Abstract
Background
There is substantial evidence that a virus homologous to mouse mammary tumor virus (MMTV) may have a role in human breast cancer. The present study indicates that those who developed breast cancer associated with an MMTV-like virus had this virus in their non-cancerous breast tissues years before the cancer developed.
Methods
Polymerase chain reaction (PCR) techniques and sequencing were used to identify MMTV-like envelope gene sequences (MMTV-like env sequences) in Australian benign breast biopsy specimens from women who several years later developed breast cancer. Murine contamination was excluded by stringent laboratory procedures, and the absence of intracisternal A particle sequences and mitochondrial cyclooxygenase sequences.
Results
MMTV-like env sequences (also called HMTV sequences to denote their source) were found in 9 of 25 breast cancer specimens (36%). Among 25 non-cancerous breast biopsies of these same patients taken 1 to 11 years earlier, six contained MMTV-like sequences (24%). Five of the six were among the nine virally-associated breast cancers. In two pairs of specimens, benign and malignant, env sequences were 97% identical.
Conclusions
The identification of MMTV (MMTV-like) sequences in breast tissues prior to the development of MMTV positive breast cancer fulfills a key criterion for a possible causal role for the MMTV-like virus in human breast cancer.
Similar content being viewed by others
Background
Mouse mammary tumor viruses (MMTV) have a well-documented causal role in mouse mammary tumors in feral and experimental mice [1]. This role of MMTV was first observed by John Bittner in 1936 [2]. These observations led to the search for a similar virus in human breast cancer. Close morphologic and immunologic similarity were repeatedly shown. More recently, near identity of molecular structure to MMTV (90-98%) has been identified in human breast cancers [3]. The MMTV-like virus in humans is also called human mammary tumor virus (HMTV) [4].
The evidence of a role for an MMTV-like virus in human breast cancer is substantial but incomplete. An essential criterion to establish a causal role of an infectious agent in any disease, including cancer, is evidence of prior infection by the suspect agent [5]. This criterion had not been investigated with respect to MMTV-like virus. MMTV-like env sequences have rarely been identified in benign breast tissues [3, 6, 7], nor in the benign breast tissue of individuals with breast cancer containing MMTV-like sequences [8]. For this reason we sought to identify MMTV-like env sequences in benign breast biopsy tissues from women who some years later developed MMTV-like positive breast cancer.
The evidence suggestive of a role for an MMTV–like virus in human breast cancer is as follows: (i) a meta-analysis of 22 studies concluded that the identification of MMTV–like gene sequences in breast cancer tissues, was associated with a 15 fold increase in breast cancer [9], (ii) MMTV-like env gene sequences were identified in 38% of US human breast tumors but were extremely uncommon in healthy breast tissues [3, 6], (iii) the near complete proviral structure of MMTV-like virus that was 95-98% homologous to MMTV has been identified in human breast tumors [4, 10], (iv) MMTV viral proteins have been identified in human breast cancer [11], (v) Wnt-1 oncogene expression is significantly higher in MMTV-like positive compared to MMTV-like negative breast cancer specimens, which parallels high Wnt-1 expression in MMTV positive mouse mammary tumors [12, 13], (vi) MMTV can infect human cells and randomly integrate its genomic information [14, 15], and produce virus particles [16] (vii) there is increased prevalence of MMTV-like viral sequences in healthy breast tissues (nil), healthy tissue adjacent to breast cancer (19%), breast hyperplasia (27%), ductal carcinoma in situ (82%) [17], (viii) the age standardized rates for breast cancers in five countries of Asia are less frequent (29–43 per 100,000) than in seven countries of Europe, the Americas, and Australia (47–92) [18]. These findings correlate with different burdens of MMTV-like infection in human breast cancers, 0-20% in Asia vs. 27-60%, in the seven countries which are associated with different prevalence of MMTV in the indigenous mouse species [18, 19]. (ix) MMTV–like sequences have been identified in milk from healthy lactating women and three fold positivity in milk from women at high risk for breast cancer [20, 21], (x) MMTV-like sequences have been identified in the saliva of 27% of healthy children, 11% of healthy adults and 57% of adults with breast cancer, which is suggestive of a human to human viral transmission [22] and (xi) MMTV-like viral sequences have been identified in breast cancers which developed in a father, mother and daughter of the same family which is suggestive of an infectious condition [23]. Overall this evidence is consistent with MMTV having similar influences in both human breast cancer and mouse mammary tumors.
If an MMTV-like virus infects human breasts, MMTV-like antibodies should be present in the sera of infected individuals since mice infected with MMTV develop high titers of MMTV specific antibodies. Indeed, MMTV antibodies in human breast cancer have been described [24, 25]. Antibodies were not identified in one recent study, however [26].
Methods
Ethics
This project was formally considered and approved by the Human Research Ethics Committees of the several participating institutions.
Twenty five patients were identified for whom both benign breast and subsequent breast cancer specimens were available from the archives of an Australian pathology service (Douglass Hanly Moir – Pathology). All the specimens were formalin fixed and paraffin mounted. Seventeen of these sets of specimens were analysed by polymerase chain reaction (PCR) techniques at the Icahn School of Medicine at Mount Sinai (ISMMS) (New York). Eight additional specimen pairs were analysed by the same PCR techniques but with the cancer tissues microdissected from the specimens at the University of Pisa (UP) (Italy). Eight of the sets of specimens were analysed in both Centers.
Detection of MMTV-like env sequences
The DNA extraction and detection of MMTV-like env sequences were performed by PCR techniques as described by Wang et al. [3]. The primer sequences used in these PCR analyses include part of the MMTV env gene, which differs from human endogenous retrovirus 10 (HERV-K10). The same PCR techniques were used in both the ISMMS and UP laboratories with the exception of microdissection of the tumor tissues, that were analysed in the UP laboratory by fluorescence nested PCR. Materials from all patients were not available due to the exhaustion of the blocks. The outcomes from each of the two laboratories are shown in Table 1.
Contamination is a well-known problem with PCR analyses. Therefore, all reagents were shown to be free of MMTV-like sequences before use. PCR products were tested for the presence of murine mitochondrial (MoMt) and genomic DNA to exclude contamination. The methods used were as described by Deligdisch et al. [27] and outlined below:
-
(i)
Detection of mouse mitochondrial DNA sequences.
A series of PCR analyses was conducted to detect MoMt contamination by the detection of cytochrome oxidase (cox-2) gene as part of the MoMt in any sample DNA in which MMTV env sequences were detected. The following primers for cox-2 were used: mt5982F (5-AGACGCACCTACGGTGAAGA-3) and mt16267R (5-AGAGTTTTGGTTCACGGAA CATGA-3). The product yields an amplicon of 286 base pairs. The semi-nested PCR was done using the primers mt16115F (5-TGCCAAACCCCAAAAACACT-3) and mt16267R, which results in a 153-bp amplicon. After transfer from the gel to a nylon membrane, the amplicon was detected by hybridization with a MoMt 32P-probe (5-GAACTAGAATTGATCAGGCAT-3).
-
(ii)
Detection of murine intracisternal A particle long terminal repeats (IAP)
IAPs are retrotransposon sequences present at the level of approximately 1000 copies of varying length per mouse genome. Amplification of the IAP sequences was carried out in PCR reactions using the following primers: forward primer (5-ATAATCTGCCGCATGAGCCAAGG-3) and reverse primer (5-AGGAAGAACACCACAGACCAGA-3) one cycle of 95 °C for 5 min, 35 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 20 s, and one cycle of 72 °C for 7 min. If present, the products of variable size, reflecting diversity of the IAP sequences, can be visualized on a 2% ethidium bromide stained agarose gel.
Results
The results from each of the two laboratories are shown in Table 1. Gaps in the results are due to lack of materials due to exhaustion of the blocks. The time between the benign breast biopsy specimen and subsequent breast cancer in the same patients varied from 1 to 11 years.
Outcomes of PCR analyses
MMTV-like env gene sequences were identified in 6 (24%) of 25 benign breast specimens and 9 (36%) of 25 breast cancer specimens. Of the 6 MMTV-like positive benign specimens, 5 later developed (MMTV-like positive) cancer.
Eight sets of benign and later breast cancer blocks were analysed in both the ISMMS and UP laboratories. Negative outcomes of PCR analyses were the same for 5 sets of blocks. MMTV sequences were identified by both laboratories in breast cancer of patient 5. MMTV sequences were identified by the ISMMS lab only (not the UP lab) in patients 4 and 23 breast cancer specimens.
Neither MoMt nor IAP DNA sequences were identified in any of the MMTV-like env positive DNAs. This indicates there was no murine DNA contamination.
Comparison of MMTV env gene sequences in benign and breast cancer specimens within the same patient
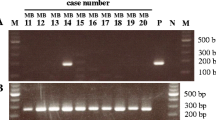
Over 97% of the MMTV-like env were identical in both the benign breast biopsy and subsequent breast cancer in 2 selected patients. These sequences are shown in Fig. 1. There are variations in approximately 3% of the sequences between the benign and subsequent breast cancers that developed several years later in the same patients. Such variations could be due to alterations in the MMTV-like genome following integration into the human genome as has been described for murine retroviruses [28] or could also occur during the PCR cycling procedures. These sequence variations indicate that contamination during PCR analyses was unlikely.
The morphological (histotype) characteristics in 8 of the 9 MMTV-like sequence positive breast cancers were similar to MMTV positive mouse (C3H strain mice) mammary tumours. These characteristics are shown in Fig. 2.
Discussion
In this study MMTV-like env gene sequences have been identified in benign breast biopsy specimens prior to the subsequent development of MMTV-like env positive breast cancer specimens in the same patient. This is consistent with prior infection by MMTV-like virus in breast tissues prior to the development of the same MMTV-like virus positive breast cancer some years later. This finding fulfils one key evidentiary criterion that MMTV-like virus may have a role in some human breast cancers.
Validity of the data
We consider the data generated in this study to be valid for the following reasons: (i) The study was conducted by PCR with primers based on MMTV-like envelope gene sequences as described by Wang et al. [13]. These MMTV env gene sequences are unique to the MMTV genome and are not present in human endogenous retrovirus sequences (HERV) and which are commonly identified in studies of the human genome. (ii) Neither MoMt nor IAP DNA sequences were identified in any one of the MMTV-like env positive DNAs. This indicates there was no murine DNA contamination. (iii) Variations in approximately 2% of the MMTV env sequences of two specimen pairs were identified. This is an indication that contamination during PCR analyses was unlikely. Such variations could be due to alterations in the MMTV genome following integration into the human genome. Variations in sequences can also occur during PCR cycling procedures.
There was one case (case 19 – Table 1) where the earlier biopsy was positive for MMTV-like sequences and the ensuing breast tumor (developing after two years) was negative. In addition the results for cases 4 and 23 are not consistent between the two laboratories. There are several possible reasons: (i) MMTV sequences identified by PCR can be inconsistent and false negatives are possible; (ii) MMTV sequences may be present in some, but not all parts of the tumor; (iii) although unlikely, changes in MMTV env sequences may have occurred. The problem of inconsistent outcomes of PCR based analyses has been considered in detail by Vinner et al. [29]. There are particular difficulties in obtaining consistent outcomes from PCR analyses of retroviruses when present in extremely low viral concentrations. While these problems, including the exhaustion of several of the materials, do not invalidate the identification of MMTV in this current study, it would be wise to replicate this study with increased numbers of patients.
It is of interest that MMTV sequences from the long terminal repeat (LTR) section of the MMTV genome have been identified in human breast cancers using Next Generation massive parallel Sequencing (NGS) [30]. These MMTV sequences were highly homologous to the reference MMTV genome based on BLAST technology [31]. These data, based on techniques very different from PCR, confirm the identification of MMTV-like gene sequences in human breast tumors and add validity to PCR based studies that were used in this current investigation. NGS techniques are not as sensitive for the identification of retroviral nucleotide sequences as PCR [29]. This is the reason for the much more frequent identification of MMTV-like nucleotide sequences by PCR compared to NGS.
The identification of MMTV-like env gene sequences in 9 (36%) of 25 Australian breast cancer specimens is a similar percentage to previous investigations of Australian breast cancers [7].
It is of considerable interest that the morphology (histotype) of 8 of 9 MMTV-like positive breast cancers was similar to the morphology of MMTV positive mouse mammary tumors. This has been previously observed by Wellings [32] and Lawson et al. [33]. If this observation is confirmed this has important implications as it provides potential evidence that MMTV infections may lead to a specific morphological type of breast cancer.
Conclusions
The aim of this project has been achieved, namely the identification of MMTV-like sequences in benign breast biopsy tissues from women who several years later developed MMTV-like positive breast cancer. The findings in this study offer an important contribution to the overall body of evidence, which links MMTV-like virus (also called HMTV) to human breast cancer and fulfills a key criterion for a possible causal role for the MMTV-like virus in human breast cancer. Although other viruses and bacteria have been found in human breast cancer, none other is known to have a similar parallel in the animal kingdom. Conclusive proof of HMTV (MMTV-like virus) as a cause of human breast cancer will open a new era for prevention and therapy [34].
Abbreviations
- BLAST technology:
-
Basic Local Alignment Search Tool is an algorithm for comparing primary biological sequence information
- cox-2 :
-
cytochrome oxidase gene
- env sequences:
-
Envelope gene sequences
- HERV-K10:
-
Human endogenous retrovirus
- HMTV:
-
Human mammary tumor virus
- IAP:
-
Intracisternal a particle long terminal repeats
- ISMMS:
-
Icahn School of Medicine at Mount Sinai (ISMMS) (New York)
- LTR:
-
Long terminal repeat section of the MMTV genome
- MMTV:
-
Mouse mammary tumor virus
- MoMt:
-
Murine mitochondrial DNA
- NGS:
-
Next Generation massive parallel Sequencing
- PCR:
-
Polymerase chain reaction
- UP:
-
University of Pisa, Italy
References
Ross SR. MMTV infectious cycle and the contribution of virus-encoded proteins to transformation of mammary tissue. J Mammary Gland Biol Neoplasia. 2008;13:299–307.
Bittner JJ. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162. doi:10.1126/science.84.2172.162.
Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, Cantarella A, Stellrecht K, Mani S, Pogo BG. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55:5173–9.
Liu B, Wang Y, Melana SM, Pelisson I, Najfeld V, Holland JF, Pogo BG. Identification of a proviral structure in human breast cancer. Cancer Res. 2001;61:1754–9.
Hill AB. The environment and disease: Association or causation? Proc R Soc Med. 1965;58:295–330.
Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH. Mouse mammary tumor virus-like env gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res. 2000;6:1273–8.
Ford CE, Tran DD, Deng YM, Rawlinson WD, Lawson JS. Mouse mammary tumour like virus prevalence in breast tumours of Australian and Vietnamese women. Clin Cancer Res. 2003;9:1118–20.
Melana SM, Holland JF, Pogo BG. Search for mouse mammary tumor virus-like env sequences in cancer and normal breast from the same individuals. Clin Cancer Res. 2001;7:283–4.
Wang F, Hou J, Shen Q, Yue Y, Xie F, Wang X, Jin H. Mouse mammary tumor virus-like virus infection and the risk of human breast cancer: a meta-analysis. Am J Transl Res. 2014;6:248–66.
Melana SM, Nepomnaschy I, Sakalian M, Abbott A, Hasa J, Holland JF, Pogo BG. Characterization of viral particles isolated from primary cultures of human breast cancer cells. Cancer Res. 2007;67:8960–5.
Melana SM, Nepomnaschy I, Hasa J, Djougarian A, Djougarian A, Holland JF, Pogo BG. Detection of human mammary tumor virus proteins in human breast cancer cells. J Virol Methods. 2010;163:157–61.
Lawson JS, Glenn WK, Salmons B, Ye Y, Heng B, Moody P, Johal H, Rawlinson WD, Delprado W, Lutze-Mann L, Whitaker NJ. Mouse mammary tumor virus-like sequences in human breast cancer. Cancer Res. 2010;70:3576–85.
Callahan R, Mudunur U, Bargo S, Raafat A, McCurdy D, Boulanger C, Lowther W, Stephens R, Luke BT, Stewart C, Wu X, Munroe D, Smith GH. Genes affected by mouse mammary tumor virus (MMTV) proviral insertions in mouse mammary tumors are deregulated or mutated in primary human mammary tumors. Oncotarget. 2012;3:1320–34.
Indik S, Günzburg WH, Salmons B, Rouault F. Mouse mammary tumor virus infects human cells. Cancer Res. 2005;65:6651–9.
Faschinger A, Rouault F, Sollner J, Lukas A, Salmons B, Günzburg WH, Indik S. Mouse mammary tumor virus integration site selection in human and mouse genomes. J Virol. 2008;82:13.
Konstantoulas C, Indik S. C3H strain of mouse mammary tumor viruses like GR strain infects human mammary epithelial cells albeit less efficiently than murine mammary epithelial cells. J Gen Virol. 2015;96:650–62.
Mazzanti CM, Al Hamad M, Fanelli G, Scatena C, Zammarchi F, Zavaglia K, Lessi F, Pistello M, Naccarato AG, Bevilacqua G. A mouse mammary tumor virus env-like exogenous sequence is strictly related to progression of human sporadic breast carcinoma. Am J Pathol. 2011;179:2083–90.
Stewart BW, Wild CP. World Cancer Report. Lyon, France: IARC, WHO; 2014.
Stewart TH, Sage RD, Stewart AF, Cameron DW. Breast cancer incidence hightest in the range of one species of house mouse, Mus domesticus. Br J Cancer. 2000;82(2):446–51.
Johal H, Ford CE, Glenn WK, Heads J, Lawson JS, Rawlinson WD. Mouse mammary tumor like virus (MMTV) sequences in breast milk from healthy lactating women. Breast Cancer Res Treat. 2011;129:149–55.
Nartey T, Moran H, Marin T, Arcaro KF, Anderton DL, Etkind P, Holland JF, Melana SM, Pogo BG. Human Mammary Tumor Virus (HMTV) sequences in human milk. Infect Agent Cancer. 2014;9:20.
Mazzanti CM, Lessi F, Armogida I, Zavaglia K, Franceschi S, Al Hamad M, Roncella M, Ghilli M, Boldrini A, Aretini P, Fanelli G, Marchetti I, Scatena C, Hochman J, Naccarato AG, Bevilacqua G. Human saliva as route of inter-human infection for mouse mammary tumor virus. Oncotarget. 2015;6:18355–63.
Etkind PR, Stewart AF, Wiernik PH. Mouse mammary tumor virus (MMTV)-like DNA sequences in the breast tumors of father, mother, and daughter. Infect Agent Cancer. 2008;3:2.
Witkin SS, Sarkar NH, Good RA, Day NK. An enzyme-linked immunoassay for the detection of antibodies to the mouse mammary tumor virus: application to human breast cancer. J Immunol Methods. 1980;32:85–91.
Day NK, Witkin SS, Sarkar NH, Kinne D, Jussawalla DJ, Levin A, Hsia CC, Geller N, Good RA. Antibodies reactive with murine mammary tumor virus in sera of patients with breast cancer: geographic and family studies. Proc Natl Acad Sci U S A. 1981;78:2483–7.
Goedert JJ, Rabkin CS, Ross SR. Prevalence of serologic reactivity against four strains of mouse mammary tumour virus among US women with breast cancer. Br J Cancer. 2006;94:548–51.
Deligdisch L, Marin T, Lee AT, Etkind P, Holland JF, Melana S, Pogo BG. Human mammary tumor virus (HMTV) in endometrial carcinoma. Int J Gynecol Cancer. 2013;23:1423–8.
Monk RJ, Malik FG, Stokesberry D, Evans LH. Direction determination of the point mutation rate of murine retroviruses. J Virol. 1992;66:3683–9.
Vinner L, Mourier T, Friis-Nielsen J, Gniadecki R, Dybaker K, Rosenberg J, Langhoff JL, Cruz DF, Fonager J, Izarzugaza JM, Gupta R, Sicheritz-Ponten T, Brunak S, Willerslev E, Nielsen LP, Hansen AJ. Investigation of Human Cancers for Retrovirus by Low-Stringency Target Enrichment and High-Throughput Sequencing. Sci Rep. 2015;5:13201.
Larsson lab – http://larssonlab.org/tcga-viruses/report_BRCA.php.
Larsson E. Personal communication. Sweden: University of Gottenburg; 2016.
Wellings SR. A hypothesis of the origin of human breast cancer from the terminal ductal lobular unit. Pathol Res Pract. 1980;166:515–35.
Lawson JS, Tran DD, Carpenter E, Ford CE, Rawlinson WD, Whitaker NJ, Delprado W. Presence of mouse mammary tumour-like virus gene sequences may be associated with specific human breast cancer morphology. J Clin Pathol. 2006;59:1287–92.
Braitbard O, Roniger M, Bar-Sinai A, Rajchman D, Gross T, Abramovitch H, La Ferla M, Franceschi S, Lessi F, Naccarato AG, Mazzanti CM, Bevilacqua G, Hochman J. A new immunization and treatment strategy for mouse mammary tumor virus (MMTV) associated cancers. Oncotarget 2016. doi: 10.18632/oncotarget.7762.
Acknowledgements
The archival specimens were provided by Professor Warick Delprado and the Douglass Hanly Moir Pathology Laboratories of Sydney, Australia.
Funding
The James and Margaret Lawson Research Fund. The T.J. Martell Foundation for Leukemia. Cancer and AIDS Research. The Faith Lynn Price Kash Family Fund. The Derald H. Ruttenberg Foundation.
Availability of data and materials
All data is available from the corresponding author.
Authors’ contributions
TN- laboratory analyses; CM – laboratory analyses; SM – laboratory analyses; WG- concepts, histology assessments, data analyses, preparation of the manuscript; GB- concepts, quality control, organisation, preparation of the manuscript; JH– concepts, quality control, preparation of the manuscript; NW- concepts, quality control, data analyses, preparation of the manuscript; JL– initial concept, identification and collection of the specimens, histological assessments, data analyses, preparation of the manuscript; BP– concepts, laboratory analyses, quality control, preparation of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This project was formally considered and approved by the Human Research Ethics Committees of the several participating institutions. The University of New South Wales Human Research Ethics Committee reference number is HREC Ref: HC11421. This study was based on de-identified archival specimens. There was no direct contact with patients.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at http://dx.doi.org/10.1186/s13027-017-0126-9.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nartey, T., Mazzanti, C.M., Melana, S. et al. Mouse mammary tumor-like virus (MMTV) is present in human breast tissue before development of virally associated breast cancer. Infect Agents Cancer 12, 1 (2017). https://doi.org/10.1186/s13027-016-0113-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13027-016-0113-6