Abstract
The prolactin receptor gene (PRLR) may contribute to polycystic ovarian syndrome (PCOS) since it plays important roles in physiological ovarian functions. PRLR-knockout mice have irregular cycles and subfertility and variants in or around the PRLR gene were associated in humans with female testosterone levels and recurrent miscarriage. We tested 40 variants in the PRLR gene in 212 Italian families phenotyped by type 2 diabetes (T2D) and PCOS and found two intronic PRLR-variants (rs13436213 and rs1604428) significantly linked to and/or associated with the risk of PCOS. This is the first study to report PRLR as a novel risk gene in PCOS. Functional studies are needed to confirm these results.
Similar content being viewed by others
Introduction
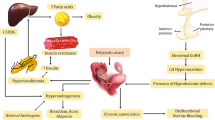
Prolactin (PRL) is well-known for its role in milk production and social bonding and less known for its role in insulin secretion and normal ovarian function [1,2,3]. PRL was shown to play a role in ovarian follicular development and maintenance of the corpus luteum [2]. These effects are mediated by the prolactin receptor (PRLR) which is encoded by the PRLR gene and whose variants are implicated in glucose homoeostasis [1] and gestational diabetes [4]. Both the PRL and the PRLR are expressed locally in the ovaries of premenopausal women controlling follicular formation and maturation as well as ovulation [5], possibly through paracrine or autocrine action [6]. The PRLR gene can potentially be implicated in the risk of polycystic ovarian syndrome (PCOS), which is a common condition characterized by anovulation, hyperandrogenism, insulin resistance, and polycystic ovaries [7]. More than one-third of PCOS patients have abnormally high PRL levels [8]. And the expression of the PRLR gene is reduced in the endometrium of women with PCOS [9] and in mouse models with PCOS-like phenotype [10], reinforcing the link between the PRL system and PCOS. High PRL levels also impair the gonadotropin-releasing hormone (GnRH) release and pulsatility [11], which then affects GnRH effects on the release of the gonadotropins follicle-stimulating hormone (FSH) and luteinizing hormone (LH) [12], which respectively stimulate ovarian follicles formation and ovulation, thereby impairing the ovulatory cycles [13]. Furthermore, PRLR-knockout mice have irregular cycles and subfertility [14] and variants in or around the PRLR gene were associated with changes in female testosterone levels [15] and recurrent miscarriage [16]. Thus, an impaired PRLR function might mediate altered effects of PRL on GnRH and the hypothalamic-pituitary-ovarian axis, reproducing the PCOS abnormalities [6]. Yet, no study has reported PRLR as a risk gene in PCOS. In this study we report for the first-time novel risk variants in the PRLR gene significantly linked and associated with the risk of PCOS in Italian families.
Materials and methods
We have tested 40 variants in the PRLR gene in 212 peninsular Italian families originally recruited for a type 2 diabetes (T2D) study and subsequently phenotyped for PCOS according to the PCOS Rotterdam diagnostic criteria (presence of at least two of these three characteristics: chronic anovulation or oligomenorrhea, clinical or biochemical hyperandrogenism, and/or polycystic ovaries) [17]. Identical twins and cases of uncertain paternity were excluded. The subjects were at least Italian for 3 generations [18].The samples were previously collected from the subjects’ whole blood and underwent the traditional phenol/chloroform DNA extraction method. Primers were chosen from the Affymetrix microarray database used that were available for the PRLR gene. The SNPs were selected based on their presence within the gene and validation in at least 3 public genetic databases. SNPs to be considered valid had to reach a quality control of at least 0.96. Random replicates from the samples were run to verify the accuracy of the results. PLINK [18] was used to detect genotyping or Mendelian errors allowing to identify any potential sample swap or paternity uncertainty or adoption case. The analyses we ran were free of any potential error. We used Pseudomarker [19] to test for 2-point parametric-linkage to and linkage disequilibrium (LD, that is linkage + association) with PCOS across the models: dominant with complete penetrance (D1), dominant with incomplete penetrance (D2), recessive with complete penetrance (R1), and recessive with incomplete penetrance (R2). Pseudomarker allows to test for linkage and LD (i.e., linkage + association) in extended pedigrees, affected siblings, trios (parents and affected child), and for association within unrelated individuals across families. Both linkage and LD under various hypotheses are tested. The LD/linkage can be considered the most important test, as it tests for both linkage and association, given the presence of linkage for the marker under analysis. Variants with p < 0.05 were considered significant. We performed functional bioinformatics analysis for the 2 PCOS-risk variants testing for disruption of transcription-factor binding (SNP2TFBS [20]), splicing (SNP-function prediction [21]) miRNA binding (mirSNP [22]), and/or regulation potential (RegulomDB [23]).
[23]Results and Discussion.
We detected two intronic PRLR-variants (rs13436213 and rs1604428) significantly linked to/in LD with PCOS across the two recessive models (R1 and R2) (p < 0.05) (Table 1). Both variants are novel and have not been linked to any of PCOS-related phenotypes (i.e., obesity, insulin resistance, T2D, metabolic syndrome, hyperglycemia, hyperandrogenism, male-pattern baldness, acne, hirsutism, infertility, oligomenorrhea, anovulation or irregular menses). The same risk allele (C) of the variant (rs1604428) was significantly linked to the risk of depression in a previous analysis of the same dataset [24] Women with PCOS have increased risk of depression [25] and the PRLR-rs1604428 variant could partially explain this association. The functional bioinformatics analysis for the 2 PCOS-risk variants in our study found that the variants intersect with repressed chromatin state and potentially negative PRLR gene expression in the uterine tissue according to the analysis done by RegulomeDB [23] which predicts the regulatory potential of variants such as transcription-factor binding and chromatin state. Interestingly, this is consistent with a previous study reporting lower PRLR-mRNA levels in the endometrium of obese patients with PCOS compared to controls [9].
Conclusion
This is the first study to report PRLR as a novel risk gene in PCOS. Functional studies are needed to confirm these results.
Data availability
The study data are available on reasonable request, and due to lacking specific patients’ consent and privacy restrictions, they are not publicly available.
References
Nteeba J, Kubota K, Wang W, Zhu H, Vivian J, Dai G et al. Pancreatic prolactin receptor signaling regulates maternal glucose homeostasis. J Endocrinol. 2019.
Le JA, Wilson HM, Shehu A, Mao J, Devi YS, Halperin J, et al. Generation of mice expressing only the long form of the prolactin receptor reveals that both isoforms of the receptor are required for normal ovarian function. Biol Reprod. 2012;86(3):86.
Baumgard LH, Hausman GJ, Sanz Fernandez MV. Insulin: pancreatic secretion and adipocyte regulation. Domest Anim Endocrinol. 2016;54:76–84.
Le TN, Elsea SH, Romero R, Chaiworapongsa T, Francis GL. Prolactin receptor gene polymorphisms are associated with gestational diabetes. Genet Test Mol Biomarkers. 2013;17(7):567–71.
Marano RJ, Ben-Jonathan N, Minireview. Extrapituitary prolactin: an update on the distribution, regulation, and functions. Mol Endocrinol. 2014;28(5):622–33.
Chang S, Copperman AB. New insights into human prolactin pathophysiology: genomics and beyond. Curr Opin Obstet Gynecol. 2019;31(4):207–11.
Rotterdam EA-SPCWG. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81(1):19–25.
Davoudi Z, Araghi F, Vahedi M, Mokhtari N, Gheisari M. Prolactin level in polycystic ovary syndrome (PCOS): an approach to the diagnosis and management. Acta Biomed. 2021;92(5):e2021291.
Paulson M, Norstedt G, Sahlin L, Hirschberg AL. Association between prolactin receptor expression and proliferation in the endometrium of obese women with polycystic ovary syndrome. Gynecol Endocrinol. 2020;36(3):226–32.
Rotgers E, Nicol B, Rodriguez K, Rattan S, Flaws JA, Yao HH. Constitutive expression of steroidogenic factor-1 (NR5A1) disrupts ovarian functions, fertility, and metabolic homeostasis in female mice. Faseb j. 2021;35(8):e21770.
Henderson HL, Townsend J, Tortonese DJ. Direct effects of prolactin and dopamine on the gonadotroph response to GnRH. J Endocrinol. 2008;197(2):343–50.
McNeilly AS. Prolactin and the control of gonadotrophin secretion in the female. J Reprod Fertil. 1980;58(2):537–49.
Son WY, Das M, Shalom-Paz E, Holzer H. Mechanisms of follicle selection and development. Minerva Ginecol. 2011;63(2):89–102.
Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, et al. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 1997;11(2):167–78.
Ruth KS, Day FR, Tyrrell J, Thompson DJ, Wood AR, Mahajan A, et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat Med. 2020;26(2):252–8.
Hanna CW, Bretherick KL, Liu CC, Stephenson MD, Robinson WP. Genetic variation within the hypothalamus-pituitary-ovarian axis in women with recurrent miscarriage. Hum Reprod. 2010;25(10):2664–71.
Rotterdam EA-SP. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod. 2004;19(1):41–7.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75.
Hiekkalinna T, Schaffer AA, Lambert B, Norrgrann P, Goring HH, Terwilliger JD. PSEUDOMARKER: a powerful program for joint linkage and/or linkage disequilibrium analysis on mixtures of singletons and related individuals. Hum Hered. 2011;71(4):256–66.
Kumar S, Ambrosini G, Bucher P. SNP2TFBS - a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res. 2017;45(D1):D139–d44.
Xu Z, Taylor JA. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Research. 2009;37(SUPPL. 2).
Liu C, Zhang F, Li T, Lu M, Wang L, Yue W et al. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genomics 2012 13:1. 2012;13(1):1–10.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7.
Amin M, Wu R, Postolache T, Gragnoli C. The prolactin receptor gene (PRLR) is linked to and associated with the comorbidity of depression and type 2 diabetes in italian families. Genes & Diseases (In Press; 2023.
Cooney LG, Dokras A. Depression and anxiety in polycystic ovary syndrome: etiology and treatment. Curr Psychiatry Rep. 2017;19(11):83.
Acknowledgements
We thank the families who participated in the study, and we thank Bios Biotech Multi-Diagnostic Health Center, Rome, Italy, for data access and for financial, medical, and laboratory staff support. This study was supported by Penn State College of Medicine. This publication was supported in part with the funds received under Nebraska Laws 2021, LB 380, Sect. 109 awarded to C.G. (PI), Creighton University School of Medicine, through the Nebraska Department of Health & Human Services (DHHS). Its contents represent the views of the authors and do not necessarily represent the official views of the State of Nebraska or DHHS.
Author information
Authors and Affiliations
Contributions
M.A. (https://orcid.org/0000-0003-2876-0784) helped with manuscript drafting and in silico analysis. C.G. (https://orcid.org/0000-0002-3873-6617) conceived and performed the study, and critically revised the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Families were recruited following the Helsinki declaration guidelines, and individuals provided written informed consent prior to participation. The Bios Ethical Committee approved this study.
Competing interests
The authors have declared that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Amin, M., Gragnoli, C. The prolactin receptor gene (PRLR) is linked and associated with the risk of polycystic ovarian syndrome. J Ovarian Res 16, 222 (2023). https://doi.org/10.1186/s13048-023-01280-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13048-023-01280-5
Keywords
- Prolactin
- PRL
- Prolactin receptor
- PRLR
- Gene
- Expression
- Polycystic ovarian syndrome
- PCOS
- Ovary
- Cortisol
- Hypothalamic-pituitary-ovarian axis
- HPO-axis
- Metabolic
- Insulin resistance
- IR
- Obesity
- Type 2 diabetes
- T2D
- Families
- Familial
- Peninsular
- Italy
- Italian
- Parametric analysis
- Linkage disequilibrium
- Association
- Single nucleotide polymorphisms
- SNP
- Risk
- Variant
- Model
- Dominant
- Recessive
- Penetrance
- Complete
- Incomplete
- Irregular menses
- Oligomenorrhea
- Subfertility
- Folliculogenesis
- Fat metabolism
- Ethnic group
- Appetite
- Energy expenditure
- Estrous cycle length
- Follicle luteinization
- Maturation
- Metabolism
- Adrenal
- Steroidogenesis
- Anovulation
- Testosterone
- Hyperandrogenism
- Hyperandrogenemia
- Endocrine
- Disorder
- Women
- Reproductive age
- Impaired glucose metabolism
- Milk production
- Social bonding
- Genotyping
- Mendelian
- PLINK
- Pseudomarker
- Testosterone
- Miscarriage
- Recurrent
- Depression
- Rotterdam diagnostic criteria
- Identical twins
- Uncertain paternity
- Generation
- Endometrium
- Obese
- RegulomeDB
- Control
- rs13436213
- rs1604428