4:
-
DNA damage inducible transcript 4
- SETX:
-
Senataxin
- WAS:
-
WASP Actin Nucleation Promoting Factor
- ARL1:
-
ADP Ribosylation Factor Like GTPase 1
- CAXII:
-
Carbonic anhydrase XII
- XBP1:
-
X-Box Binding Protein 1
- VEGF:
-
Vascular endothelial growth factor
- PI3K:
-
Phosphatidylinositol-3-kinase
- MAPK:
-
Mitogen-activated protein kinase
- WASP:
-
Wiskott-Aldrich syndrome protein
References
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67.
Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22(1):61.
Jing X, Yang F, Shao C, Wei K, Xie M, Shen H, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18(1):157.
Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl). 2015;3:83–92.
Pastorekova S, Ratcliffe PJ, Pastorek J. Molecular mechanisms of carbonic anhydrase IX-mediated pH regulation under hypoxia. BJU Int. 2008;101(Suppl 4):8–15.
Bernardi R, Gianni L. Hallmarks of triple negative breast cancer emerging at last? Cell Res. 2014;24(8):904–5.
Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508(7494):103–7.
Semenza GL. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–14.
Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92(12):5510–4.
Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270(3):1230–7.
Tan EY, Yan M, Campo L, Han C, Takano E, Turley H, et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer. 2009;100(2):405–11.
Sadlecki P, Bodnar M, Grabiec M, Marszalek A, Walentowicz P, Sokup A, et al. The role of Hypoxia-inducible factor-1 α, glucose transporter-1, (GLUT-1) and carbon anhydrase IX in endometrial cancer patients. Biomed Res Int. 2014;2014: 616850.
Oh S, Kim H, Nam K, Shin I. Glut1 promotes cell proliferation, migration and invasion by regulating epidermal growth factor receptor and integrin signaling in triple-negative breast cancer cells. BMB Rep. 2017;50(3):132–7.
Airley RE, Loncaster J, Raleigh JA, Harris AL, Davidson SE, Hunter RD, et al. GLUT-1 and CAIX as intrinsic markers of hypoxia in carcinoma of the cervix: relationship to pimonidazole binding. Int J Cancer. 2003;104(1):85–91.
Trastour C, Benizri E, Ettore F, Ramaioli A, Chamorey E, Pouysségur J, et al. HIF-1alpha and CA IX staining in invasive breast carcinomas: prognosis and treatment outcome. Int J Cancer. 2007;120(7):1451–8.
Chia SK, Wykoff CC, Watson PH, Han C, Leek RD, Pastorek J, et al. Prognostic significance of a novel hypoxia-regulated marker, carbonic anhydrase IX, in invasive breast carcinoma. J Clin Oncol. 2001;19(16):3660–8.
Thike AA, Yong-Zheng Chong L, Cheok PY, Li HH, Wai-Cheong Yip G, Huat Bay B, et al. Loss of androgen receptor expression predicts early recurrence in triple-negative and basal-like breast cancer. Mod Pathol. 2014;27(3):352–60.
Yeong J, Thike AA, Lim JC, Lee B, Li H, Wong SC, et al. Higher densities of Foxp3(+) regulatory T cells are associated with better prognosis in triple-negative breast cancer. Breast Cancer Res Treat. 2017;163(1):21–35.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.
Hedlund EE, McDonald PC, Nemirovsky O, Awrey S, Jensen LDE, Dedhar S. Harnessing induced essentiality: targeting carbonic anhydrase IX and angiogenesis reduces lung metastasis of triple negative breast cancer xenografts. Cancers (Basel). 2019;11(7):1002.
Li Y, Wang H, Oosterwijk E, Selman Y, Mira JC, Medrano T, et al. Antibody-specific detection of CAIX in breast and prostate cancers. Biochem Biophys Res Commun. 2009;386(3):488–92.
Brennan DJ, Jirstrom K, Kronblad A, Millikan RC, Landberg G, Duffy MJ, et al. CA IX is an independent prognostic marker in premenopausal breast cancer patients with one to three positive lymph nodes and a putative marker of radiation resistance. Clin Cancer Res. 2006;12(21):6421–31.
Colpaert CG, Vermeulen PB, Fox SB, Harris AL, Dirix LY, Van Marck EA. The presence of a fibrotic focus in invasive breast carcinoma correlates with the expression of carbonic anhydrase IX and is a marker of hypoxia and poor prognosis. Breast Cancer Res Treat. 2003;81(2):137–47.
Chen Z, Ai L, Mboge MY, Tu C, McKenna R, Brown KD, et al. Differential expression and function of CAIX and CAXII in breast cancer: a comparison between tumorgraft models and cells. PLoS ONE. 2018;13(7): e0199476.
Kappler M, Taubert H, Schubert J, Vordermark D, Eckert AW. The real face of HIF1α in the tumor process. Cell Cycle. 2012;11(21):3932–6.
Li Y, Patel SP, Roszik J, Qin Y. Hypoxia-driven immunosuppressive metabolites in the tumor microenvironment: new approaches for combinational immunotherapy. Front Immunol. 2018;9:1591.
Yehia L, Boulos F, Jabbour M, Mahfoud Z, Fakhruddin N, El-Sabban M. Expression of HIF-1α and markers of angiogenesis are not significantly different in triple negative breast cancer compared to other breast cancer molecular subtypes: implications for future therapy. PLoS ONE. 2015;10(6): e0129356.
Harris BH, Barberis A, West CM, Buffa FM. Gene expression signatures as biomarkers of tumour hypoxia. Clin Oncol (R Coll Radiol). 2015;27(10):547–60.
Chu CY, Jin YT, Zhang W, Yu J, Yang HP, Wang HY, et al. CA IX is upregulated in CoCl2-induced hypoxia and associated with cell invasive potential and a poor prognosis of breast cancer. Int J Oncol. 2016;48(1):271–80.
Shafee N, Kaluz S, Ru N, Stanbridge EJ. PI3K/Akt activity has variable cell-specific effects on expression of HIF target genes, CA9 and VEGF, in human cancer cell lines. Cancer Lett. 2009;282(1):109–15.
Pinto JA, Rolfo C, Raez LE, Prado A, Araujo JM, Bravo L, et al. In silico evaluation of DNA damage inducible transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci Rep. 2017;7(1):1526.
Tirado-Hurtado I, Fajardo W, Pinto JA. DNA damage inducible transcript 4 gene: the switch of the metabolism as potential target in cancer. Front Oncol. 2018;8:106.
Becherel OJ, Yeo AJ, Stellati A, Heng EY, Luff J, Suraweera AM, et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013;9(4): e1003435.
Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57(4):636–47.
Izdebska M, Zielińska W, Hałas-Wiśniewska M, Grzanka A. Involvement of actin and actin-binding proteins in carcinogenesis. Cells. 2020;9(10):2245.
Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, et al. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature. 2018;559(7712):61–6.
Hurst V, Shimada K, Gasser SM. Nuclear actin and actin-binding proteins in DNA repair. Trends Cell Biol. 2019;29(6):462–76.
Luoto KR, Kumareswaran R, Bristow RG. Tumor hypoxia as a driving force in genetic instability. Genome Integr. 2013;4(1):5.
Yang L, Forker L, Irlam JJ, Pillay N, Choudhury A, West CML. Validation of a hypoxia related gene signature in multiple soft tissue sarcoma cohorts. Oncotarget. 2018;9(3):3946–55.
Lu L, Tai G, Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol Biol Cell. 2004;15(10):4426–43.
Parker AL, Teo WS, McCarroll JA, Kavallaris M. An emerging role for tubulin isotypes in modulating cancer biology and chemotherapy resistance. Int J Mol Sci. 2017;18(7):1434.
Nami B, Wang Z. Genetics and expression profile of the tubulin gene superfamily in breast cancer subtypes and its relation to taxane resistance. Cancers (Basel). 2018;10(8):274.
Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24(2):213–28.
Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov. 2008;7(2):168–81.
Wang B, Zhao Q, Zhang Y, Liu Z, Zheng Z, Liu S, et al. Targeting hypoxia in the tumor microenvironment: a potential strategy to improve cancer immunotherapy. J Exp Clin Cancer Res. 2021;40(1):24.
Acknowledgements
We like to thank the team at the Cancer Epigenome research laboratory at National Cancer Centre for the processing and running of the NanoString experiments in our study.
Funding
This research was supported by the Transition Award from the Singapore National Medical Research Council (NMRC/TA/0041/2015) and by the SingHealth Duke-NUS Pathology Academic Clinical Programme (ACP). BL is part of the SIgN Immunomonitoring platform (supported by a BMRC IAF 311006 grant, BMRC transition funds #H16/99/b0/011, BMRC IAF-PP H1901a0024 grant and NRF SIS NRF2017_SISFP09 grant).
Author information
Authors and Affiliations
Contributions
JI and TPH conceived the study while JI, AAT, HYL, and JPSY planned and supervised the study. CHCO, JCTL, JXL performed the laboratory experiments and analysis as well as data extraction and data quality check. HHL provided the statistical analysis while BL conducted the bioinformatic analysis. CHCO, DYL, and JI interpreted the data and contributed to the manuscript with input from all authors. All the authors provided critical feedback on the analysis and manuscript done. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethics approval was obtained from The Centralized Institutional Review Board of SingHealth, Singapore and consent was waived for this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest. Prof Tan Puay Hoon declares non-financial competing interest as she is on the Breast Cancer Research Journal editorial board.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Document contains supplementary tables and figures mentioned in the manuscript
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ong, C.H.C., Lee, D.Y., Lee, B. et al. Hypoxia-regulated carbonic anhydrase IX (CAIX) protein is an independent prognostic indicator in triple negative breast cancer. Breast Cancer Res 24, 38 (2022). https://doi.org/10.1186/s13058-022-01532-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13058-022-01532-0