Abstract
Background
Noninvasive prenatal testing (NIPT) based on cell-free DNA in maternal circulation has been accepted worldwide by the clinical community since 2011 but limitations, such as maternal malignancy and fetoplacental mosaicism, preclude its full replacement of invasive prenatal diagnosis. We present a novel silicon-based nanostructured microfluidics platform named as “Cell Reveal™” to demonstrate the feasibility of capturing circulating fetal nucleated red blood cells (fnRBC) and extravillous cytotrophoblasts (EVT) for cell-based noninvasive prenatal diagnosis (cbNIPD).
Methods
The “Cell Reveal™” system is a silicon-based, nanostructured microfluidics using immunoaffinity to capture the trophoblasts and the nucleated RBC (nRBC) with specific antibodies. The automated computer analysis software was used to identify the targeted cells through additional immunostaining of the corresponding antigens. The identified cells were retrieved for whole genome amplification for subsequent investigations by micromanipulation in one microchip, and left in situ for subsequent fluorescence in situ hybridization (FISH) in another microchip. When validation, bloods from pregnant women (n = 24) at gestational age 11–13+6 weeks were enrolled. When verification, bloods from pregnant women (n = 5) receiving chorionic villus sampling or amniocentesis at gestation age 11+4–21 weeks with an aneuploid or euploid fetus were enrolled, followed by genetic analyses using FISH, short tandem repeat (STR) analyses, array comparative genomic hybridization, and next generation sequencing, in which the laboratory is blind to the fetal genetic complement.
Results
The numbers of captured targeted cells were 1–44 nRBC/2 ml and 1–32 EVT/2 ml in the validation group. The genetic investigations performed in the verification group confirmed the captured cells to be fetal origin. In every 8 ml of the maternal blood being blindly tested, both fnRBC and EVT were always captured. The numbers of captured fetal cells were 14–22 fnRBC/4 ml and 1–44 EVT/4 ml of maternal blood.
Conclusions
This report is one of the first few to verify the capture of fnRBC in addition to EVT. The scalability of our automated system made us one step closer toward the goal of in vitro diagnostics.
Similar content being viewed by others
Background
Noninvasive prenatal testing (NIPT) that uses cell-free DNA (cfDNA) in maternal circulation for fetal aneuploidy detection had already achieved widespread recognition and adoption by the clinician community worldwide since 2011 [1, 2]. On the other hand, the progress of cell-based noninvasive prenatal diagnosis (cbNIPD) is relatively not so promising or stagnant until very recently [3,4,5,6,7,8]. Scarcity of fetal cells in the maternal circulation poses a great hurdle to the progress of cbNIPD when compared with much more robust cfDNA-based NIPT. The cfDNA-based testing was conducted through the robust maximal parallel sequencing methods by utility of the high-sensitive, high-throughput, rapid-evolving platforms called next generation sequencing (NGS) technologies which can discriminate the trivial differences between the maternal blood who carry the euploid fetuses and those who carry the aneuploidy fetuses. The NIPT was successfully validated for common fetal chromosomal numerical disorders such as trisomy 13, 18, and 21 [1]. Recently some service providers claimed the repertoire of NIPT can be expanded to all autosomes, and even microdeletion syndromes [9], which is controversial. Most published statements, consensus, or recommendation from the professional societies now consider using NIPT to detect fetal microdeletion syndromes is not recommended [2, 10]. However, cfDNA-based screening heavily relied upon bioinformatics protected by intellectual property which is less easily accessible and thus mainly dominated by the commercial service providers, and had revolutionarily changed the landscape of prenatal diagnosis [2, 11]. Meanwhile, cfDNA-based tests need innovative algorithms to analyze the NGS data, and it is now well known that origins of the cfDNA, in addition to those from maternal, are from the placenta (trophoblasts) instead of from the fetus proper, indicating that fetoplacental mosaicism (namely, the chromosome complements of the fetus and the placenta are different), is an unarguable source of false-negatives and false-positives with the current NIPT [12, 13].
Since 2014, we have developed our in-house patent protected algorithms for cfDNA NIPT (called GWNS™) and the resolution, in some cases, can be even enhanced to a 3.21 Mb microduplication by simply using 20 M reads shallow-sequencing with 12.5% of fetal DNA fraction [14,15,16]. However, we also noticed the problem of fetoplacental mosaicism [12, 15, 17] and thus re-focused back our effort to cbNIPD since 2015. We believed with the utility of cbNIPD the issue of fetoplacental mosaicism in noninvasive prenatal diagnosis can be better tackled, if we can capture both cells from the fetal and placental origins, namely, the fetal nucleated red blood cells (fnRBC) and the extravillous cytotrophoblasts (EVT).
The major difficulty of cbNIPD is the extreme scarcity of fetal cells. It is estimated, by the best recent reports, there were only 1–45 cells per 30 ml maternal blood. And most of the previous efforts captured EVT only, instead of fnRBC, which can truly represent the fetal genome [5]. How to find a feasible method to capture and enrich the fetal cells from the maternal blood become the major challenges of cbNIPD [5, 8]. However, the same technology, if being developed, can be used to capture not only the circulating fetal cells (CFC) but also other cells, including the circulating tumor cells (CTC), and the technologies devised for CTC are numerous: including PCR-based, flow cytometry, laser scanning cytometry, FDA-cleared Cell Search (Veridex, New Jersey, USA), EPISPOT assay [18], and microchip (microfluidics/lab-on-a-chip)-based technologies [19]. Among microchip technologies, the methods had been used to isolate single cells included immune-affinity, immune-magnetic, and size-based methodologies [20]. We selected microchip-based method by using immune-affinity approach which utilizes a microfluidic device (called PicoBioChip, the manufacturing flowchart of the chip was shown in Fig. 1) coated with antibodies which can capture the corresponding antigens on the targeted cells, due to the strength of the semiconductor industry in Taiwan, to devise our novel automated platform Cell Reveal™ to explore the feasibility of cbNIPD to capture CFC (including fnRBC and EVT), firstly by using a group of pregnant women at GA 11–13+6 weeks to demonstrate the capability of capturing nucleated RBC (nRBC) and EVT, followed by another group of pregnant woman to verify the captured cells are indeed fetal origin by subsequent genetic investigations, such as fluorescence in situ hybridization (FISH), array comparative genomic hybridization (aCGH), and NGS. The results of cbNIPD are compared with paralleled cfDNA NIPT, and confirmed with invasive procedures using karyotyping, aCGH, and if necessary, NGS. The aim of this small pilot series of proof-of-principle study is to demonstrate the feasibility of our platform with a performance comparable or even superior to the capture efficiencies of other designs previously reported in the literature (which ranged from <1 to 3–5 fetal cells per ml of maternal blood) [4, 6, 8]. Meanwhile, the need of manual handling is reduced to a minimum in our automated system.
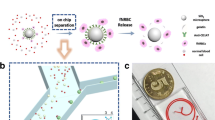
a Flowchart of the PicoBioChip manufacture: 1. standard cleaning, 2. photolithography, 3. Ag deposition, 4. etching, 5. Ag and photoresist removal, and 6.surface modification. b The porous morphology on the PicoBioChip with a “nano-on-nano” structure. c Conceptual illustration of how an PicoBioChip can be employed to achieve significantly enhanced capture of targeted cell
Results
Circulating fnRBC and EVT captured by PicoBioChip
The scanning electron microscope (SEM) micrograph of the PicoBioChip is shown in Fig. 2a. The micrograph demonstrated that the PicoBioChip surface morphology is composed of patterned nanostructure with the same dimension and space, so that circulating cells could be captured by the chip surface (Fig. 2b). The processes of cells capture were automatically performed on a Cell Reveal™ system. By using a fluorescence microscope, the nRBC and EVT can be unequivocally distinguished from the background packed with the WBC from the maternal origin (Fig. 3).
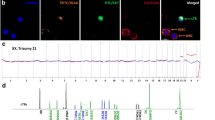
Discrimination of a-e fnRBC and f-j EVT from maternal white blood cells (WBC) by fluorescence microscope. The fnRBC and EVT can be recognized by different antibodies labeled with TRITC (GPA and HLA-G) or FITC (CD71 and CK7). The maternal leukocytes can be recognized by antibody labeled with CY5 (CD45)
Capture efficiency
In every 4 ml of the maternal blood used for validation, circulating nRBC and EVT were always captured in all the 24 pregnant women with capture efficiencies as 1–44 nRBC/2 ml and 1-32EVT/ 2 ml (Additional file 1: Table S1). In every 8 ml of the maternal blood used for verification, circulating fnRBC (please refer to the below sections that the nRBCs we captured are indeed fetal origin) and EVT were always captured for all five pregnant women examined (Table 1). A total of 150 fetal cells (fnRBC + EVT) were successfully captured. The numbers of captured fetal cells were: 14–22 cells per 4 ml of maternal blood for fnRBC and 1–44 cells per 4 ml of maternal blood for EVT. The overall capture efficiency of the novel system is estimated as 2.38–7.25 fetal cells (fnRBC + EVT) per ml of maternal blood per individual (Table 2).
Fluorescence in situ hybridization (FISH)
Interphase FISH for the captured fetal cells from the blood of pregnant women with a fetus of trisomy 13, trisomy 18, or trisomy 21 revealed correct diagnoses in all cases. The number of fnRBC and EVT examined ranged from one to ten for each case (Table 1). FISH for the trisomy 13 revealed nuc ish(RB1/D13S1195/D13S1155/D13S915x3, D21S270/D21S1867/D21S337/D21S1425/D21S1444/D21S341x2), for the trisomy 18 revealed nuc ish(D18Z1x3,DXZ1x2), and for the trisomy 21 revealed nuc ish(RB1/D13S1195/D13S1155/D13S915x2,D21S270/D21S1867/D21S337/D21S1425/D21S1444/D21S341x3) (Fig. 4).
Fluorescent in situ hybridization (FISH) for the captured fnRBC from 3 pregnant women with an aneuploid fetus of a trisomy 13, b trisomy 18, and c trisomy 21. In a and c, chromosome 13 was identified by a panel of probes (RB1, D13S1195, D13S1155, D13S915) in green and chromosome 21 was identified by a panel of probes (D21S270, D21S1867, D21S337, D21S1425, D21S1444, D21S341) in orange. In b, chromosome 18 was identified by a probe (D18Z1) in aqua and chromosome X was identified by a probe (DXZ1) in green
Whole genome amplification (WGA)
All pooled captured cells underwent WGA successfully except those the total numbers of cells were too few (namely, less than 4 cells) to reach the amplified threshold for subsequent molecular genetic analyses by short tandem repeat (STR) analysis, aCGH, and NGS. Overall, fnRBC WGA from all the five cases and EVT WGA from two cases were obtained (Table 1). The WGA products were 50 μl in total with a concentration ranged from 290 to 844 ng/μl.
Short tandem repeat (STR) analysis
STR analyses were performed for the WGA DNA from captured fetal cells and maternal leukocytes as well as the DNA from the abortus tissue (if available). The results demonstrated the captured fnRBC and/or EVT are indeed fetal origin in all the five cases examined. For each case, there are 4–8 informative STR makers containing non-maternal alleles that are feasible to distinguish the fetal cells from the maternal cells (Table 3).
Array comparative genomic hybridization (aCGH) and next generation sequencing (NGS)
Both of aCGH and NGS analyses were performed for the captured fnRBC from the five cases and all the cases were correctly diagnosed. The results of aCGH are comparable with that of NGS (Fig. 5), and are consistent with the karyotyping results.
Array comparative genomic hybridization (aCGH) and next generation sequencing (NGS) for the same whole genome amplification (WGA) products of captured fnRBC from 5 pregnant women. WGA product from Promega male DNA (Promega, Wisconsin, USA) was used as reference. The aCGH were performed with GenetiSure Pre-Screen Array Kit 8x60K (Agilent Technologies, CA, USA) on a G4900DA SureScan microarray scanner (Agilent Technologies). NGS was performed using Ion PGM Hi-Q Sequencing Kit with Ion 316 chip (Thermo Fisher Scientific, California, USA) on the Ion Torrent PGM Instrument (Thermo Fisher Scientific) platform. Aneuploidy chromosomes are indicated by arrows. Aneuploidy chromosomes are indicated by arrows. The results of aCGH are comparable with that of NGS
Discussion
The quest to search for a true noninvasive prenatal diagnosis had been the Holy Grail of prenatal diagnosis since 1969 [5]. The major hurdle is the scarcity of fetal cells in maternal circulation, and therefore contributed to the soaring cost of the technologies involved in the enrichment and isolation. Most previous reports found that the majority of nucleated red blood cells isolated from the maternal blood are actually maternal origin instead of fetal [5] whereas a recent published study did isolate fnRBC, confirmed by using chromosome Y-specific FISH [21]. Another group explored the monoclonal antibodies specific for fnRBC in addition to those specific for nucleated RBCs (including both fetal nucleated RBC and nucleated RBC from adult bone marrow origin) such as CD36, CD71, glycophorin-A, antigen-i, and galactose [22]. Unfortunately, it is not commercially available for such monoclonal antibodies claimed to be fetal specific. It is evident that our Cell Reveal™ system with PicoBioChip can capture both the fnRBC and EVT with capture efficiencies superior to or at least comparable to the previous reports (19–58 cells/8 ml v.s. 1–45 cells/30 ml maternal blood), especially all the NRBCs being captured in the verification group by our system are fnRBC. Therefore the feasibility of our platform to tackle the primary hurdle, the scarcity of fetal cells and the difficulty to successfully enrich and capture the very few fetal cells from a limited amount of maternal blood (i.e. 8 ml), is demonstrated. The importance to capture not only the trophoblasts (which is considered placental origin), but also the fnRBC is that the fetoplacental mosaicism can be overcome, otherwise cbNIPD adds not much additional value when being compared with the extremely successful cfDNA-based NIPT, since only fnRBC can genuinely represent the fetal origin instead of placenta origin, which NIPT has already managed [1, 23]. The nucleated red blood cells isolated through our platform are confirmed to be fetal origin by the subsequent analyses including STR analysis, FISH, aCGH, and NGS. Meanwhile, cbNIPD may clearly delineate which co-twin is affected (or much more uncommon, both co-twins are affected) by aneuploidy when the result of cfDNA-based NIPT showed high risk for aneuploidy in twin pregnancies.
In this proof-of-principle pilot study, we only demonstrated the feasibility of our Cell Reveal™ platform to detect fetal aneuploidy by using common trisomies (trisomy 13, 18, and 21). We admit it will be more persuasive if we included some genuine fetoplacental mosaicism cases as some of the cases we published before, and this will be included in our future studies [15, 17, 24]. Some researchers proposed that cbNIPD on EVT can be used to detect de novo copy number variations which can only be reliably diagnosed by microarray, since at the moment the claims by some commercial service providers to expand the repertoire of cfDNA-based NIPT to include microdeletions/microduplications are not widely endorsed by the academic community [5, 10]. However, the errors that may be introduced by the indispensable WGA procedure had also been noticed recently from the experience obtained from the studies regarding preimplantation genetic screening. More and more reports were published since 2015 that euploid babies were born after transferring the aneuploid embryos into the womb [25] and the consistency of PGS across the laboratories adopting different genotyping technologies is questioned [26,27,28,29]. Thus, we considered aCGH and NGS can be used but should be interpreted with caution because WGA is the necessary step before using these technologies. A better capture efficiency to capture more cells and group them together for the subsequent analyses will be very helpful to minimize the errors introduced by WGA. In our laboratory, the previous experience on PGD/PGS made us only did WGA on at least four cells, therefore we did not do WGA on the EVT captured on Case no. 1, 4, 5 whereas we did WGA on the nRBC we captured on all the five cases in the verification group (Table 1). Remarkably, most previous reports in the literature regarding cbNIPD using different methods such as immunoaffinity by magnetic enrichment [8] or NanoVelcro microchip [30], or isolation by size of tumor/trophoblast cells (ISET) [31] only successfully isolated trophoblasts instead of fnRBC. However, it is remarkable that ISET was reported to be able to isolate living cells [32].
On the other hand, it is now better known the trend of the variation of the fetal DNA fractions during the whole gestation, as well as the possible confounding factors, since the fetal fraction is one of the major factors affecting the accuracy of NIPT [10, 33]. Needless to say, such similar large-scale studies are mandatory for cbNIPD in order to better understand the variation as well as the confounding factors affecting the numbers of both types of the fetal cells (fnRBC and EVT), to facilitate its wider acceptance of clinical utility. The future studies will be ideal if including some fetoplacental mosaicism cases as well as twin pregnancies with one or two aneuploidy cases to better demonstrate its feasibility to supplement the current cfDNA-based NIPT. Lastly, another critical issue affecting the uptake of cbNIPD by the clinical community in the future is the cost. A detailed cost-effective analysis is needed in the future for cbNIPD as it has been done in NIPT for fetal aneuploidy [34]. Nevertheless, this platform has the potential to be used for capturing the circulating tumor cells (CTC) as well [32]. It is arguably that the nRBC captured in the validation group (n = 24) may both include the fetal and the maternal nRBC (these cells are released from the adult bone marrow) since we did not use the fetal specific monoclonal antibodies such as those recognize the Epsilon hemoglobin [35]. However, the subsequent genetic investigations we performed (including FISH, aCGH, STR, and NGS) had verified the captured cells in the verification group (n = 5) are indeed fetal origin.
To the best of our knowledge, this report is one of the very few studies on the successful use of circulating fetal cells for noninvasive prenatal diagnosis. The strength of our study is that all the processes of cell capture are automatic which can be performed on a single individual case and completed within 15 h (Additional file 2: Table S2). The captured cells are available for a variety of genetic testing, such as FISH, aCGH and NGS. Overall, the turnaround time of the cbNIPD is less than 2 weeks, similar to that of NIPT as performed in our laboratory.
Conclusion
We demonstrated our silicon-based nanostructured microfluidics “The Cell Reveal™ system” can capture both the fnRBC and EVT. The scalability is greatly enhanced with the automation of the whole system, which may render cbNIPD from mainly a laboratory-developed-test (LDT) conducted only in a limited number of core laboratories, into an in-vitro-diagnostics (IVD) that can be applied in many research and clinical sites.
Methods
Samples
During 2016–2017, 24 women who carried the singleton pregnancy and received the first trimester serum screening for Down syndrome at GA 11–13 + 6 weeks or who decided to receive NIPT, was asked to donate blood sample to be used for validation. For each individual, 4 ml additional blood was stored in the BD vacutainer® with ACD solution A (Becton, Dickinson and Company, New Jersey, USA) for cbNIPD. When verification, at 2017 another five pregnant women carrying the singleton pregnancy at first or second trimester who decided to receive invasive procedures (chorionic villus sampling or amniocentesis) were recruited as a research basis to receive paralleled cfDNA testing (i.e. NIPT) and cbNIPD after informed consents (with the approved protocol CCH-IRB-141219) were signed. For each individual, approximately 20 ml of venous blood were collected. The blood was taken and stored in the Streck Cell-Free DNA BCT® (Streck, Nebraska, USA) for NIPT (12 ml) and in the BD vacutainer® with ACD solution A (Becton, Dickinson and Company, New Jersey, USA) for cbNIPD (8 ml). A total of 3 pregnant women who had singleton pregnancy affected with fetal aneuploidy were recruited, including trisomy 13 (n = 1), trisomy 18 (n = 1), and trisomy 21 (n = 1) fetuses respectively. Meanwhile, two women carrying the euploid fetuses (46,XX, n = 1 and 46,XY, n = 1) were also enrolled. The pregnant women were enrolled at first or second trimester ranged from gestational age 11+4 to 21 weeks (Table 1). It is noteworthy that the examiners of the cbNIPD lab have no prior knowledge of the karyotyping results, namely, they were blind to the results to avoid ascertainment bias. The recruitment of patients, collection of samples, and conduct of research projects, were approved by the Ethical Commitees of the medical institutions where the samples were collected (the Taiwan Adventist Hospital, Taipei, Taiwan, and the Changhua Christian Hospital, Changhua, Taiwan).
PicoBioChip manufacture
The PicoBioChip is a Si nanostructure with a porous morphology that is fabricated using the metal-assisted chemical etching (MACE) technology. The fabrication sequence is described as followed: the starting materials are p-type (100) silicon wafers which followed standard cleaning procedures to remove environmental contaminants. The pattern of the PicoBioChip is defined by standard photolithographic techniques. The Ag film is deposited onto the silicon wafer in a HF/AgNO3 mixture solution, and the wafers are etched in a HF/H2O2mixture solution. Then, after the etching step and the Ag film removal, a Si nanostructure with a porous morphology is formed that is a “nano-on-nano” structure. To enhance the capturing effect, the potential targeted cells are pre-labeled with biotinylated antibodies and the PicoBioChip surface is made from a streptavidin material which has a specific binding interaction with biotin. The streptavidin-biotin is the strongest non-covalent biological interaction currently known. Via streptavidin-biotin interaction, biotinylated antibodies can be conjugated, enabling a high efficiency for targeted cells capture. The manufacturing flowchart, nano-on-nano structure and capture conception of the PicoBioChip are shown in Fig. 1.
Circulating fetal cells captured by cell reveal™ system with PicoBioChip
The whole blood sample (8 ml) is flown through the automated Cell Reveal™ system and then CFC are captured by PicoBioChips. For each run of test, four PicoBioChips were used: two for fnRBC capture and two for EVT capture. The antibodies used for primary capture of circulating fetal cells are CD71+ for fnRBC and EpCAM+ for EVT. PicoBioChips are examined using a fluorescence microscope equipped with a built-in automatic inspection and image analysis system, called the Cell Analysis Tool (CytoAurora CAT™), to filter out images of maternal white blood cells (WBC) for further analyses. The fnRBC and EVT can therefore be targeted, identified and enumerated. Image analyses with the count-in/filter-out criteria for different cell types are CD71(+)/GPA+(glycophorin-A)/CD45−/DAPI+ for fnRBC and CK7+(Cytokeratin-7)/HLA-G(+)/CD45−/DAPI+ for EVT, according to literatures and our in-house optimization [6, 21, 22, 35,36,37,38,39,40]. Namely, we first used one antibody to capture fnRBC and EVT separately, and then using other antibodies to stain the captured cells. Hence, the fnRBC were primarily captured by CD71 and then stained with CD71 and GPA, whereas the EVT were primarily captured by EpCAM and then stained with CK7 and HLA-G. Namely, we utilized dual antibodies (CD71 and GPA) to delineate the fnRBC and triple antibodies (EpCAM, CK7, HLA-G) to delineate EVT. It is noteworthy that in the validation group (n = 24) to validate the capture efficiency, only 4 ml maternal blood was used, in which 2 ml was for fnRBC (or more strictly, nRBC) and 2 ml was for EVT in each case. Only the five pregnant women enrolled for verification had 8 ml maternal blood to be withdrawn and used for cbNIPD.
Fluorescence in situ hybridization (FISH)
FISH was performed directly on one PicoBioChip capturing for fnRBC and one chip for EVT. Prior to hybridization, the formaldehyde on PicoBioChips were treated by 10 mM sodium citrate at 90 °C for 20 min, followed by being immersed in 0.1% Triton-X at room temperature for 10 min, then followed by serial washes of 0.2 N HCl at 25 °C for 20 min, purified water (double distilled) at 25 °C for 3 min and 2X SSC at 25 °C for 3 min, and an immersion of Vysis pretreatment solution (1 N NaSCN) (Abbott, IL, USA) at 25 °C overnight. Then, the PicoBioChips were deposited in purified water at 25 °C for 1 min, 2X SSC at 25 °C for 5 min (repeated two times), pepsin solution (10 μl 10% Pepsin / 40 ml 0.01 N HCl) at 37 °C for 3 min and 2X SSC at 25 °C for 5 min (repeated two times). Finally, the PicoBioChips were immersed in 70% ethanol at 4 °C for 1 min, 85% ethanol at 4 °C for 1 min and 100% ethanol at 4 °C for 1 min, and dried at 50 °C for 5 min. Interphase FISH for chromosome 13, 18 and 21 on captured fnRBC and EVT was then conducted using Aquarius® FAST FISH Prenatal kit (Cytocell, Cambridge, UK). For hybridization experiment, the PicoBioChips were dehydrated in an ethanol series and hybridized overnight in a moist chamber at 37 °C. The chips were washed for 2 min in 0.4X SSC at 70 °C and for 5 min in 4X SSC, 0.1% Tween 20 at room temperature and blocked in 4X SSC, 3% bovine serum albumin (BSA), 0.1% Tween 20 at 37 °C for 30 min. The hybridization signal was detected with Nikon-Ni-E microscope system (Nikon, Tokyo, Japan). Chromosomes were counterstained with 0.125 μg/ml DAPI in Antifade (Vysis, Illinois, USA). FISH analyses were performed using the Aquarius® FAST FISH Prenatal kit (Cytocell). The chromosome 13 probe for RB1, D13S1195, D13S1155, and D13S915, the chromosome 18 probe for centromere of chromosome 18(D18Z1), the chromosome 21 probe for D21S270, D21S1867, D21S337, D21S1425, D21S1444, and D21S341, and the chromosome X probe for centromere of chromosome X (DXZ1) were labeled with green, aqua, orange, and green fluorophores, respectively.
Retrieval of captured cells by PicoBioChip
The captured fnRBC and EVT are separately released by with capillary micropipette from PicoBioChips which are destined for DNA analyses. The location of captured cells-on-chip is acquired by the CytoAurora CAT™. Capillary micropipette crashes the chip’s nano structure of the target captured cells. The captured cells on the chip surface are followed by capillary micropipette picking up, which allows captured cells to escape from chip to be released for sequential analyses.
Whole genome amplification (WGA)
The captured fetal cells retrieved from the same PicoBioChip are pooled. The fnRBC and EVT were subjected separately to WGA, with 1.8 μg/μl BSA serving as the blocking agent to reduce the surface interaction from the silicon debris. WGA was performed using REPLI-g Single Cell Kit (Qiagen, Hilden, Germany) and following the manufacturer’s instructions. Amplified DNA was purified using the QIAamp DNA Blood Mini Kit (Qiagen). The DNA purities and concentrations were examined by Qubit fluorometer (Thermo Fisher Scientific, Delaware, USA) and Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific).
Short tandem repeat (STR) analysis
STR analysis was performed to confirm that the circulating cells captured and WGA DNA of fnRBC and EVT are indeed from fetuses instead of maternal origin. GenomeLab Human STR Primer Set kit (Beckman Coulter, California, USA) containing 12 primer pairs to amplify 11 STR loci and one gender-specific locus (Table 2) was used to analyze patterns of the STR by capillary electrophoresis according with the supplier’s protocol. PCR products were run on GenomeLab™ GeXP Genetic Analysis System (Beckman Coulter). FRAGMENTS application program (Beckman Coulter) was used for data collection and allele sizing.
Array comparative genomic hybridization (aCGH)
Approximately 1000 ng of WGA DNA was subjected to aCGH by GenetiSure Pre-Screen Array Kit 8x60K (Agilent Technologies, CA, USA), following the manufacturer’s instructions. The image on a chip was acquired with a G4900DA SureScan microarray scanner (Agilent Technologies, CA, USA) and analyzed with Agilent CytoGenomics software (Agilent Technologies) for chromosome gain or loss. Aberrations were detected by using default setting.
Next generation sequencing (NGS)
Approximately 1000 ng of WGA DNA was used for library construction using Ion Xpress Plus gDNA Fragment Library Preparation Kit Set (Thermo Fisher Scientific, California, USA) and following the manufacturer’s instructions. The quantity of library was determined using Qubit dsDNA HS assay kits (Thermo Fisher Scientific) with Qubit fluorometer (Thermo Fisher Scientific). The template-positive Ion Sphere Particles were generated using Ion PGM Hi-Q Template Kits (Thermo Fisher Scientific) with the Ion OneTouch 2 Instrument (Thermo Fisher Scientific) and then enriched with the Ion OneTouch ES Instrument (Thermo Fisher Scientific). Sequencing was performed on the Ion Torrent PGM Instrument (Thermo Fisher Scientific) platform using the Ion PGM Hi-Q Sequencing Kit and Ion 316 chip (Thermo Fisher Scientific). Analysis of the WGA product being sequenced was performed by using the cloud-based the Ion Reporter™ Server System (https://ionreporter.thermofisher.com/ir/).
Abbreviations
- aCGH:
-
Array comparative genomic hybridization
- BSA:
-
Bovine serum albumin
- cbNIPD:
-
Cell-based noninvasive prenatal diagnosis
- CFC:
-
Circulating fetal cells
- cfDNA:
-
Cell-free DNA
- CTC:
-
Circulating tumor cells
- EVT:
-
Extravillous cytotrophoblasts
- FISH:
-
Fluorescence in situ hybridization
- fnRBC:
-
Fetal nRBC
- GWNS:
-
Genome wide normalized score
- MACE:
-
Metal-assisted chemical etching
- NGS:
-
Next generation sequencing
- NIPT:
-
Noninvasive prenatal testing
- nRBC:
-
Nucleated red blood cells
- SEM:
-
Scanning electron microscope
- STR:
-
Short tandem repeat
- WBC:
-
White blood cells
- WGA:
-
Whole genome amplification
References
Chitty LS, Lo YM. Noninvasive prenatal screening for genetic diseases using massively parallel sequencing of maternal plasma DNA. Cold Spring Harb Perspect Med. 2015;5:a023085.
Society for Maternal-Fetal Medicine, (SMFM) with the assistance of Norton ME, Biggio JR, Kuller JA, Blackwell SC. The role of ultrasound in women who undergo cell-free DNA screening. Am J Obstet Gynecol. 2017;216:B2–7.
Hatt L, Brinch M, Singh R, Møller K, Lauridsen RH, Schlütter JM, et al. A new marker set that identifies fetal cells in maternal circulation with high specificity. Prenat Diagn. 2014;34(11):1066–72.
Schlütter JM, Kirkegaard I, Petersen OB, Larsen N, Christensen B, Hougaard DM, et al. Fetal gender and several cytokines are associated with the number of fetal cells in maternal blood--an observational study. PLoS One. 2014;9(9):e106934.
Beaudet AL. Using fetal cells for prenatal diagnosis: history and recent progress. Am J Med Genet C Semin Med Genet. 2016;172(2):123–7.
Breman AM, Chow JC, U'Ren L, Normand EA, Qdaisat S, Zhao L, et al. Evidence for feasibility of fetal trophoblastic cell-based noninvasive prenatal testing. Prenat Diagn. 2016;36(11):1009–19.
Calabrese G, Fantasia D, Alfonsi M, Morizio E, Celentano C, Guanciali Franchi P, et al. Aneuploidy screening using circulating fetal cells in maternal blood by dual-probe FISH protocol: a prospective feasibility study on a series of 172 pregnant women. Mol Genet Genomic Med. 2016;4(6):634–40.
Kølvraa S, Singh R, Normand EA, Qdaisat S, van den Veyver IB, Jackson L, et al. Genome-wide copy number analysis on DNA from fetal cells isolated from the blood of pregnant women. Prenat Diagn. 2016;36(12):1127–34.
Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 2015;212(3):332. e1-9
Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the AmericanCollege of medical genetics and genomics. Genet Med. 2016;18(10):1056–65.
Agarwal A, Sayres LC, Cho MK, Cook-Deegan R, Chandrasekharan S. Commercial landscape of noninvasive prenatal testing in the United States. Prenat Diagn. 2013;33(6):521–31.
Grati FR, Malvestiti F, Ferreira JC, Bajaj K, Gaetani E, Agrati C, et al. Fetoplacental mosaicism: potential implications for false-positive and false-negative noninvasive prenatal screening results. Genet Med. 2014;16(8):620–4.
Meck JM, Kramer Dugan E, Matyakhina L, Aviram A, Trunca C, Pineda-Alvarez D, et al. Noninvasive prenatal screening for aneuploidy: positive predictive values based on cytogenetic findings. Am J Obstet Gynecol. 2015;213(2):214. e1-5
Yeang CH, Ma GC, Hsu HW, Lin YS, Chang SM, Cheng PJ, et al. Genome-wide normalized score: a novel algorithm to detect fetal trisomy 21 during non-invasive prenatal testing. Ultrasound Obstet Gynecol. 2014;44(1):25–30.
Cheng HH, Ma GC, Tsai CC, WJ W, Lan KC, Hsu TY, et al. Confined placental mosaicism of double trisomies 9 and 21: discrepancy between non-invasive prenatal testing, chorionic villus sampling and postnatal confirmation. Ultrasound Obstet Gynecol. 2016;48(2):251–3.
WJ W, Ma GC, Lin YS, Yeang CH, Ni YH, Li WC, et al. Detection of 22q11.2 microduplication by cell-free DNA screening and chromosomal microarray in fetus with multiple anomalies. Ultrasound Obstet Gynecol. 2016;48(4):530–2.
WJ W, Ma GC, Lee MH, Chen YC, Chen M. Normal prenatal ultrasound findings reflect outcome in case of trisomy 14 confined placental mosaicism developing after preimplantation genetic diagnosis. Ultrasound Obstet Gynecol. 2017;50:128–30.
Ramirez JM, Fehm T, Orsini M, Cayrefourcq L, Maudelonde T, Pantel K, et al. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin Chem. 2014;60(1):214–21.
Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P. Technologies for single-cell isolation. Int J Mol Sci. 2015;16(8):16897–919.
Sahmani M, Vatanmakanian M, Goudarzi M, Mobarra N, Azad M. Microchips and their significance in isolation of circulating tumor cells and monitoring of cancers. Asian Pac J Cancer Prev. 2016;17(3):879–94.
Byeon Y, Ki CS, Han KH. Isolation of nucleated red blood cells in maternal blood for non-invasive prenatal diagnosis. Biomed Microdevices. 2015;17(6):118.
Zimmermann S, Hollmann C, Stachelhaus SA. Unique monoclonal antibodies specifically bind surface structures on human fetal erythroid blood cells. Exp Cell Res. 2013;319(17):2700–7.
Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372(17):1589–97.
Chen M, Yeh GP, Shih JC, Wang BT. Trisomy 13 mosaicism: study of serial cytogenetic changes in a case from early pregnancy to infancy. Prenat Diagn. 2004;24(2):137–43.
Greco E, Minasi MG, Fiorentino F. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med. 2015;373(21):2089–90.
Gleicher N, Vidali A, Braverman J, Kushnir VA, Barad DH, Hudson C, et al. International PGS consortium study group. Accuracy of preimplantation genetic screening (PGS) is compromised by degree of mosaicism of human embryos. Reprod Biol Endocrinol. 2016;14(1):54.
Orvieto R. Preimplantation genetic screening- the required RCT that has not yet been carried out. Reprod Biol Endocrinol. 2016;14(1):35.
Orvieto R, Gleicher N. Should preimplantation genetic screening (PGS) be implemented to routine IVF practice? J Assist Reprod Genet. 2016;33(11):1445–8.
Tortoriello DV, Dayal M, Beyhan Z, Yakut T, Keskintepe L. Reanalysis of human blastocysts with different molecular genetic screening platforms reveals significant discordance in ploidy status. J Assist Reprod Genet. 2016;33(11):1467–71.
Hou S, Chen JF, Song M, Zhu Y, Jan YJ, Chen SH, et al. Imprinted NanoVelcro microchips for isolation and characterization of circulating fetal trophoblasts: toward noninvasive prenatal diagnostics. ACS Nano. 2017; https://doi.org/10.1021/acsnano.7b03073.
Mouawia H, Saker A, Jais JP, Benachi A, Bussières L, Lacour B, et al. Circulating trophoblast cells provide genetic diagnosis in 63 fetuses at risk for cystic fibrosis or spinal muscular atrophy. Reprod BioMed Online. 2012;25(5):508–20.
Laget S, Broncy L, Hormigos K, Dhingra DM, BenMohamed F, Capiod T, et al. Technical insights into highly sensitive isolation and molecular characterization of fixed and live circulating tumor cells for early detection of tumor invasion. PLoS One. 2017;12(1):e0169427.
Ma GC, WJ W, Lee MH, Lin YS, Chen M. The use of low molecular weight heparin reduced the fetal fraction and rendered the cell-free DNA testing for fetal trisomy 21 false negative. Ultrasound Obstet Gynecol. 2017; https://doi.org/10.1002/uog.17473.
Sinkey RG, Odibo AO. Cost-effectiveness of old and new technology for aneuploidy screening. Clin Lab Med. 2016;36(2):237–48.
Sørensen MD, Gonzalez Dosal R, Jensen KB, Christensen B, Kølvraa S, Jensen UB, et al. Epsilon haemoglobin specific antibodies with applications in noninvasive prenatal diagnosis. J Biomed Biotechnol. 2009;2009:659219.
Iacono KT, Brown AL, Greene MI, Saouaf SJ. CD147 immunoglobulin superfamily receptor function and role in patheology. Exp Mol Pathol. 2007;83(3):283–95.
Nagao K, Zhu J, Heneghan MB, Hanson JC, Morasson MI, Tessarollo L, et al. Abnormal placental development and early embryonic lethality in EpCAM-null mice. PLoS One. 2009;4(12):e8543.
Hackmon R, Pinnaduwage L, Zhang J, Lye SJ, Geraghty DE, Dunk CE. Definitive class I human leukocyte antigen expression in gestational placentation: HLA-F, HLA-E, HLA-C, and HLA-G in extravillous trophoblast invasion on placentation, pregnancy, and parturition. Am J Reprod Immunol. 2017;77(6):e1264377.
Schreier S, Sawaison P, Udomsanpetch R, Triampo W. Advances in rare cell isolation: an optimization and evaluation study. J Transl Med. 2017;15:6.
Kanda E, Yura H, Kitagawa M. Practicability of prenatal testing using lectin-based enrichment of fetal erythroblasts. J Obstet Gynecol Res. 2016;42(8):918–26.
Acknowledgements
The authors would like to thank Dr. Wan-Ju Wu, Department of Obstetrics and Gynecology, Changhua Christian Hospital, Taiwan, and Dr. Wei-Cheng Hsu, Mr. Sheng-Wen Chen and Mr. Hsin-Cheng Ho of Cytoaurora Biotechnolgies, Inc., Hsinchu Science Park, Hsinchu, Taiwan, for their assistance in manuscript preparation. The “Cell Reveal™” system has been granted Taiwanese patent no. M539091.
Funding
Part of the study (the cfDNA part of the experiments) was funded by a grant (MOST 104–2314-B-371-009-MY3) from the Ministry of Science and Technology, Executive Yuan, Taiwan to MC.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
CEH, GCM, MC designed the study. HJJ and MC recruited the patients. CEH, GCM, WHL, DJL, YSL did the experiments and performed the analyses. CEH, GCM, and MC wrote the manuscript. HJJ, NG, HFC, and FMCC provided critical review of the manuscript. MC revised the final version. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study is a retrospective cohort study. The institutional review board (IRB) of Changhua Christian Hospital had approved the study (CCH-IRB-141219).
Consent for publication
All authors had consented the submission and publication of the paper.
Competing interests
CEH is the founder, CEO and holds equity of the Cytoaurora Biotechnologies Inc., Hsinchu Science Park, Hsinchu, Taiwan. All other authors declare no conflict of interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
The numbers of nRBC and EVT captured in the 24 validated cases. (DOCX 17 kb)
Additional file 2: Table S2.
The timeframe of the fetal cell capture from maternal blood by Cell RevealTM system. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Huang, CE., Ma, GC., Jou, HJ. et al. Noninvasive prenatal diagnosis of fetal aneuploidy by circulating fetal nucleated red blood cells and extravillous trophoblasts using silicon-based nanostructured microfluidics. Mol Cytogenet 10, 44 (2017). https://doi.org/10.1186/s13039-017-0343-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-017-0343-3