Abstract
Background
Fabry disease (FD, OMIM #301500) is an X-linked lysosomal disorder caused by the deficiency of α-galactosidase A (α-GalA), encoded by the GLA gene. Among more than 1100 reported GLA mutations, few were deep intronic mutations which have been linked to classic and cardiac variants of FD.
Methods and results
We report a novel hemizygous deep intronic GLA mutation (IVS4+1326C>T) in a 33-year-old Chinese man with a mild α-GalA deficiency phenotype involving isolated proteinuria and predominant globotriaosylceramide deposits in podocytes. IVS4+1326C>T, which appears to be the first deep intronic GLA mutation associated with renal variant of FD, was identified by Sanger sequencing the entire GLA genomic DNA sequence of the patient’s peripheral mononuclear blood lymphocytes (PBMCs). Further sequencing of cDNA from PBMCs of the patient revealed a minor full-length GLA transcript accounting for about 25% of total GLA transcript, along with two major aberrantly spliced GLA transcripts encoding mutant forms of α-GalA with little enzyme activity characterized by in vitro α-GalA overexpression system in the HEK293T cells. Thus, the combined clinical phenotype, genetic analysis and functional studies verified the pathogenicity of IVS4+1326C>T.
Conclusions
The identification of IVS4+1326C>T establishes a link between deep intronic GLA mutation and the renal variant of FD, which extends the mutation spectrum in GLA gene and justifies further study of how IVS4+1326C>T and potentially other deep intronic GLA mutations contribute to Fabry podocytopathy through aberrant splicing. Future studies should also assess the true incidence of IVS4+1326C>T in patients with different variants of FD, which may improve early genetic diagnosis to allow timely treatment that can prevent disease progression and improve survival.
Similar content being viewed by others
Introduction
Fabry disease (FD) is a rare X-linked lysosomal disorder characterized by deficiency of α-galactosidase A (α-GalA), encoded by the GLA gene. α-GalA deficiency results in intracellular accumulation of neutral glycosphingolipids, primarily globotriaosylceramide (GL-3) and its deacylated derivative globotriaosylsphingosine (Lyso-GL-3) inside of lysosomes, leading to multi-organ involvement. Fabry nephropathy is a hallmark of FD and in untreated FD patients of classic phenotype with absent or negligible α-GalA activity (< 1% residual enzyme activity, REA), end stage renal disease (ESRD) usually occurs by the time they reach their 40 s [1] if the disease goes untreated with enzyme replacement therapy (ERT) with α-GalA or chaperone therapy, the early initiation of which can slow or stop renal deterioration.
Screenings for FD have shown that FD prevalence in male patients with chronic kidney disease (CKD) or ESRD is 0.48–1.69% [2]. Thus, FD needs to be considered in the differential diagnosis of any patient with unexplained CKD, as FD can present as ‘renal variant’, with renal disease as the only or prominent manifestation [3]. Having varying levels of residual α-GalA activity, FD patients with renal variants usually lack glycosphingolipid accumulation in microvascular endothelium and thus the classic manifestations such as angiokeratoma, acroparesthesia and hypohydrosis are absent, which often leads to a delay in diagnosis.
Mutation analysis of the GLA gene is a valuable tool for screening and diagnosis of FD. Renal variant of FD reported so far has been linked to numerous GLA missense mutations (p.Ala37Thr [4]; p.Met42Leu [5]; p.Glu66Lys [6]; p.Glu66Gln [3]; p.Ile91Thr [4]; p.Arg112His [4, 7]; p.Phe113Leu [8]; p.Ala143Thr [9]; p.Arg196Thr [4]; p.Pro205Ser [4]; p.Pro210Ser [10]; p.Phe229Val [11]; p.Met290Val [4]; p.Arg356Gly [4]; p.Gly360Ser [12]) and two frameshift mutations (p.Leu344fs*31 [13]; p.Lys426Argfs*24 [14]). In this study, we reported the identification of a novel deep intronic GLA mutation (IVS4+1326C>T) in a 33-year-old Chinese man with a mild α-GalA deficiency phenotype involving isolated proteinuria and predominant GL-3 accumulation in podocytes. IVS4+1326C>T, which appears to be the first deep intronic GLA mutation associated with renal variant of FD, was identified by analyzing the full length GLA genomic DNA sequence from the peripheral mononuclear blood lymphocytes (PBMCs) of the patient. Molecular studies were further performed to investigate how IVS4+1326C>T may contribute to renal variant of FD.
Methods
Participants and ethical statement
The proband patient was admitted to Xin Hua Hospital in 2018 with proteinuria two years. Written informed consent was obtained from the involved subjects and all procedures were performed in accordance with the Declaration of Helsinki. The study was performed under the approval of the Ethics Committee from Xin Hua Hospital, Shanghai Jiao Tong University School of Medicine (approval No.XHEC-D-2020-002).
DNA and RNA isolation
Blood samples were collected from the patient, his family members and normal controls using test tubes containing EDTA anticoagulant. Genomic DNA was extracted from PBMCs using DNA Midi Kit (Qiagen, Milano, Italy) according to the manufacturer’s protocol. Total RNA in PBMCs was collected in Tempus™ blood RNA tubes (Biotek, Canada) and isolated with Tempus™ Spin RNA Isolation kit (Biotek, Canada) according to the manufacturer’s protocol.
Genomic DNA sequencing analysis
The full length genomic DNA of GLA were amplified by nested polymerase chain reaction (PCR). Primers are designed according to the genomic GLA reference sequence (NCBI reference sequence NM_000169) listed in Additional file 1: Table 1. Sanger sequencing was performed to verify the sequences.
Whole exome sequencing (WES)
Whole-exome sequencing was performed on the patient as described [15], and the WES data were evaluated for causative mutations in GLA and other genes associated with inherited podocytopathies [16]. The non-neutral variant p.Arg229Gln in the NPHS2 gene and the high-risk alleles G1 and G2 in the APOL1 gene were manually inspected.
Reverse transcription polymerase chain reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR)
To evaluate the range of transcripts produced from the GLA gene, total RNA was extracted from PBMCs from the patient, his mother, and healthy controls as described above. First-strand cDNA synthesis was performed using the Hifair™ 1st Strand cDNA Synthesis SuperMix Kit for qPCR (Yeasen, Shanghai, China). Then PCR was performed using the PrimerSTAR MAX DNA Polymerase (TaKaRa, Beijing, China), and amplicons were visualized on 1.5% agarose gels and sequenced on an ABI PRISM 3100 Genetic Analyzer. Primer sequences were listed in Additional file 1: Table 1.
To quantify the effect of IVS4+1326C>T on the expression of GLA mRNA level in PBMCs, qRT-PCR were performed using 2 pairs of cross-exon primers (listed in Additional file 1: Table 1) and Green Master Mix (Yeasen, Shanghai, China) to specifically amplify the full-length transcript of GLA, and qRT-PCR was normalized to that of GAPDH. Data was analyzed using software Geneious version 10.2.3 (Biomatters, Auckland, New Zealand).
Plasmid construction
Full-length human GLA cDNA was subcloned into the PHAGE vector at the BamHI and SalI sites, upstream from a hemagglutinin (HA) tag. The resulting PHAGE-GLA plasmid was then subjected to multiple PCR using mutated primers (listed in Additional file 1: Table 1) and PrimerSTAR MAX DNA Polymerase (TaKaRa Bio, Beijing, China) to give rise to mutant PHAGE-GLA plasmids encoding aberrantly spliced transcripts of GLA. All constructs were verified by sequencing.
Cell culture and transfection
HEK293T cells were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin at 37 °C with 5% CO2. HEK293T cells were transfected with the wild-type PHAGE-GLA plasmid or mutant PHAGE-GLA plasmids using Lipofectamine 2000 (Invitrogen, California, USA) according to the manufacturer’s instructions. Transfected cells were incubated for 24–48 h post transfection and proteins were analyzed by Western blot analysis.
Western blot analysis
Cell lysates were prepared using Cell Lysis Buffer (Beyotime, Shanghai, China). Protein (10–40 µg) was fractionated on 8–12.5% SDS–polyacrylamide gels and transferred to 0.22-µm polyvinylidene fluoride membranes. After 1 h of incubation at room temperature in 5% milk, the membrane was incubated with the primary antibody for 2 h followed by incubation with the appropriate secondary antibodies for 2 h. Antibody binding was detected using the ECL Plus Western Blotting System (GE, Buckinghamshire, UK). GAPDH was detected as an internal control. The primary antibodies were mouse monoclonal anti-HA antibody (1:2000, Cell Signal Technology, Massachusetts, USA) and rabbit polyclonal anti-GAPDH antibody (1:1000, Cell Signal Technology, Massachusetts, USA).
α-Galactosidase (α-GalA) enzyme assay
Activities of α-GalA in cell lysates of HEK293T cells transfected with the wild-type or mutant PHAGE-GLA plasmids were measured using the Micro α-Galactosidase Assay Kit (Solarbio, Beijing, China) according to the manufacturer’s instructions. To correct for endogenous α-GalA activity in HEK293T cells, a mock control plasmid was also transfected and the level of α-GalA activity in lysates from these cells were subtracted from those in mutant- or wild-type-transfected cells.
Statistical analysis
Continuous data that were normally distributed were expressed as mean ± standard deviation. Intergroup differences were assessed for significance using ANOVA with Bonferroni-Holm post-test correction or Student’s t test, as appropriate. Differences associated with P < 0.05 were considered statistically significant.
Results
Clinical description
A 33-year-old Chinese man was referred to Xin Hua Hospital in 2018 with proteinuria two years prior to renal biopsy. On admission, his blood pressure and pulse rate were in the normal range. Laboratory data on admission showed that the serum creatinine level was 0.85 mg/dL, blood urea nitrogen level was 15 mg/dL and 24-h proteinuria was 1100 mg. The following serum test results were normal or negative: complement testing, assay for anti-neutrophil cytoplasmic antibody, assay for anti-DNA antibody, hepatitis serology, and protein electrophoresis.
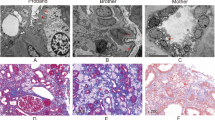
A renal biopsy was performed and eight glomeruli were observed by light microscopy. Global sclerosis was identified in two glomeruli, while the other six showed foamy changes in the podocytes (Fig. 1A), and one glomerulus had segmental sclerosis (Fig. 1B). Vacuolation was present in some tubular epithelial cells, 10% of which showed tubulointerstitial atrophy. No significant alterations in blood vessels were observed. Immunofluorescence findings were unremarkable. Electron microscopy revealed myelin-like inclusions predominantly in the cytoplasm of podocytes (Fig. 1C) with 40% foot process fusion rate. The patient denied ever taking any medicine affecting the activity of lysosomal enzymes, such as amiodarone, chloroquine or tamoxifen and he also denied pain in the extremities, acroparesthesia, hypohydrosis, angiokeratomas, or gastrointestinal disturbance. Measurement of α-GalA activity in patient peripheral blood leukocytes revealed reduced α-GalA activity (27.9 nmol/mL/h/mg; normal, > 37.0 nmol/mL/h/mg). Plasma Lyso-GL-3 was elevated (2.49 ng/mL; normal, 0.24–0.86 ng/mL).
A–C Kidney biopsies of the patient. A Tissue was stained with periodic acid-Schiff reagent, revealing two glomeruli with enlarged and vacuolated podocytes. Magnification, 400×. B Tissue was stained with periodic acid silver methenamine, showing one glomerulus with segmental sclerosis. Magnification, 400×. C Electron micrograph showing abundant, electron-dense myelin structures within the cytoplasm of podocytes. Magnification, 5000×
Genetic analysis
As the clinical and pathological findings of the patient highly suspicious for Fabry podocytopathy, we performed WES using DNA from the patient's PBMCs to look for mutation in the exonic and flanking intronic regions of GLA as well as other genes associated with genetic podocytopathy without identifying any candidate variant. Next, the full length GLA genomic DNA from the patient's PBMCs were amplified by nested PCR followed by Sanger sequencing, which revealed a rare hemizygous mutation at nucleotide 9738 (IVS4+1326C>T) (Fig. 2A). IVS4+1326C>T has not been registered in the established FD-specific database such as International Fabry Disease Genotype–Phenotype Database (dbFGP) or broader genetic variants databases such as Human Gene Mutation Database (HGMD), ClinVar Database or Leiden Open Variation Database (LOVD). Thus, IVS4+1326C>T appeared to be the first deep intronic GLA mutation associated with renal variant of FD as all other renal variant associated hemizygous GLA mutations identified in PubMed were located in the exonic regions of GLA gene (Fig. 2B). Family cascade genotyping revealed a heterozygous IVS4+1326C>T mutation in the patient’s mother (Fig. 2A), who did not present any FD-related symptoms or positive laboratory findings at the age of 55. The patient’s younger brother and sister didn’t have the IVS4+1326C>T mutation and we didn’t manage to find other male or female patient relatives from the patient’s maternal side.
A Genomic sequencing of the patient, his mother and a normal control (NC). The patient was hemizygous and the mother heterozygous for the GLA mutation IVS4+1326C>T. B Schematic of the GLA gene indicating the relative position of the seven exons and showing the position of IVS4+1326C>T and the other 19 renal variant associated hemizygous GLA mutations. We only include renal variant associated GLA mutations reported in the database of PubMed with thorough investigation of all patient organs. Mutations identified only in renal variant of FD are shown in black letters. Mutations also identified in cardiac variant of FD are shown in green letters (p.Ala37Thr [17]; p.Glu66Gln [18]; p.Ile91Thr [19]; p.Arg112His [20]; p.Phe113Leu [21]; p.Ala143Thr [20]; p.Pro205Ser [22]). Mutations also identified in FD patients with classical phenotype are shown in red letters (p.Arg356Gly [23]; p.Lys426Argfs*24 [24])
RT-PCR of GLA exons 3 and 6 in mRNAs from the patient’s PBMCs revealed a minor full-length GLA transcript (392 bp) as well as two major aberrant splicing transcripts of 414 bp and 277 bp (Fig. 3A). The relative ratio of the three transcripts was 1:1.6:1.4. qRT-PCR showed that the level of full-length GLA mRNA from PBMCs of the patient was significantly reduced compared to that of normal controls (Fig. 3B). Further cDNA sequencing showed that the longer aberrant transcript lost a 35-bp sequence at the 5’ end of exon 4 as well as incorporated the 57-bp pseudoexon from intron 4 [25] (△4q_35bp_▼4_57bp), resulting in a premature stop codon (PTC) at residue 202 (p.Cys202*). The shorter aberrant transcript lost a 62-bp sequence at the 5’ end of exon 3 and a 51-bp sequence at the 5’ end of exon 5 (△3q_62bp_△5q_51bp), resulting in a frameshift that generated a PTC at residue 164 (p.Gly163Leufs*2) (Fig. 3C, D).
A Reverse transcription-PCR analysis of mRNAs from peripheral blood leukocytes of three normal controls (NC1-3) and the patient. In addition to a minor expression of the full length GLA transcript (a: 392 bp), the patient showed two major aberrant GLA transcripts (b: 414 bp and c: 277 bp). The relative ratio of the three transcripts was quantified by means of examing their grey scale using ImageJ (http://imagej.nih.gov/ij/). B qRT-PCR analysis using 2 pairs of cross-exon primers were performed on total RNA from peripheral blood leukocyte of three normal controls and the patient for the quantification of GLA full-length mRNA transcript. Levels were normalized to the amount of GAPDH. Data represented the mean ± SD of triplicate experiments. C Schematic of normal and aberrant splicing patterns. D Sanger sequencing of the full-length 392-bp transcript (a), the 414-bp aberrant transcript (b) and the 277-bp aberrant transcript (c). E: 57-bp pseudoexon; M: molecular weight markers; q: donor-site shift followed by the number of bp that are skipped or included; wt: wild type; △x: skipping of all or part of exon x; ▼: inclusion of intronic sequence in transcript
Functional experiments
To further assess the activity of the proteins encoded by the two aberrantly spliced GLA transcripts produced by IVS4+1326C>T, we transfected the PHAGE-GLA mutant constructs containing △4q_35bp_▼4_57bp or △3q_62bp_△5q_51bp into HEK293T cells in culture. Wild-type PHAGE-GLA construct and the mutant PHAGE-GLA construct only containing ▼4_57bp were transfected as controls. The full length and mutant protein bands of 52, 26, 23 and 19 kD were detected by western blot analysis (Fig. 4A). The α-GalA activities of HEK293T cells transfected with mutant PHAGE-GLA constructs containing the aberrantly spliced transcripts produced by IVS4+1326C>T were less than 20% of that of wild-type PHAGE-GLA construct transfected cells (Fig. 4B).
A Western blot analysis of cell lysates of HEK293T cells transfected with mutant PHAGE-GLA constructs containing △4q_35bp_▼4_57bp (b) or △3q_62bp_△5q_51bp (c). Expression of wild-type PHAGE-GLA construct (a) or the mutant PHAGE-GLA construct containing ▼4_57bp in HEK293T cells was performed as controls. A hemagglutinin (HA) monoclonal antibody was used for the detection of α-GalA. GAPDH was used as the loading control. The wild-type and mutant protein of 52, 26, 23 and 19 kDa were detected. B α-GalA activity (nmol/h/104 cells) in HEK293T cells transfected with wild-type or mutant PHAGE-GLA constructs was expressed as the mean ± SD of triplicate experiments. A mock control plasmid was also transfected and the level of α-GalA activity in lysates from these cells (0.09 ± 0.01 nmol/h/104 cell) were subtracted from those in mutant- or wild-type-transfected cells
Treatment and follow-up
Based on the combined kidney biopsy findings, biochemical and molecular analysis, as well as the lack of other FD-related organ involvement as ophthalmological examination, echocardiography, and brain magnetic resonance imaging showed no evidence of FD, the patient was diagnosed with a renal variant of FD. Thus, the patient began taking 75 mg irbesartan per day. Enzyme replacement therapy was not covered by the patient’s medical insurance and so was not provided. At the most recent follow-up, nearly three years after diagnosis, the patient showed normal renal function and 24-h proteinuria of 0.8–1.0 g.
Discussion
Among more than 1100 GLA mutations described in the HGMD, few were intronic mutations that are ‘deep’, i.e. located more than 20 bp away from the exon–intron junction [25,26,27,28]. It is presumed that ‘missing mutations’ might reside within the regions that are not covered by routine exonic DNA sequencing whereas sequencing the entire GLA gene sequence using traditional Sanger sequencing or the high-throughput sequencing technology such as long-read sequencing nanopore sequencing [27] may help their detection. Besides, deep intronic GLA mutations sometimes can be difficult to detect from RNA samples if insufficient RNA is available or if aberrant RNAs are unstable and have been degraded. IVS4+1326C>T identified in our study, to the best of our knowledge, is the first association between the deep intronic GLA mutation and renal variant of FD.
Recent studies have supported the pivotal role of podocyte in the development and progression of Fabry nephropathy. Among all renal cell types, GL-3 mainly accumulated in podocytes, which have the slowest turnover rate of the renal cell populations and GL-3 deposits already exist in the majority of normoalbuminuric young classic Fabry patients and was shown to increase with age and directly correlated with proteinuria [29]. Besides, podocytes are relatively resistant to ERT as GL-3 deposits can persist post ERT [30]. In vitro studies generating transient and stable GLA knockout FD podocyte cell lines [31, 32] have shown dysregulated cellular signaling pathways in human podocytes. IVS4+1326C>T identified in our case was associated with mild α-GalA enzyme deficiency that manifested predominantly as Fabry podocytopathy. One recent study also found that slightly decreased α-GalA activity may be associated with patients with focal segmental glomerulosclerosis [33]. However, residual α-GalA activity analyzed in peripheral leukocytes of our patient of 75% of normal activity was much higher than the cut-off value for defining a ‘mild’ GLA mutation (≥ 20% of mean normal level of α-GalA activity) [34]. Although such high REAs were also observed in other renal variant patients associated with GLA missense mutations (Met42Leu [5]; Pro210Ser [10]) and the renal involvement was limited to podocyte pathology, the true threshold level of REA below which could induce podocyte injury remains to be elucidated and the identification of other patients carrying IVS4+1326C>T could help further verify the mild α-GalA deficiency phenotype of Fabry podocytopathy caused by IVS4+1326C>T.
The high residual α-GalA activity associated with IVS4+1326C>T may attribute to the expression of a minor full-length GLA transcript as well as the two aberrantly spliced GLA transcripts encoding deficient α-GalA as our in vitro study found that each had about 25% of expressed wile-type α-GalA activity. More severe enzymatic phenotypes were observed for two previously reported deep intronic GLA mutations in intron 4. IVS4 + 919G > A was first described in a Japanese man with the late-onset cardiac variant of FD [25], and it was later found to be prevalent among Taiwanese patients with hypertrophic cardiomyopathy [35]. IVS4 + 919G > A lies close to the 5’ splice site of the GLA pseudoexon, so it disrupts the binding of the nuclear ribonucleoproteins A1 and A2/B1 to the exon splicing silencer that overlaps with the 5’ splice site and that normally prevents the 57-bp pseudoexon inclusion [36]. Consequently, IVS4 + 919G > A induces the overexpression of an aberrantly spliced GLA mRNA (less than 5% of the total GLA mRNA in normal controls whereas up to 90% of the total GLA mRNA in patient carrying IVS4 + 919G > A) that retains the 57-bp pseudoexon, which introduced a PTC predicted to code for a shorter protein devoid of enzyme activity. The remainder of the normal GLA mRNA was transcribed to result in about 10% of normal α-GalA activity. Later on, IVS4 + 861C > T was identified in an Italian family presenting with the classic phenotype of FD and nearly no REA [26]. IVS4 + 861C > T lies 5 bp upstream of the alternative 3’ splice site of the 57-bp pseudoexon, and it is predicted to create a new exon splicing enhancer that overexpresses the aberrant transcript containing the 57-bp pseudoexon predominantly and produces negligible full-length GLA transcript. The three deep intronic GLA mutations with prominent clinical heterogeneity highlight the contribution of aberrant splicing of GLA transcripts to FD and also implies that approaches to restore normal splicing of GLA transcripts may benefit patients carrying deep intronic GLA mutations.
Patients diagnosed with renal variant of FD have variable speed of renal progression. Some arrive at ESRD already in their 40 s, such as those with FD involving the mutations p.Phe113Leu [8] or p.Ala143Thr [9] while others may presented as isolated proteinuria detected by accident at an advanced age, such as patients with FD involving mutations p.Met42Leu [5] or p.Phe229Val [11]. Genotype and phenotype variations also manifest in that GLA mutations associated with renal variant of FD have been identified in FD patients with the cardiac variant or the classic phenotype (Fig. 2B). How genotype influences disease phenotype remains unclear and likely to be complex, probably depending on interactions among GLA, other potential modifying genes, environmental factors, and in the case of women, skewed X inactivation. The presence of other renal disease superimposed upon Fabry nephropathy may also modify prognosis. As late-onset FD sometimes involves both the renal and cardiovascular system [37], we cannot fully exclude concomitant latent cardiomyopathy since we did not perform endomyocardial biopsy in our patient. Besides, some organ involvement may be delayed in ‘later-onset’ phenotypes of FD. As our patient’s disease progresses, he may develop cardiac and cerebrovascular symptoms since he was only 36 years old at the most recent follow-up.
Conclusion
We identified a novel deep intronic GLA mutation (IVS4+1326C>T) in a 33-year-old Chinese man with a mild α-GalA deficiency phenotype of Fabry podocytopathy and establishes a link between deep intronic GLA mutation and the renal variant of FD. Our research extends the mutation spectrum in GLA gene and justifies future studies of how IVS4+1326C>T and potentially other deep intronic GLA mutations contribute to Fabry podocytopathy through aberrant splicing. Future studies should assess the true incidence of the IVS4+1326C>T mutation in patients with different variants of FD, which may improve genetic diagnosis and personalized treatment.
Availability of data and materials
Data cannot be made publicly available due to ethical reasons. In the area of rare diseases, information about the diagnosis in combination with personal information may compromise anonymity and confidentiality of the participants. The Ethics Committee from Xin Hua Hospital, Shanghai Jiao Tong University School of Medicine assessed our research project beforehand. The ethics vote allows sharing data with eligible researchers but we do not have approval to share the data publicly. Researchers interested in getting access to the data should feel free to contact the corresponding author (linfujun@xinhuamed.com.cn).
References
Ortiz A, Cianciaruso B, Cizmarik M, Germain DP, Mignani R, Oliveira JP, Villalobos J, Vujkovac B, Waldek S, Wanner C, et al. End-stage renal disease in patients with Fabry disease: natural history data from the Fabry Registry. Nephrol Dial Transplant. 2010;25(3):769–75.
Nagata A, Nasu M, Kaida Y, Nakayama Y, Kurokawa Y, Nakamura N, Shibata R, Hazama T, Tsukimura T, Togawa T, et al. Screening of Fabry disease in patients with chronic kidney disease in Japan. Nephrol Dial Transplant. 2021;37(1):115–25.
Nakao S, Kodama C, Takenaka T, Tanaka A, Yasumoto Y, Yoshida A, Kanzaki T, Enriquez AL, Eng CM, Tanaka H, et al. Fabry disease: detection of undiagnosed hemodialysis patients and identification of a “renal variant” phenotype. Kidney Int. 2003;64(3):801–7.
Pan X, Ouyang Y, Wang Z, Ren H, Shen P, Wang W, Xu Y, Ni L, Yu X, Chen X, et al. Genotype: a crucial but not unique factor affecting the clinical phenotypes in Fabry disease. PLoS ONE. 2016;11(8):e0161330.
Rosenthal D, Lien YH, Lager D, Lai LW, Shang S, Leung N, Fervenza FC. A novel alpha-galactosidase a mutant (M42L) identified in a renal variant of Fabry disease. Am J Kidney Dis. 2004;44(5):e85-89.
Cybulla M, Schaefer E, Wendt S, Ling H, Kröber SM, Hövelborn U, Schandelmaier S, Rohrbach R, Neumann HP. Chronic renal failure and proteinuria in adulthood: Fabry disease predominantly affecting the kidneys. Am J Kidney Dis. 2005;45(5):e82-89.
Maruyama H, Miyata K, Mikame M, Taguchi A, Guili C, Shimura M, Murayama K, Inoue T, Yamamoto S, Sugimura K, et al. Effectiveness of plasma lyso-Gb3 as a biomarker for selecting high-risk patients with Fabry disease from multispecialty clinics for genetic analysis. Genet Med. 2019;21(1):44–52.
Vigneau C, Germain DP, Larmet D, Jabbour F, Hourmant M. Screening for Fabry disease in male patients with end-stage renal disease in western France. Nephrol Ther. 2021;17(3):180–4.
Merta M, Reiterova J, Ledvinova J, Poupetová H, Dobrovolny R, Rysavá R, Maixnerová D, Bultas J, Motán J, Slivkova J, et al. A nationwide blood spot screening study for Fabry disease in the Czech Republic haemodialysis patient population. Nephrol Dial Transplant. 2007;22(1):179–86.
Lin CJ, Chien YH, Lai TS, Shih HM, Chen YC, Pan CF, Chen HH, Hwu WL, Wu CJ. Results of Fabry disease screening in male pre-end stage renal disease patients with unknown etiology found through the platform of a chronic kidney disease education program in a Northern Taiwan Medical Center. Kidney Blood Press Res. 2018;43(5):1636–45.
Turkmen K, Guclu A, Sahin G, Kocyigit I, Demirtas L, Erdur FM, Sengül E, Ozkan O, Emre H, Turgut F, et al. The prevalence of fabry disease in patients with chronic kidney disease in Turkey: the TURKFAB study. Kidney Blood Press Res. 2016;41(6):1016–24.
Shimohata H, Ogawa Y, Maruyama H, Hirayama K, Kobayashi M. A renal variant of Fabry disease diagnosed by the presence of urinary mulberry cells. Intern Med. 2016;55(23):3475–8.
Choi JS, Kim CS, Park JW, Bae EH, Ma SK, Choi YD, Kim GH, Yoo HW, Kim SW. A novel small insertion mutation, C.1030_1031ins (T) in α-galactosidase A leads to renal variant fabry disease. Ren Fail. 2012;34(3):390–3.
Al-Salam S, Chaaban A, Al-Jasmi F, Amann K, Abouchacra S. Renal variant of Fabry disease with sporadic GLA gene mutation: role of early renal biopsy. Clin Kidney J. 2012;5(5):416–9.
Lin FJ, Yao L, Hu XQ, Bian F, Ji G, Jiang GR, Gale DP, Ren HQ. First identification of PODXL nonsense mutations in autosomal dominant focal segmental glomerulosclerosis. Clin Sci (Lond). 2019;133(1):9–21.
Identification of genetic causes for sporadic steroid-resistant nephrotic syndrome in adults. Kidney Int. 2018;94(5):1013–22.
Sadick N, Thomas L. Cardiovascular manifestations in Fabry disease: a clinical and echocardiographic study. Heart Lung Circ. 2007;16(3):200–6.
Zhao L, Tian Z, Fang Q. Diagnostic accuracy of cardiovascular magnetic resonance for patients with suspected cardiac amyloidosis: a systematic review and meta-analysis. BMC Cardiovasc Disord. 2016;16:129.
Nordin S, Kozor R, Baig S, Abdel-Gadir A, Medina-Menacho K, Rosmini S, Captur G, Tchan M, Geberhiwot T, Murphy E, et al. Cardiac phenotype of prehypertrophic fabry disease. Circ Cardiovasc Imaging. 2018;11(6):e007168.
Niemann M, Rolfs A, Störk S, Bijnens B, Breunig F, Beer M, Ertl G, Wanner C, Weidemann F. Gene mutations versus clinically relevant phenotypes: lyso-Gb3 defines Fabry disease. Circ Cardiovasc Genet. 2014;7(1):8–16.
Smirnova A, Di Toro A, Giuliani L, Tagliani M, Urtis M, Favalli V, Arbustini E. Renal and brain complications in GLA p.Phe113Leu Fabry disease. Eur J Med Genet. 2020;63(2):103703.
Cakar N, Barman H. Evaluation of echocardiographic findings of fabry patients: a single center experience. Ann Med Res. 2020;27(2):678–81.
Duro G, Zizzo C, Cammarata G, Burlina A, Burlina A, Polo G, Scalia S, Oliveri R, Sciarrino S, Francofonte D, et al. Mutations in the GLA gene and LysoGb3: is it really Anderson-Fabry disease? Int J Mol Sci. 2018;19(12):3726.
Yasuda M, Shabbeer J, Osawa M, Desnick RJ. Fabry disease: novel alpha-galactosidase A 3’-terminal mutations result in multiple transcripts due to aberrant 3’-end formation. Am J Hum Genet. 2003;73(1):162–73.
Ishii S, Nakao S, Minamikawa-Tachino R, Desnick RJ, Fan JQ. Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am J Hum Genet. 2002;70(4):994–1002.
Filoni C, Caciotti A, Carraresi L, Donati MA, Mignani R, Parini R, Filocamo M, Soliani F, Simi L, Guerrini R, et al. Unbalanced GLA mRNAs ratio quantified by real-time PCR in Fabry patients’ fibroblasts results in Fabry disease. Eur J Hum Genet. 2008;16(11):1311–7.
Nowak A, Murik O, Mann T, Zeevi DA, Altarescu G. Detection of single nucleotide and copy number variants in the Fabry disease-associated GLA gene using nanopore sequencing. Sci Rep. 2021;11(1):22372.
Higuchi T, Kobayashi M, Ogata J, Kaneshiro E, Shimada Y, Kobayashi H, Eto Y, Maeda S, Ohtake A, Ida H, et al. Identification of cryptic novel α-galactosidase A gene mutations: abnormal mRNA splicing and large deletions. JIMD Rep. 2016;30:63–72.
Najafian B, Tøndel C, Svarstad E, Gubler MC, Oliveira JP, Mauer M. Accumulation of globotriaosylceramide in podocytes in Fabry nephropathy is associated with progressive podocyte loss. J Am Soc Nephrol. 2020;31(4):865–75.
Thurberg BL, Rennke H, Colvin RB, Dikman S, Gordon RE, Collins AB, Desnick RJ, O’Callaghan M. Globotriaosylceramide accumulation in the Fabry kidney is cleared from multiple cell types after enzyme replacement therapy. Kidney Int. 2002;62(6):1933–46.
Liebau MC, Braun F, Höpker K, Weitbrecht C, Bartels V, Müller RU, Brodesser S, Saleem MA, Benzing T, Schermer B, et al. Dysregulated autophagy contributes to podocyte damage in Fabry’s disease. PLoS ONE. 2013;8(5): e63506.
Jehn U, Bayraktar S, Pollmann S, Van Marck V, Weide T, Pavenstädt H, Brand E, Lenders M. α-galactosidase a deficiency in Fabry disease leads to extensive dysregulated cellular signaling pathways in human podocytes. Int J Mol Sci. 2021;22(21):256.
Hasbal NB, Caglayan FB, Sakaci T, Ahbap E, Koc Y, Sevinc M, Ucar ZA, Unsal A, Basturk T. Unexpectedly high prevalence of low alpha-galactosidase A enzyme activity in patients with focal segmental glomerulosclerosis. Clinics (Sao Paulo). 2020;75: e1811.
Lukas J, Scalia S, Eichler S, Pockrandt AM, Dehn N, Cozma C, Giese AK, Rolfs A. Functional and clinical consequences of novel α-galactosidase a mutations in Fabry disease. Hum Mutat. 2016;37(1):43–51.
Hsu TR, Sung SH, Chang FP, Yang CF, Liu HC, Lin HY, Huang CK, Gao HJ, Huang YH, Liao HC, et al. Endomyocardial biopsies in patients with left ventricular hypertrophy and a common Chinese later-onset Fabry mutation (IVS4 + 919G > A). Orphanet J Rare Dis. 2014;9:96.
Palhais B, Dembic M, Sabaratnam R, Nielsen KS, Doktor TK, Bruun GH, Andresen BS. The prevalent deep intronic c.639+919 G>A GLA mutation causes pseudoexon activation and Fabry disease by abolishing the binding of hnRNPA1 and hnRNP A2/B1 to a splicing silencer. Mol Genet Metab. 2016;119(3):258–69.
Lin HY, Huang CH, Yu HC, Chong KW, Hsu JH, Lee PC, Cheng KH, Chiang CC, Ho HJ, Lin SP, et al. Enzyme assay and clinical assessment in subjects with a Chinese hotspot late-onset Fabry mutation (IVS4+919G→A). J Inherit Metab Dis. 2010;33(5):619–24.
Acknowledgements
Not applicable.
Funding
This work was supported by a Clinical Research Study Grant from the Shanghai Hospital Development Center (SHDC12018X07), a Research Grant from the Shanghai Municipal Health Commission (201940255), a Research Grant from the Youth Science and Technology Innovation Studio of Shanghai Jiao Tong University School of Medicine, a Research Grant from the Interdisciplinary Innovative Talent Training Program of Shanghai Jiao Tong University and a Research Grant from the Shanghai Scientific and Technological Committee (19411968000).
Author information
Authors and Affiliations
Contributions
XTD and XZ drafted the initial manuscript and performed the molecular experiments. FJL and XXP were responsible for diagnosis, recruitment, clinical characterization and blood collection for all subjects studied. FJL,WL and GRJ were responsible for the concept and design of the study as well as analyzed and interpreted the data. FJL review the final manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from the involved subjects and all procedures were performed in accordance with the Declaration of Helsinki. The study was performed under the approval of the Ethics Committee from Xin Hua Hospital, Shanghai Jiao Tong University School of Medicine (approval no. XHEC-D-2020-002). All participants gave informed consent.
Consent for publication
All authors agreed to the submission of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Primer sequences for genomic DNA sequencing ,RT-PCR, qRT-PCR and PHAGE-GLA plasmid construction.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Dai, X., Zong, X., Pan, X. et al. Identification and functional characterization of the first deep intronic GLA mutation (IVS4+1326C>T) causing renal variant of Fabry disease. Orphanet J Rare Dis 17, 237 (2022). https://doi.org/10.1186/s13023-022-02377-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13023-022-02377-8