Abstract
Background
A male individual with a karyotype of 46,XX is very rare. We explored the genetic aetiology of an infertility male with a kayrotype of 46,XX and SRY negative.
Methods
The peripheral blood sample was collected from the patient and subjected to a few genetic testing, including chromosomal karyotyping, azoospermia factor (AZF) deletion, short tandem repeat (STR) analysis for AMELX, AMELY and SRY, fluorescence in situ hybridization (FISH) with specific probes for CSP 18/CSP X/CSP Y/SRY, chromosomal microarray analysis (CMA) for genomic copy number variations(CNVs), whole-genome analysis(WGA) for genomic SNV&InDel mutation, and X chromosome inactivation (XCI) analysis.
Results
The patient had a karyotype of 46,XX. AZF analysis showed that he missed the AZF region (including a, b and c) and SRY gene. STR assay revealed he possessed the AMELX in the X chromosome, but he had no the AMELY and SRY in the Y chromosome. FISH analysis with CSP X/CSP Y/SRY showed only two X centromeric signals, but none Y chromosome and SRY. The above results of the karyotype, FISH and STR analysis did not suggest a Y chromosome chimerism existed in the patient's peripheral blood. The result of the CMA indicated a heterozygous deletion with an approximate size of 867 kb in Xq27.1 (hg19: chrX: 138,612,879–139,480,163 bp), located at 104 kb downstream of SOX3 gene, including F9, CXorf66, MCF2 and ATP11C. WGA also displayed the above deletion fragment but did not present known pathogenic or likely pathogenic SNV&InDel mutation responsible for sex determination and development. XCI assay showed that he had about 75% of the X chromosome inactivated.
Conclusions
Although the pathogenicity of 46,XX male patients with SRY negative remains unclear, SOX3 expression of the acquired function may be associated with partial testis differentiation of these patients. Therefore, the CNVs analysis of the SOX3 gene and its regulatory region should be performed routinely for these patients.
Similar content being viewed by others
Background
46,XX sex reversal is a disease caused by the abnormalities of ovarian development, characterized by 46,XX karyotype and male phenotype, often referred to as 46,XX male [1]. 46,XX male is very rare, and the incidence rate in male newborns is about 1/20,000 [2]. 46,XX male is often classified into 46,XX(SRY+) male or 46,XX(SRY-) male according to the presence or absence of SRY gene in his genome. 46,XX(SRY-) males are only a minority of 46,XX males, as the percentage of 46,XX(SRY +) males caused by SRY translocation and Y chromosome chimerism is more than 90% [3,4,5]. There are some causes of 46,XX(SRY-) males previously reported, such as the dose change or mutation of SOX9 gene [6, 7], loss of function mutation of ovarian stimulating gene WNT4 or RSPO1 gene [8, 9], heterozygous mutation of NR5A1 gene [10, 11]. Furthermore, the arrangement of the SRY-box transcription factor 3 (SOX3) gene was considered as a cause of this disease [12]. We will proceed with the molecular genetic identification of a 46,XX(SRY-) male patient admitted to our clinic. The research results will enrich the theoretical knowledge and guide the clinical treatment of this kind of patient.
Methods
Subject
A 31-years-old patient, heigh 166 cm and weigh 52.5 kg, went to our clinic due to primary infertility. Physical examination showed a male appearance, a thin beard, Adam's apple, two broad bean-size of testicles and an average size of the penis. No sperm was found in three routine semen tests. Hormone test results were follow: Testosterone: 1.75 ng/mL(reference value range(RVR): 2.80–8.00), progesterone: 0.11 ng/mL(RVR: 0.20–1.40), prolactin: 208.44uIU/mL(RVR: 86.00–324.00), estradiol: 5.00 pg/mL(RVR:27.10–52.20), luteinizing hormone: 29.32mIU/mL(RVR: 1.70–8.60), follicle-stimulating hormone 37.88mIU/mL(RVR: 1.50–12.40). The patient had no siblings and denied family history. His parents are not consanguineous, had no abnormal phenotype, and refused the laboratory testing and physical examination.
Specimen preparation and DNA extraction
5 mL of venous blood was collected from patients with heparin sodium and EDTA-Na2 anticoagulant tubes, respectively, and ready for use. According to the manufacturer’s protocols, genomic DNA was extracted from EDTA-Na2 anticoagulated blood using QIAamp DNA Mini Kit (QIAGEN Company). DNA was qualified when the concentration was more than 30 ng/uL, the OD260/280 value between 1.8 to 2.0 determined by ultraviolet spectrophotometer Nanodrop 1C (Thermo Fisher Scientific).
Chromosome karyotype analysis
Lymphocytes of heparin sodium anticoagulated blood were cultured, harvested, and prepared for microscope slides before Giemsa staining according to conventional cell culture methods. Zeiss karyotype analysis system (Karl Zeiss, Germany) was adopted for chromosome count and karyotype analysis, the same as previous studies [13].
Azoospermia factor (AZF) detection
Multiplex amplified was performed with AZF detection kit (Yaneng corp.), then 2.0% agarose electrophoresis and imaging, according to manufacturer’s instructions.
Short tandem repetition(STR) analysis
STR sites were performed by multiplex fluorescence quantitative PCR amplification with Devyser compact v3 kit (Devyser AB, Sweden). The amplification condition was 95 ℃ for 15 min; 94 ℃ 30 s, 58 ℃ 1 min 30 s, 72 ℃ 1 min 30 s, 27 cycles;72 ℃ for 30 min. The amplified products were subjected to capillary electrophoresis with AB 3500Dx gene analyzer, and the electrophoresis data were analyzed by GeneMapper software. The fluorescence peaks of AMELX in Xp22.2, AMELY in Yp11.2 and SRY in Yp11.31 were used to evaluate the patient’s sex chromosomes. The experimental method was referred to in the previous report [14].
Fluorescence in situ hybridization (FISH) analysis
Lymphocytes in EDTA-Na2 anticoagulant blood were isolated by lymphocyte separation solution and hybridized by CSP 18/CSP X/CSP Y probe (Jin Pujia corp.), using the same method as previously reported [15]. Meanwhile, the metaphase cells harvested from lymphocyte culture were co-hybridized with CSP X/CSP Y probe (Jinpuga Company) and SRY probe (Abbott Company). The MIX-1 was prepared by SRY hybridization buffer and SRY probe at a ratio of 9:1, and the MIX-2 was made with CSP hybridization buffer and CSP X/CSP Y centromeric probe at a ratio of 4:1, and then added the MIX-1 and MIX-2 to the metaphase cells loaded on the glass slide. The Glass slide was denaturated at 78 ℃ for 10 min and hybridized at 42 ℃ for more than 16 h. Refer to reagent instruction for the experimental operation. A fluorescence microscope observed the fluorescence signal of hybridization.
Chromosomal microarray analysis (CMA) analysis
500-1000 ng of patient DNA and the equivalent amount of reference DNA was taken for simultaneous experiments. After digestion, the labelled patient sample was mixed with the reference sample and co-hybridized to SurePrint G3 CGH + SNP (180 K) chip. Agilent DNA Microarray Scanner was used to scan the fluorescence signals after the slides were washed. Agilent Feature Extraction Software extracted the data from the images(.tif) and converted it to log-ratios data. Agilent CytoGenomics software was used to analyze CNV. Agilent Technologies provide the reagents, chips, instruments and analytical software. Refer to the instructions for specific methods. CMA analysis mainly adopts some online databases such as OMIM (https://omim.org/), DGV (http://dgv.tcag.ca/dgv/), Decipher (https://decipher.sanger.ac.uk/), ClinGen (https://wwW.clinicalgenome.org/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Whole genome analysis (WGA) analysis
Illumina HiSeq PE150 high-throughput dual-terminal sequencing was performed after random interruption into tiny fragments of DNA, terminal repair, phosphorylation, a-tail addition, connector and library construction. Quality control was carried out on the raw sequencing data to obtain high-quality clean data; Then, fastp software [16] was used for comparative analysis of clean data and human reference genome sequence, and data such as sequencing depth and coverage of the target region were counted and obtained Bam files. Finally, SNP/InDel was detected and annotated based on Bam files to obtain all mutation information. The Haplotyper tool of Sentieo software [17] was used to detect SNP and InDel mutation, and ANNOVAR software [18] was used to annotate the mutation results accompanied by multiple databases (such as dbSNP, 1000G, ESP6500, HGMD, OMIM). Meanwhile, CNVkit software was used to analyze CNV [19].
X chromosome inactivation detection
The sample DNA of undigested and digested by HpaII, which methylation-sensitive restriction enzyme, was amplified by androgen receptor (AR) gene-specific primers and capillary electrophoresis subsequently. B2M was used as the reference gene, and the digestion proceeded overnight in a 37 ℃ water bath. Multiplex PCR amplified the samples before and after enzyme digestion, respectively. As reported in the literature [20], FAM fluorescein was added to the 5 'end of the forward primer [21]. PCR reaction conditions followed: 95 ℃ for 5 min; 28 cycles of 95 ℃ for 45 s, 58 ℃ for 30 s, 72 ℃ for 30 s; 72 ℃ for 7 min. PCR products were subjected to capillary electrophoresis. XCI ratio was calculated according to formula (d1/u1)/(d1/u1 + d2/u2), and XCI bias was confirmed if the XCI ratio > 70% [22, 23].
Results
Result of cytogenetic analysis
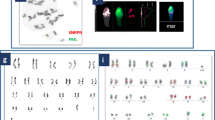
The patient's karyotype was 46,XX, as shown in Fig. 1.
Result of AZF detection
The electrophoresis result of the patient showed neither SRY nor AZF bands (including sY84 and sY86 of AZFa, sY127 and sY134 of AZFb, sY254 and sY255 of AZFc) (The electrophoregram did not show).
Result of STR analysis
The STR result of the patient showed a fluorescence peak of AMELX but not that of the AMELY and SRY, as shown in Fig. 2.
Capillary electrophoresis diagram of STR of the 46,XX (SRY-) male patient. The STR result of the patient showed a fluorescence peak of AMELX but not that of AMELY and SRY. The AMELX, AMELY and SRY represent the loci of Xp22.2, Yp11.2 and Yp11.31, respectively. The solid arrow indicated a fluorescence peak of a specific amplicon, while the hollow arrow indicated no
Result of FISH analysis
FISH result of interphase cells hybridized by CSP 18/CSP X/CSP Y probes showed two 18 and two X fluorescence signals but no Y signal, as shown in Fig. 3a. Another FISH result of metaphase cells hybridized with CSP X/CSP Y/SRY probes showed two green signals of X chromosome centromere, but not that of SRY and Y chromosome, as shown in Fig. 3b.
a FISH image of interphase cells of the patient detected with the CSP 18, CSP X, and CSP Y probes. The blue signal represented chromosome 18, while the green represented chromosome X, as indicated by the arrows. b FISH image of metaphase cells of the patient hybridized with the CSP X, CSP Y and SRY probes. The green represented chromosome X, as indicated by the arrows
Results of Y chromosome chimerism analysis
A single Y chromosome was not found in 200 cells in karyotype analysis. Moreover, Neither AMELY and SRY fluorescence peaks were detected in the STR analysis results, nor Y chromosome and SRY fluorescence signals were shown in the FISH result. A comprehensive evaluation of the above detection results showed no Y chromosome chimerism in the patient's peripheral blood.
Results of CMA analysis
Taking 46,XX female genomes as a standard reference, the patient's Xq27.1 (hg19: chrX: 138,612,879–139,480,163 bp) had about 867 Kb of heterozygotic deletion, and the deletion region was located at 104 Kb downstream of the SOX3 gene, including F9, CXorf66, MCF2, ATP11C four protein-coding genes, as shown in Fig. 4a and b.
a CMA results of the 46,XX (SRY-) male patient. There was about 867 kb heterozygous deletion in Xq27.1 (hg19: chrX: 138,612,879–139,480,163 bp), as the line indicated. b Schematic diagram of deletion region of the 46,XX (SRY-) male patient. The deletion region (hg19: chrX: 138,612,879–139,480,163 bp) is located at 104 kb downstream of the SOX3 gene in Xq27.1. The dotted line indicated the deletion region of 867 kb
Results of WGA analysis
The WGA results showed no pathogenic or likely pathogenic SNV&InDel variant related to sexual development, which can clearly explain the patient phenotype; meanwhile, the results also showed about 892 kb heterozygous deletion (hg19: chrX: 138,609,392–139,501,392) in Xq27.1.
Results of XCI analysis
The XCI ratio of the patient was about 75% (as shown in Fig. 5), which indicated he had a non-random inactivation of the X chromosome.
The X chromosome inactivation results of the 46,XX (SRY-) male patient. The ordinate and abscissa represent fluorescence intensity and fragment length. The figures near the fluorescence peaks indicate the height of fluorescence peaks. The black arrow indicates the amplified products of the reference gene. After complete digestion, there is no amplified products peak (as shown below). The red and green arrows indicate the two alleles of the AR gene in the X chromosomes. X chromosome inactivation ratio was calculated according to the formula (d1/u1)/(d1/u1 + d2/u2)*100%. This patient's X chromosome inactivation ratio was about 75%, which was non-random. d1: the height of the higher peak after enzyme digestion, u1: the height of the undigested peak, which corresponds to d1; d2: the height of the shorter peak after digestion; u2: the height of the undigested peak, which corresponds to d2
Comparison of the clinical phenotypes of 46,XX SRY-negative male patients with CNV of SOX3
Table 1 summarises the clinical features of 46,XX males with SRY-negative individuals involved in the CNV of SOX3. The patients all had the typical male appearance and showed common abnormal phenotypes, including spermatogenous testicular dysplasia, because they were absent from the entire Y chromosome. Among the seven patients, five patients (case 1, 3, 4, 5, 6) had microduplications spanning the entire SOX3 gene, another two patients, including case 2 and our patient, had microdeletions near the SOX3 gene, which were speculated to play a regulatory role for SOX3 expression. Our patient showed a CNV near the SOX3 gene in CMA, and WGA ruled out other SNV&InDel mutations associated with sex determination and development. Meanwhile, he had a skewed X chromosome inactivation, which was not inconsistent with case 1.
Discussion
The SRY gene is recognized as the best TDF candidate gene. As long as the SRY gene exists in the individual's genome, male gonadal development will occur even without the Y chromosome; this is the primary mechanism of 46,XX(SRY +) male pathogenesis. Therefore, we should pay more attention to the Y chromosome chimerism in 46,XX(SRY-) males and rule out as much as possible. The Y chromosome was not found in our patient's peripheral blood derived from the mesoderm through several experimental analyses. However, the possibility of hidden Y chromosome mosaicism in other tissues developed from the endoderm or ectoderm cannot be excluded completely. Of course, it is not easy to find that mosaicism exists in the gonad tissue due to its unavailability.
The rearrangement of the SOX3 gene has confirmed the causes of some 46,XX(SRY-) male individuals in previous reports [12, 24]. SOX3 gene is one of 20 SOX(SRY-related HMG-box) gene family members. Stevanovic et al. cloned the SOX3 gene and identified its location at Xq27.1 in 1993 [25]. Its sequence is highly conserved among different species. The SOX3 gene is ancestral to the SRY gene [12, 26], so it has high homology with SRY and other SOX family genes. SOX3 gene consists of a single exon and contains an HMG box, encoding 446 amino acids of a transcription factor sox3 protein expressed in vertebrate embryos' central nervous system [27]; it plays a vital role in the pituitary, craniofacial and adrenal development. The variation of the SOX3 gene is associated with X-linked mental retardation, growth hormone deficiency, X-linked hypothyroidism, 46,XX male sex reversal, and other diseases [12, 28,29,30,31].
Loss-of-function of SOX3 gene mutation did not cause the abnormality of sex determination in mice and humans [32]. However, studies in transgenic mice had shown that in-situ expression of SOX3 in bipotent gonads resulted in up-regulation of Sox9 expression, testicular induction and XX male sex reversal. Moalem S et al. provided evidence of the duplication of the SOX3 in XX bipotent gonads causing the acquisition of the SOX3 function [33], which was related to partial testis differentiation in XX mice lacking the SRY gene. Moreover, overexpression of SOX3, synergistically expression with SF1, up-regulated SOX9 stimulated gonad development into testis in XX mice [12]. The SRY gene was derived from regulatory region mutation of the SOX3 gene and expressed in the early gonad. The data of transgenic mice indicated that SOX3 and SRY were interchangeable in sex determination function.
The variation types of the SOX3 gene associated with 46,XX males were all the CNV in previous reports. To date, 6 cases of 46,XX males related to the SOX3 CNV have been reported. In some cases, the copy number duplication of the SOX3 gene resulted in changing gene product dose [33,34,35]; among other cases, it was speculated that the CNV had a position effect on the SOX3 gene expression because the CNV did not contain the SOX3 gene but was only close to the region of the SOX3 gene, or the breakpoint of CNV fell in the regulatory region of SOX3 [12, 36]. CMA showed that our patient had a heterozygous deletion of about 867 kb in Xq27.1 (hg19: chrX: 138,612,879–139,480,163 bp), which was located at 104 kb downstream of the SOX3 gene, including F9, CXorf66, MCF2 and ATP11C; Meanwhile, whole-genome sequencing also found an 892 kb heterozygosity deletion in Xq27.1 (hg19: chrX: 138,609,392–139,501,392), and no SNV&InDel mutation associated with abnormal sex determination and development. No similar report was found in the DGV database about this deletion area, and no sexual reversal phenotype was reported in Decipher and ClinVar databases. Similar to previous reports [12, 34], we speculated that the deletion region might involve the regulation region of the SOX3 gene, leading to determination and differentiation of male testis through weakening inhibition of SOX3 and increasing expression SOX3 [12, 24, 35].
Similar to the two adults patients reported by Sutton E et al. [12], the main features of our patient are azoospermia and infertility [37] because of a deletion of the entire Y chromosome. Microspermatocentesis is not recommended for this patient because his testis produces no sperm, and the plan of donated sperm was introduced to him.
Since the deletion of our patient existed in the X chromosome, the XCI factor affecting the clinical significance should be considered. The XCI is a dose-compensation mechanism in XX individuals. It usually occurs in early embryonic development as one of the X chromosomes in a woman is inactivated, with only one paternal or maternal chromosome being expressed in each cell of the female individual. In general, XCI is random, i.e., the inactivation ratio of the two X chromosomes in females is 50%: 50% [38]. In 46,XX (SRY+) males, some studies [21, 39] showed a high degree of XCI bias (greater than 90%). Their phenotype differed with the variable inactivation of the X chromosome carrying the SRY gene [40, 41]. Here, we conducted the XCI analysis of the peripheral blood in 46,XX male(SRY-) patient for the first time. The XCI ratio was about 75%, a moderate XCI bias. We speculate that the expression of positive selection of the deficient X chromosomes, which contained the deleted fragment, results in the development of male gonads of the patient, similar to previous reports [42]. The XCI ratio of gonads tissues may differ from peripheral blood [22]. However, it cannot be accurately known because the sample is inaccessible. We could not know the origin of the Xq27.1 microdeletion in the downstream region of the SOX3 gene of the patient because the relevant results of the patient's parents were not available. We have a tentative assumption that the CNV might regulate the SOX3 gene expression, thus leading to the patient with a karyotype of 46,XX SRY-negative developing into a male. The pathogenicity of CNV has not been confirmed, and this is our upcoming research task to be performed.
At present, the pathogenesis of SRY positive 46,XX male patients is relatively straightforward. However, for SRY negative 46,XX male, the molecular mechanism, signalling pathway, and genetic regulation are remain unknown, and the diagnosis and treatment are still relatively complex. Sutton et al. [12] identified three arrangements containing or adjacent to the SOX3 gene, accounting for 19% (3/16) in 16 SRY-negative 46,XX male patients. Subsequently, several case reports indicated that SOX3 was a critical pathogenic factor of these patients. Therefore, it is crucial to conduct the CNV determination of the SOX3 gene in all 46,XX(SRY-) males. It is noteworthy that the current and reported SOX3 duplications or deletions are below the detection threshold of conventional karyotype and can be found using CMA. Therefore, CMA analysis is routinely recommended to the CNV of SOX3, and high-throughput sequencing such as WGA can simultaneously proceed to exclude other SNV/INDEL mutations. In addition, the XCI analysis of these patients also should be considered.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
References
Lee PA, Houk CP, Ahmed SF, Hughes IA. Consensus statement on management of intersex disorders. Int Consen Conf Intersex Pediatr. 2006;118(2):e488-500. https://doi.org/10.1542/peds.2006-0738.
Delachapelle A, Hortling H, Niemi M, Wennstroem J. XX sex chromosomes in a human male first case. Acta Med Scand. 1964;175(SUPPL 412):25–8. https://doi.org/10.1111/j.0954-6820.1964.tb04630.x.
Andersson M, Page DC, de la Chapelle A. Chromosome Y-specific DNA is transferred to the short arm of X chromosome in human XX males. Science. 1986;233(4765):786–8. https://doi.org/10.1126/science.3738510.
Ahmad A, Siddiqui MA, Goyal A, Wangnoo SK. Is 46XX karyotype always a female. BMJ Case Rep. 2012. https://doi.org/10.1136/bcr-2012-006223.
Inoue H, Nomura M, Yanase T, Ichino I, Goto K, Ikuyama S, et al. A rare case of 46, XX true hermaphroditism with hidden mosaicism with sex-determining region Y chromosome-bearing cells in the gonads. Intern Med. 1998;37(5):467–71. https://doi.org/10.2169/internalmedicine.37.467.
Kim GJ, Sock E, Buchberger A, Just W, Denzer F, Hoepffner W, et al. Copy number variation of two separate regulatory regions upstream of SOX9 causes isolated 46, XY or 46, XX disorder of sex development. J Med Genet. 2015;52(4):240–7. https://doi.org/10.1136/jmedgenet-2014-102864.
Mengen E, Kayhan G, Kocaay P, Uçaktürk SA. A duplication upstream of SOX9 associated with SRY Negative 46, XX ovotesticular disorder of sex development: a case report. J Clin Res Pediatr Endocrinol. 2020;12(3):308–14. https://doi.org/10.4274/jcrpe.galenos.2019.2019.0101.
Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, Valentini S, et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38(11):1304–9. https://doi.org/10.1038/ng1907.
Mandel H, Shemer R, Borochowitz ZU, Okopnik M, Knopf C, Indelman M, et al. SERKAL syndrome: an autosomal-recessive disorder caused by a loss-of-function mutation in WNT4. Am J Hum Genet. 2008;82(1):39–47. https://doi.org/10.1016/j.ajhg.2007.08.005.
Igarashi M, Takasawa K, Hakoda A, Kanno J, Takada S, Miyado M, et al. Identical NR5A1 missense mutations in two unrelated 46, XX individuals with testicular tissues. Hum Mutat. 2017;38(1):39–42. https://doi.org/10.1002/humu.23116.
Bashamboo A, Donohoue PA, Vilain E, Rojo S, Calvel P, Seneviratne SN, et al. A recurrent p.Arg92Trp variant in steroidogenic factor-1 (NR5A1) can act as a molecular switch in human sex development. Hum Mol Genet. 2016;25(16):3446–53. https://doi.org/10.1093/hmg/ddw186.
Sutton E, Hughes J, White S, Sekido R, Tan J, Arboleda V, et al. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest. 2011;121(1):328–41. https://doi.org/10.1172/JCI42580.
Liu Y, Kong XD, Wu QH, Li G, Song L, Sun YP. Karyotype analysis in large-sample infertile couples living in Central China: a study of 14965 couples. J Assist Reprod Genet. 2013;30(4):547–53. https://doi.org/10.1007/s10815-013-9964-6.
Masoudzadeh N, Teimourian S. Comparison of quantitative fluorescent polymerase chain reaction and karyotype analysis for prenatal screening of chromosomal aneuploidies in 270 amniotic fluid samples. J Perinat Med. 2019;47(6):631–6. https://doi.org/10.1515/jpm-2019-0069.
Song Y, Chen Q, Zhang Z, Hou H, Zhang D, Shi Q. Effects of age on segregation of the X and Y chromosomes in cultured lymphocytes from Chinese men. J Genet Genomics. 2009;36(8):467–74. https://doi.org/10.1016/S1673-8527(08)60136-8.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884-884i890. https://doi.org/10.1093/bioinformatics/bty560.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. https://doi.org/10.1093/bioinformatics/btp324.
García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28(20):2678–9. https://doi.org/10.1093/bioinformatics/bts503.
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Ling Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. https://doi.org/10.1101/gr.129684.111.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51(6):1229–39.
Bouayed Abdelmoula N, Portnoi MF, Keskes L, Recan D, Bahloul A, Boudawara T, et al. Skewed X-chromosome inactivation pattern in SRY positive XX maleness: a case report and review of literature. Ann Genet. 2003;46(1):11–8. https://doi.org/10.1016/s0003-3995(03)00011-x.
Sharp A, Robinson D, Jacobs P. Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet. 2000;107(4):343–9. https://doi.org/10.1007/s004390000382.
Minks J, Robinson WP, Brown CJ. A skewed view of X chromosome inactivation. J Clin Invest. 2008;118(1):20–3. https://doi.org/10.1172/JCI34470.
Mizuno K, Kojima Y, Kamisawa H, Moritoki Y, Nishio H, Nakane A, et al. Elucidation of distinctive genomic DNA structures in patients with 46, XX testicular disorders of sex development using genome wide analyses. J Urol. 2014;192(2):535–41. https://doi.org/10.1016/j.juro.2014.02.044.
Stevanović M, Lovell-Badge R, Collignon J, Goodfellow PN. SOX3 is an X-linked gene related to SRY. Hum Mol Genet. 1993;2(12):2013–8. https://doi.org/10.1093/hmg/2.12.2013.
Foster JW, Graves JA. An SRY-related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis-determining gene. Proc Natl Acad Sci USA. 1994;91(5):1927–31. https://doi.org/10.1073/pnas.91.5.1927.
Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, et al. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122(2):509–20.
Lagerström-Fermér M, Sundvall M, Johnsen E, Warne GL, Forrest SM, Zajac JD, et al. X-linked recessive panhypopituitarism associated with a regional duplication in Xq25-q26. Am J Hum Genet. 1997;60(4):910–6.
Laumonnier F, Ronce N, Hamel BC, Thomas P, Lespinasse J, Raynaud M, et al. Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet. 2002;71(6):1450–5. https://doi.org/10.1086/344661.
Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36(3):247–55. https://doi.org/10.1038/ng1309.
Solomon NM, Ross SA, Forrest SM, Thomas PQ, Morgan T, Belsky JL, et al. Array comparative genomic hybridisation analysis of boys with X-linked hypopituitarism identifies 3.9 Mb duplicated critical region at Xq27 containing SOX3. J Med Genet. 2007;44(4):e75. https://doi.org/10.1136/jmg.2007.049049.
Laronda MM, Jameson JL. Sox3 functions in a cell-autonomous manner to regulate spermatogonial differentiation in mice. Endocrinology. 2011;152(4):1606–15. https://doi.org/10.1210/en.2010-1249.
Moalem S, Babul-Hirji R, Stavropolous DJ, Wherrett D, Bägli DJ, Thomas P, et al. XX male sex reversal with genital abnormalities associated with a de novo SOX3 gene duplication. Am J Med Genet A. 2012;158A(7):1759–64. https://doi.org/10.1002/ajmg.a.35390.
Grinspon RP, Nevado J, Mori-Alvarez L, Rey GD, Castera R, Venara M, et al. 46,XX ovotesticular DSD associated with a SOX3 gene duplication in a SRY-negative boy. Clin Endocrinol (Oxf). 2016;85(4):673–5. https://doi.org/10.1111/cen.13126.
Tasic V, Mitrotti A, Riepe FG, Kulle AE, Laban N, Polenakovic M, et al. Duplication of The SOX3 Gene in an Sry-negative 46, XX male with associated congenital anomalies of kidneys and the urinary tract: case report and review of the literature. Balkan J Med Genet. 2019;22(1):81–8. https://doi.org/10.2478/bjmg-2019-0006.
Bowl MR, Nesbit MA, Harding B, Levy E, Jefferson A, Volpi E, et al. An interstitial deletion-insertion involving chromosomes 2p25.3 and Xq27.1, near SOX3, causes X-linked recessive hypoparathyroidism. J Clin Invest. 2005;115(10):2822–31. https://doi.org/10.1172/JCI24156.
Vetro A, Ciccone R, Giorda R, Patricelli MG, Mina ED, Forlino A, et al. XX males SRY negative: a confirmed cause of infertility. J Med Genet. 2011;48(10):710–2. https://doi.org/10.1136/jmedgenet-2011-100036.
Amos-Landgraf JM, Cottle A, Plenge RM, Friez M, Schwartz CE, Longshore J, et al. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet. 2006;79(3):493–9. https://doi.org/10.1086/507565.
Kusz K, Kotecki M, Wojda A, Szarras-Czapnik M, Latos-Bielenska A, Warenik-Szymankiewicz A, et al. Incomplete masculinisation of XX subjects carrying the SRY gene on an inactive X chromosome. J Med Genet. 1999;36(6):452–6.
McElreavey K, Rappaport R, Vilain E, Abbas N, Richaud F, Lortat-Jacob S, et al. A minority of 46, XX true hermaphrodites are positive for the Y-DNA sequence including SRY. Hum Genet. 1992;90(1–2):121–5. https://doi.org/10.1007/BF00210754.
McElreavey K, Vilain E, Abbas N, Herskowitz I, Fellous M. A regulatory cascade hypothesis for mammalian sex determination: SRY represses a negative regulator of male development. Proc Natl Acad Sci USA. 1993;90(8):3368–72. https://doi.org/10.1073/pnas.90.8.3368.
Fechner PY, Rosenberg C, Stetten G, Cargile CB, Pearson PL, Smith KD, et al. Nonrandom inactivation of the Y-bearing X chromosome in a 46, XX individual: evidence for the etiology of 46,XX true hermaphroditism. Cytogenet Cell Genet. 1994;66(1):22–6. https://doi.org/10.1159/000133656.
Acknowledgements
The authors thank all staff of Department of Medical Genetics and Prenatal Diagnosis for their assistance.
Funding
This work was supported by the Science and Technology Innovation Funds (CXZD01-2020) of Sichuan Provincial Maternity and Child Health Care Hospital.
Author information
Authors and Affiliations
Contributions
Shengfang Qin conceived and designed the study, interpreted the data and drafted the manuscript. Xueyang Wang enrolled the patients, performed genetic counseling and provided clinical information. Jin Wang performed the laboratory detection and experimental data acquisition. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The research was approved by the Institutional Committee for the Protection of Human Subjects (Institutional Review Board of Sichuan Provincial Maternity and Child Health Care Hospital, 20201113–98), and the patient signed the informed consent.
Consent for publication
The patient had provided his consent for publication.
Conflict of interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Qin, S., Wang, X. & Wang, J. Identification of an SRY-negative 46,XX infertility male with a heterozygous deletion downstream of SOX3 gene. Mol Cytogenet 15, 2 (2022). https://doi.org/10.1186/s13039-022-00580-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-022-00580-7