Abstract
Background
This study was aimed to explore the role of long noncoding RNA C17orf91 and its potential mechanisms in ovarian cancer development.
Results
To assess its role in ovarian cancer, microarray datasets (GSE14407, GSE30587, and GSE17260) in Gene Expression Omnibus (GEO) were utilized to assess the expression and clinical significance of C17orf91 in ovarian cancer. Next, loss-of-function studies were performed to establish the role of C17orf91 and the underlying mechanisms in ovarian cancer development. It was found that elevated expression of C17orf91 was observed in omental metastases when compared with matched primary ovarian tumors(GSE30587, P = 0.016). Moreover, Log Rank analysis revealed that increased expression of C17orf91 was associated with shorter progression free survival(PFS)(HR = 1.90(1.19-3.03), P = 0.008). Overall survival(OS) also showed a similar trend, but did not reach statistical significance(HR = 1.75(0.97-3.13), P = 0.061). Loss-of-function studies further demonstrated that C17orf91 repression impaired migration, invasion and viability of ovarian cancer cells, and downregulated the pro-metastatic gene, MYC, at both mRNA and protein level.
Conclusion
Collectively, our findings revealed that C17orf91 was a potential prognostic marker and functioned as an oncogene in ovarian cancer. It remains to be seen whether modulation of C17orf91 expression will cause phenotypic changes in vivo.
Similar content being viewed by others
Background
Ovarian cancer is the most lethal gynecological cancer and the majority of the patients are not diagnosed until advanced stages. Over the past decades, little improvement in overall survival has been achieved despite advances in chemotherapeutic agents [1]. Better understanding of the mechanisms involved in ovarian cancer development is of great importance in overcoming this malignancy.
Compelling evidence have indicated a large-scale regulatory network generated by noncoding RNAs, including microRNAs(miRNAs) and long noncoding RNAs(lncRNAs). Integrated analyses of the cancer genome and transcriptome have identified profound alterations in noncoding genes [2, 3]. Although numerous studies have helped unveiling the functions of miRNAs in various cancers, only a small number of functional lncRNAs have been thoroughly characterized to date. So far, lncRNAs have been involved in the regulation of various cellular processes, including but not limited to cell growth, cell cycle, apoptosis and motility [4]. C17orf91, also known as MIR22HG or MGC14376, is a long non-coding RNA(lncRNA) located on chromosome 17p13 [5, 6]. It is the host gene of miR-22 [5, 6]. Previous studies have linked miR-22 to a great number of activities, such as tumorigenesis, epigenetic modification, embryonic development [7]. However, the function of its host gene C17orf91 has not yet been defined in any conditions, including cancer [8, 9].
In this study, we tried to analyze C17orf91 expression and its clinical significance in ovarian cancer and to explore its role in ovarian cancer development.
Methods
GEO datasets
The publicly available microarray datasets used in our study is described in GEO database(GSE14407, GSE30587 and GSE17260). GSE14407 dataset was used to explore the differential expression of C17orf91 (probe ID: 214696_at) between normal ovarian surface epithelium(n = 12) and primary ovarian cancers(n = 12). GSE30587 dataset was used to determine the differential expression of C17orf91 (probe ID: 8011193) between paired primary ovarian cancer tissues(n = 9) and their corresponding omental metastases(n = 9). Relationship between C17orf91 expression (probe ID: A_23_P49610) and ovarian cancer outcome was determined using the gene expression data in 110 ovarian cancer patients(GSE17260).
Cell lines
Human serous ovarian cancer cell Hey was originally obtained from M.D. Anderson Cancer Center (Houston, TX) and cultured in RPMI medium 1640 plus 10 % fetal bovine serum with penicillin/streptomycin. 293 T cell line was originally obtained from the cell bank of Chinese Academy of Science (Shanghai) and cultured in DMEM plus 10 % fetal bovine serum with penicillin/streptomycin.
Constructs
PLKO-Scramble-shRNA was purchased from Addgene(Addgene #1864). PLKO-C17orf91-shRNA plasmids were constructed according to PLKO.1 protocol. The sequences of C17orf91 shRNA were obtained from The RNAi Consortium (TRC, MISSION® TRC shRNA library, Sigma) and shown in Additional file 1: Table S1.
Lentiviral infection
For producing lentiviral particles, 293 T cells were seeded in 3.5-cm dishes at a density of 4 × 105 per dish and transfected with PLKO-Scramble-shRNA/PLKO-C17orf91-shRNA, as well as packing plasmids psPAX2 (0.75 μg) (Addgene#12260) and pMD2.G (0.25 μg) (Addgene #12259) using Attractene Transfection Reagent (QIAGEN). Forty-eight hours post transfection, virus-containing medium (2 ml) was harvested, filtered, mixed with 2 ml of freshsly prepared medium, supplemented with 8 μg/ml polybrene (Sigma) and added to 3 × 105 Hey cells seeded in a 3.5-cm dish the day before. Puromycin (2 μg/ml) was supplemented 48 h after infection. The cells were selected for 2 days and then used for various assays.
QPCR
Briefly, reverse transcription reactions were carried out using RevertAidTM First Strand cDNA Synthesis Kit (Fermentas). QPCR was performed using SYBR® Premix Ex Taq™ II (Perfect Real Time) Kit (TaKaRa, Dalian, China) in ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Quantitative analysis was performed using Comparative CT method. The relative expression of each gene was normalized to the expression of GAPDH. Primers used in present study were shown in Additional file 1: Table S1.
Western blot assay
Whole cell extracts were prepared in chilled RIPA lysis buffer(Beyotime, China). 30 μg of lysates protein were separated by SDS-PAGE using a 10 % polyacrylamide gel and transferred to 0.45 μm PVDF membrane(Millipore). Membranes were blocked with 5 % non-fat milk in PBS containing 0.05 % Tween-20, blotted with MYC antibody(1:1000, CST) or GAPDH antibody(1:3000, beyotime) overnight, followed by goat anti-mouse antibody. The images were scanned by LAS4000 device.
Measurement of cell migration and invasion
RTCA CIM-Plate was used to explore the effect of C17orf91 on migration and invasion of ovarian cancer cells. Migration and invasion assays were performed according to protocols supplied by the manufacturers. For invasion assay, the inserts were coated with BD Matrigel (B.D. Biosciences). Briefly, 4 × 104 Scramble-shRNA- and C17orf91-shRNA-transfected Hey cells were added into the RTCA CIM-Plate and monitored by xCELLigence (Roche) for 36 h.
Measurement of cell viability
RTCA E-plate was used to explore the effect of C17orf91 on cell viability. Briefly, 2 × 103 Scramble-shRNA- and C17orf91-shRNA-transfected Hey cells were added into the RTCA E-Plate and monitored by xCELLigence (Roche) for 96 h.
Statistical analysis
All data were analyzed with SPSS statistic software (SPSS 16.0). Student’s t test (two-tailed) was used to compare two groups. The Log-rank test was used to determine the relationship between C17orf91 expression and clinical outcomes(progression free survival and overall survival). The Kaplan-Meier method was used to generate survival curves. P-value <0.05 was considered statistically significant.
Results
Differential expression of C17orf91 between OSE, primary tumors and omental metastases
To explore the clinical significance of C17orf91 in ovarian cancer development, we first determined the differential C17orf91 expression between serous ovarian cancer tissues and normal ovarian surface epithelium(OSE) with resort to the GEO datasets. It was found that C17orf91 expression appeared to be downrelulated in ovarian cancer tissues compared with OSE, though without statistical significance(GSE14407, Fig. 1a, P = 0.078). However, elevated expression of C17orf91 was observed in omental metastases when compared with matched primary ovarian tumors(GSE30587, Fig. 1b, P = 0.016).
Differential expression of C17orf91 between OSE, primary tumors and omental metastases. a C17orf91 expression appeared to be decreased in primary tumors compared with OSE, but do not reach statistical significance(GSE14407). b Paired t-test shows that C17orf91 expression was significantly elevated in omental metastases relative to corresponding primary tumors(GSE30587)
The prognostic value of C17orf91 expression in ovarian cancer
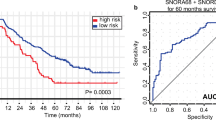
Next, we assessed the prognostic value of C17orf91 expression by taking advantage of the microarray dataset (GSE17260) in Gene Expression Omnibus. Interestingly, Log Rank (Mantel-Cox) analysis revealed that increased expression of C17orf91 was associated with shorter progression free survival(PFS) (Fig. 2a, HR = 1.90(1.19-3.03), P = 0.008). Overall survival(OS) also showed a similar trend, but did not reach statistical significance(Fig. 2b, HR = 1.75(0.97-3.13), P = 0.061).
The prognostic value of C17orf91 expression in ovarian cancer(GSE17260). a Log Rank (Mantel-Cox) analysis revealed that increased expression of C17orf91 was associated with shorter progression free survival(PFS). b Overall survival(OS) also showed a similar trend, but did not reach statistical significance. “Low” and “High” were classified according to the C17orf91 expression level. The median expression value for C17orf91 was used as the cutoff point
Oncogenic role of C17orf91 in ovarian cancer
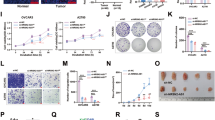
To explore the potential role of C17orf91 in ovarian cancer, we performed loss-of-function studies in Hey cells. Stable C17orf91-shRNA clones as well as their respective control clones were generated and examined for C17orf91 mRNA by Real-time RT-PCR (Fig. 3a). Interestingly, C17orf91 inhibition significantly decreased migration, invasion and viability of Hey cells (Fig. 3b–3d).
C17orf91 reglulated MYC expression in ovarian cancer
To further investigate the mechanism by which C17orf91 elicited its oncogenic role, we explored whether C17orf91 could regulate key pro-metastatic genes, such as KLF8 [10], MYC [11], SNAI2 [12], TWIST1 [13], ZEB1 [14] and ZEB2 [15] in ovarian cancer. It was found that C17orf91 repression could reduce the expression of KLF8, MYC and SNAI2 at mRNA level (Fig. 4a). Subsequent western blot assay was focused on MYC partly because it was the most obviously altered gene. It was found that C17orf91 downregulation also decreased the protein level of MYC (Fig. 4b).
Discussion
C17orf91, also known as MIR22HG, was a stress responsive lncNRA [16]. Previous studies showed that stress responsive lncRNAs might be linked to cancer progression [17–19]. Knockdown of LSINCT5 significantly impaired the proliferation of both breast and ovarian cancer cells [17]. Another stress responsive lncRNA which might also be involved in cancer progression is PRINS(psoriasis susceptibility-related RNA gene induced by stress). It was found that PRINS played a protective role in cells exposed to stress, and moreover, elevated PRINS expression in the epidermis might contribute to psoriasis susceptibility [18]. Further studies revealed that PRINS could regulate the expression of G1P3, an inferno-inducible gene with anti-apoptotic effects in cancer cells [19]. In this study, we also demonstrated that the stress responsive lncRNA C17orf91 could regulate the migration, invasion and viability of ovarian cancer cells.
Additionally, we characterized the clinical significances of C17orf91 in ovarian cancer tissues by taking advantages of publicly available microarray datasets in GEO database. It was noticeable that C17orf91 expression appeared to be downregulated in ovarian cancer tissues compared with OSE though without statistical significance. Indeed, such findings were not only limited in our study. For example, the well-known pro-metastatic gene miR-10b was found to be downregulated in primary breast tumors compared with normal breast tissues [20, 21]. Our findings, together with findings from other laboratories, have indicated that oncogenes are not always upregulated in cancer tissues when compared with corresponding normal tissues. This may also be applicable for tumor suppressors which are commonly downregulated in cancer tissues relative to normal controls.
Previous studies indicated that decreased C17orf91 expression was associated with a worse prognosis in breast and lung cancer patients [8, 9, 22]. However, our data revealed that increased C17orf91 expression correlated with shorter PFS and OS. These discrepant findings might be related to the fact that the prognostic value of C17orf91 was cancer specific. Mechanismly, we demonstrated that C17orf91 repression could downregulate the MYC expression at both mRNA and protein level, indicating that the oncogenic role elicited by C17orf91 might be partly mediated by the induction of MYC expression.
Conclusion
In conclusion, our findings revealed that C17orf91 was a potential prognostic marker and functioned as an oncogene in ovarian cancer. It remains to be seen whether modulation of C17orf91 expression will cause phenotypic changes in vivo.
References
Coleman MP, Forman D, Bryant H, Butler J, Rachet B, Maringe C, Nur U, Tracey E, Coory M, Hatcher J, McGahan CE, Turner D, Marrett L, Gjerstorff ML, Johannesen TB, Adolfsson J, Lambe M, Lawrence G, Meechan D, Morris EJ, Middleton R, Steward J, Richards MA. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995–2007 (the International Cancer Benchmarking Partnership): an analysis of population-based cancer registry data. Lancet. 2011;377:127–38.
Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc HK, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905.
Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24.
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–87.
Li X, Liu J, Zhou R, Huang S, Huang S, Chen XM. Gene silencing of MIR22 in acute lymphoblastic leukaemia involves histone modifications independent of promoter DNA methylation. Br J Haematol. 2010;148:69–79.
Wang J, Xiang G, Zhang K, Zhou Y. Expression signatures of intragenic miRNAs and their corresponding host genes in myeloid leukemia cells. Biotechnol Lett. 2012;34:2007–15.
Xiong J. Emerging roles of microRNA-22 in human disease and normal physiology. Curr Mol Med. 2012;12:247–58.
Patel JB, Appaiah HN, Burnett RM, Bhat-Nakshatri P, Wang G, Mehta R, Badve S, Thomson MJ, Hammond S, Steeg P, Liu Y, Nakshatri H. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 2011;30:1290–301.
Gyorffy B, Schafer R. Biomarkers downstream of RAS: a search for robust transcriptional targets. Curr Cancer Drug Targets. 2010;10:858–68.
Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res. 2007;67:7184–93.
Cho KB, Cho MK, Lee WY, Kang KW. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 2010;293:230–9.
Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, Cogdell D, Nykter M, Broaddus R, Rodriguez-Aguayo C, Lopez-Berestein G, Liu J, Shmulevich I, Sood AK, Chen K, Zhang W. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23:186–99.
Li CW, Xia W, Huo L, Lim SO, Wu Y, Hsu JL, Chao CH, Yamaguchi H, Yang NK, Ding Q, Wang Y, Lai YJ, LaBaff AM, Wu TJ, Lin BR, Yang MH, Hortobagyi GN, Hung MC. Epithelial-mesenchymal transition induced by TNF-alpha requires NF-kappaB-mediated transcriptional upregulation of Twist1. Cancer Res. 2012;72:1290–300.
Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, Zur HA, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95.
Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907.
Tani H, Torimura M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem Biophys Res Commun. 2013;439:547–51.
Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505.
Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y, Dobozy A, Kemeny L, Szell M. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–67.
Szegedi K, Sonkoly E, Nagy N, Nemeth IB, Bata-Csorgo Z, Kemeny L, Dobozy A, Szell M. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19:269–78.
Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8.
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70.
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z, Zhang LW. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20:2285–95.
Acknowledgements
We thank Dr. Shanhui Liang of Cancer Hospital, Fudan University, for his technical assistance.
Funding
This study is sponsored by grants from Shanghai Sailing Program (No. 16YF1401100), National Natural Science Foundation of China (No.81172456), and Natural Science Foundation of Shanghai (No.124119a5502).
Availability of data and material
The data and material supporting the conclusions of this article are included within the article.
Authors’ contributions
Conception and design: JL, HY, XL. Development of methodology: JL, HY, MX, XL. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): JL, XL. Writing, review, and/or revision of the manuscript: JL, XL. Study supervision: XL. All authors read and approved the final manuscript.
Competing interests
The authors declare that there are no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1:
Primers used in present study. (XLS 17 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, J., Yu, H., Xi, M. et al. Long noncoding RNA C17orf91 is a potential prognostic marker and functions as an oncogene in ovarian cancer. J Ovarian Res 9, 49 (2016). https://doi.org/10.1186/s13048-016-0258-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13048-016-0258-3