Abstract
Objective
To identify the pathogenic mechanism of the c.244G>T mutation in NR5A1 gene found in a Chinese patient with 46, XY disorders of sex development (DSD). Subjects and methods: Genomic DNA was extracted from a Chinese 46, XY DSD patient. Targeted next-generation and Sanger sequencing were performed to investigate and validate the gene mutation causing 46, XY DSD, respectively. In silico tools were used to predict the pathogenicity of the variant. Dual luciferase reporter gene assay and minigene splicing reporter assay were used to identify the pathogenicity of the variant.
Results
A novel heterozygous variant, c.244G>T (p.Ala82Ser), in NR5A1 gene was detected in the 46, XY DSD patient. Four of five silico tools predicting pathogenicity of missense variants indicated that the variant was pathogenic. However, in vitro functional study showed that p.Ala82Ser did not affect the transcriptional activity of NR5A1. In silico tools predicting the potential splicing loci revealed that c.244G>T led to aberrant splicing of NR5A1 RNA. Minigene splicing reporter assay confirmed that c.244G>T resulted in the deletion of exon2 or deletion of 19 nucleotides in 3′ end of exon2.
Conclusions
Mutation of c.244G>T in NR5A1 results in 46, XY DSD by inducing abnormal splicing of NR5A1 RNA instead of amino acid substitution of NR5A1.
Similar content being viewed by others
Introduction
46, XY disorders of sex development (DSD) is characterized by ambiguous external male or female genitalia with a 46, XY karyotype [1]. Depending on the etiology of the disease, 46, XY DSD is divided into disorders of gonadal development, disorders of androgen synthesis or action, and other defects, including severe hypospadias and cloacal exstrophy [2]. Chromosomal aberrations and gene mutations have been identified as the underlying causes of the disease. Currently, more than 60 genes have been linked to 46, XY DSD [3]. Specifically, mutations in NR5A1 are detected in about 10%-20% of 46, XY DSD patients.
Nuclear receptor subfamily 5 group A member 1 (NR5A1), also known as steroidogenic factor 1 (SF1), is a member of nuclear receptor family. It plays a crucial role in transcriptional regulation of genes involved in steroidogenesis, gonadal development, and reproduction [4]. NR5A1 gene is located on chromosome 9q33.3 and contains seven exons with one noncoding exon [5]. Mutations in NR5A1 gene commonly cause a wide range of phenotypes, including primary adrenal deficiency, disorders of sex development (DSD), male factor infertility, primary ovarian insufficiency, and spleen anomalies [6, 7]. More than 180 mutations in NR5A1 have been reported [7], and the most common phenotype of NR5A1 gene mutations is 46, XY DSD.
Nowadays, gene sequence analysis has widely been used to diagnose inherited diseases, leading to the identification of thousands of rare variants. Interpreting the pathogenicity of these variants has proven to be a big challenge. Common approaches tend to explicate the clinical significance of gene variants based on the substitution of amino acids. However, recent studies have shown that exonic single-nucleotide variants can affect RNA splicing [8,9,10]. Hence, the effects on both the amino acid substitution and RNA splicing need to be considered during the analysis of the pathogenicity of exonic single-nucleotide variants.
Here, we reported an exonic single-nucleotide variant of NR5A1 gene that caused 46, XY DSD by inducing abnormal splicing of NR5A1 RNA instead of amino acid substitution.
Results
Patient and genetic analysis
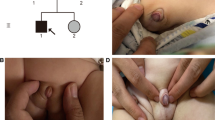
A 16-year-old girl was referred to our hospital because of clitoris hypertrophy and primary amenorrhea. The patient was born on the due date after an uneventful pregnancy. At birth, the external genitalia showed a female phenotype. At the age of 12 years, hirsutism and deepening of the voice were noted. She was admitted to the local hospital for karyotype testing, which showed a 46, XY karyotype. For further treatment, the patient was referred to our hospital. On physical examination, the patient’s breast showed Tanner stage 1, and external genitalia showed clitoris hypertrophy with a male phenotype with pubic hair of Tanner stage 6. There was neither consanguineous marriage nor similar disease in the family.
Hormone levels were as follows: luteinizing hormone, 35.64 IU/L; follicle-stimulating hormone, 93.91 IU/L; serum testosterone, 7.74 nmol/L (reference: 6.07–27.1 nmol/L); estradiol, 73.4 pmol/L (reference: 0–172.5 pmol/L); adrenocorticotrophic hormone, 33.5 pg/mL (0–46 pg/mL); and serum cortisol, 423.4 nmol/L (reference: 110.6–616.4 nmol/L). After discussing with the parents, the patient decided to continue in a female gender and underwent laparoscopic bilateral gonadectomy. No Müllerian structure was found and bilateral gonads were located in the pelvic cavity. Histology of the left and right gonad revealed dysplastic testicular tissue.
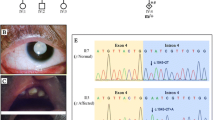
Next-generation sequencing revealed a novel heterozygous NR5A1 variant, c.244G>T, which had led to the change of the amino acid at the position 82 from Ala to Ser; Sanger sequencing confirmed the variant (Fig. 1). The variant of c.244G>T was absent in the dbSNP, ExAC, gnomAD, and Ensembl database.
In silico analysis of NR5A1 variants
The amino acid, Ala82, is highly conserved (Fig. 2). Functional predictions of the missense mutation by Sift, SNAP, SNP&GO, Polyphen2, MutPred, and REVEL are shown in Table 1. Sift, SNAP, SNP&GO, Polyphen2, and REVEL predicted the variant as deleterious, while MutPred predicted it as a natural variant.
In silico tools, HSF and Ex Skip, predicted that c.244G>T affected NR5A1 RNA splicing, while ESE Finder indicted a negative result. The splicing score calculated by MaxEntScan and Splice site score calculation is shown in Table 1.
Functional studies
Dual luciferase reporter gene assay
The transactivation activity of the c.244G>T mutants compared with that of the wild type was 118%, meaning that it was comparable to the wild type (Fig. 3), while the positive control G35E mutant showed a partial transactivation activity of 34%.
Minigene splicing reporter assay
The RT-PCR amplification products of c.244G>T mutant in minigene showed three fragments of different lengths by electrophoresis. One was of similar length to that in the wild type, while the other two were shorter (Fig. 4a). Sanger sequencing of the longest product showed that the wild type sequence came from the normal splicing (Fig. 4b). The sequencing of the medium product showed a loss of 19 nucleotide base pairs (GTGGGGATGCGCCTGGAAG) in 3′ end of exon2 (Fig. 4c), whereas the shortest product revealed a skipping of exon2 (Fig. 4d).
Minigene splicing reporter assay for the c.244G>T mutant. a RT-PCR products of minigene. 1, 2, 3 WT minigene products; 4, 5, 6 c.244G>T mutant minigene products. b Sanger sequencing of RT-products of WT; the line denotes the missing sequence in c. c Sanger sequencing of RT-products of the medium size RT-PCR products of c.244G>T mutant; the arrow indicates the breakpoint of the deletion sequence. d Sanger sequencing of RT-products of the shortest RT-PCR product of c.244G>T mutant
Discussion
In this study, a novel NR5A1 variant, c.244G>T (Ala82Ser), was detected in a Chinese patient with 46, XY DSD. The Ala82 is in the A-box of NR5A1, which is essential for stabilizing DNA binding by interacting with the DNA minor groove of target genes [11]. Four of five silico tools predicted that Ala82Ser was deleterious. In contrast, in vitro functional studies showed that Ala82Ser did not affect the transcriptional activity of NR5A1 to CYP11A1. However, minigene splicing reporter assay showed that c.244G>T led to abnormal splicing.
Over the last decade, as new DNA sequencing technologies have widely been used in clinical practice, the knowledge on novel variants associated with diseases has accumulated. About 50% of these variants are missense variants [12], and clarification of the significance of rare missense variants has been difficult. Generally, bioinformatic algorithms are adopted to analyze the pathogenicity of the variants. However, previous studies have reported the false-positive and false-negative rates of the prediction tools [13,14,15,16]. Thusberg et al. [13] reported that the accuracy of Sift, SNAP, SNP&GO, Polyphen2, and MutPred was 0.65, 0.72, 0.82, 0.71, and 0.81, respectively. Zou et al. [17] reported that using Polyphen2 to predict 24 pathogenic mutations classified two of the mutations as benign. Thus, functional studies are needed to identify the pathogenicity of variants. Similarly, in our study, the prediction results of in silico tools and the results of functional studies were in disagreement. Namely, while four of five in silico tools predicted Ala82Ser as a deleterious mutation, in vitro studies showed that mutant NR5A1 (Ala82Ser) had similar transcriptional activity to CYP11A1 compared with wild-type NR5A1.
Recent studies have shown that approximately 15%-50% of exon single-nucleotide variants can affect protein function by changing RNA splicing rather than by substituting amino acid [18, 19]. Ito et al. predicted the effect of 2390 rare synonymous and missense mutations reported in the database on RNA splicing and found that 196 mutations might affect the splicing process; minigene reporter assays confirmed that 53 mutations affect splicing [20]. Thus, considering that the typical clinical manifestations of the patient and the next-generation sequencing did not reveal other suspected pathogenic mutations, we further conducted minigene splicing reporter assay. The result showed that the mutation had led to another two types of RNA splicing, resulting in the alteration of the structure of NR5A1.
The classical splice sites of human genes are located at exon–intron boundaries, including a 5′ splice donor and a 3′ splice acceptor sequence [21]. The 5′ splice donor usually contains a 9-bp sequence with a classical GT dinucleotide, while the 3′ splice acceptor contains a 23-bp sequence with an AG dinucleotide [22]. Variants occurring within the 9- or 23-bp of the splice sites have been known to interrupt normal splicing process and lead to exon skip [23]. In this study, c.244G>T was found in the 5′ splice donor sequence of the exon2 in NR5A1, and the minigene splicing reporter assay confirmed that c.244G>T resulted in the deletion of exon2 or deletion of 19 nucleotides in 3′ end of exon2 by disrupting the splice donor, thereby altering the structure of NR5A1.
In conclusion, mutation of c.244G>T in NR5A1 results in 46, XY DSD by causing abnormal splicing of NR5A1 RNA rather than substitution of amino acid.
Patients and methods
Patients
Patients and methods have been described in detail previously [24]. Briefly, 60 patients with 46, XY DSD were admitted to the Endocrinology Department of Peking Union Medical College Hospital, Beijing between January 2010 and October 2015. The clinical data and laboratory test results were collected in detail.
Mutational analysis
Targeted next-generation sequencing and Sanger sequencing were used to identify and validate the gene mutation, respectively. The gene panel (NimblegenSeqCap EZ system, Roche, Basel, Switzerland) was designed to capture all exons and 50-bp flanking intron sequences of 83 DSD-related genes [24]. A variant was recognized as an underlying disease-causing variant when it was not found in dbSNP, ExAC, GnomAD, and Ensemble database and in 500 Chinese controls, or alternatively, when the allele frequency was found to be less than 0.001 in the database. The amino acid sequence alignment of human NR5A1 with that of other 14 species was performed using UniProt.
Dual-Luciferase reporter gene assay
Mutant NR5A1 eukaryotic expression vectors containing the c.244G>T variant and p.G35E (positive control) were generated by site-directed mutagenesis (Transgene, China). Luciferase reporter vectors comprising NR5A1-responsive promoters of human CYP11A1 in pGL3-basic (Promega, USA) were constructed to study the effect of gene variant on the transcriptional activity of NR5A1.
Transient gene expression assays for the assessment of NR5A1 function were performed in 96-well plates with the use of human embryonic kidney (HEK) 293 T cells and a Dual-Luciferase reporter assay system (Promega, USA). Wild-type or mutant NR5A1 expression vectors (2 ng/well) were co-transfected with reporters containing NR5A1-responsive promoters (CYP11A1) (100 ng/well). Luciferase assays were performed with the use of a luminometer reader (Berthold Centro XS3 LB 960, Germany). All data were standardized for Renilla activity, and the results are shown as mean ± standard error of mean of three independent experiments, each performed at least in triplicate.
In silico analysis
To predict the pathogenicity of the variant, five in silico tools were used, including Sift (http://sift.jcvi.org/), SNAP (http://rostlab.org/services/snap), SNP&GO (http://snps-and-go.biocomp.unibo.it/snps-and-go), Polyphen2 (http://genetics.bwh.harvard.edu/ pph2), MutPred (http://mutpred.mutdb.org), and REVEL (https://sites.google.com/site/revelgenomics/about).
Predictions of the presence of exonic splice enhancer or silencer sequences were made using Human Splicing Finder (HSF) (http://www.umd.be/HSF3/index.html) and ESE Finder (http://krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process = home). Ex SKIP (http://ex-skip.img.cas.cz/) was used to determine whether the variant leads to skipping of the exon. Splice scores for the natural and cryptic donor and acceptor splice sites were determined using the MaxEntScan (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) and Splice Site Score Calculation (http://rulai.cshl.edu/new_alt_exon_db2/HTML/score.html).
Minigene splicing reporter assay
Exon1 with 125 flanking 5′ UTR bases, intron1, and exon2 with 120 flanking nucleotides of the NR5A1 gene were amplified from the genomic DNA of the patient and a healthy control (total product size: 577 bp) by polymerase chain reaction (PCR), and then the PCR products were inserted between the BamHI and MluI restriction sites of the minigene vector to construct mutant and wild-type pCAS2 minigene vector [9], respectively.
Transfection of pCAS2 minigene constructs into the human embryonic kidney (HEK) 293 T cells was performed in 12-well plates. Forty-eight hours after the transfection, the cells were lysed and total RNA was extracted using the Total RNA Kit (OMEGA, USA). Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using the PrimeScript™ RT reagent kit (Takara, Japan). PCR amplification was performed by 2 × Hieff™ PCR Master Mix (Yeasen) using the pCAS2-RT-F and pCAS2-RT-R primers. Reverse-transcription (RT) PCR products of different lengths were separated by agarose gel electrophoresis (2%), purified, and subjected to Sanger sequencing.
The sequencing results were compared with the NR5A1 DNA sequence (NG_008176.1) and cDNA sequence (NM_004959.5) to determine whether the c.244G>T affected the splicing of NR5A1 RNA.
Availability of data and materials
Not applicable.
References
Ahmed SF, Bashamboo A, Lucas-Herald A, McElreavey K. Understanding the genetic aetiology in patients with XY DSD. Br Med Bull. 2013;106:67–89.
Hughes IA, Houk C, Ahmed SF, Lee PA. Consensus statement on management of intersex disorders. J Pediatr Urol. 2006;2:148–62.
Audi L, Ahmed SF, Krone N, Cools M, McElreavey K, Holterhus PM, et al. Genetics in endocrinology: approaches to molecular genetic diagnosis in the management of differences/disorders of sex development (DSD): position paper of EU COST Action BM 1303 ‘DSDnet.’ Eur J Endocrinol. 2018;179:R197-r206.
El-Khairi R, Achermann JC. Steroidogenic factor-1 and human disease. Semin Reprod Med. 2012;30:374–81.
Hoivik EA, Lewis AE, Aumo L, Bakke M. Molecular aspects of steroidogenic factor 1 (SF-1). Mol Cell Endocrinol. 2010;315:27–39.
Colson C, Aubry E, Cartigny M, Remy AA, Franquet H, Leroy X, et al. SF1 and spleen development: new heterozygous mutation, literature review and consequences for NR5A1-mutated patient’s management. Clin Genet. 2017;92:99–103.
Suntharalingham JP, Buonocore F, Duncan AJ, Achermann JC. DAX-1 (NR0B1) and steroidogenic factor-1 (SF-1, NR5A1) in human disease. Best Pract Res Clin Endocrinol Metab. 2015;29:607–19.
Goursaud C, Mallet D, Janin A, Menassa R, Tardy-Guidollet V, Russo G, et al. Aberrant splicing is the pathogenicity mechanism of the p.Glu314Lys Variant in CYP11A1 Gene. Front Endocrinol (Lausanne). 2018;9:491.
Geng X, Liu Y, Ren X, Guan Y, Wang Y, Mao B, et al. Novel NTRK1 mutations in Chinese patients with congenital insensitivity to pain with anhidrosis. Mol Pain. 2018;14:1744806918781140.
Fraile-Bethencourt E, Valenzuela-Palomo A, Diez-Gomez B, Acedo A, Velasco EA. Identification of eight spliceogenic variants in BRCA2 Exon 16 by minigene assays. Front Genet. 2018;9:188.
Lin L, Achermann JC. Steroidogenic factor-1 (SF-1, Ad4BP, NR5A1) and disorders of testis development. Sex Dev. 2008;2:200–9.
Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, et al. The human gene mutation database: 2008 update. Genome Med. 2009;1:13.
Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–68.
Gnad F, Baucom A, Mukhyala K, Manning G, Zhang Z. Assessment of computational methods for predicting the effects of missense mutations in human cancers. BMC Genomics. 2013;14(Suppl 3):S7.
Rodrigues C, Santos-Silva A, Costa E, Bronze-da-Rocha E. Performance of in silico tools for the evaluation of UGT1A1 missense variants. Hum Mutat. 2015;36:1215–25.
Luxembourg B, D’Souza M, Körber S, Seifried E. Prediction of the pathogenicity of antithrombin sequence variations by in silico methods. Thromb Res. 2015;135:404–9.
Zou M, Baitei EY, Alzahrani AS, Parhar RS, Al-Mohanna FA, Meyer BF, et al. Mutation prediction by PolyPhen or functional assay, a detailed comparison of CYP27B1 missense mutations. Endocrine. 2011;40:14–20.
Soukarieh O, Gaildrat P, Hamieh M, Drouet A, Baert-Desurmont S, Frebourg T, et al. Exonic splicing mutations are more prevalent than currently estimated and can be predicted by using in silico tools. PLoS Genet. 2016;12:e1005756.
Baralle D, Buratti E. RNA splicing in human disease and in the clinic. Clin Sci (Lond). 2017;131:355–68.
Ito K, Patel PN, Gorham JM, McDonough B, DePalma SR, Adler EE, et al. Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc Natl Acad Sci USA. 2017;114:7689–94.
Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–48.
Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucl Acids Res. 1987;15:7155–74.
Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, et al. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–8.
Yu B, Liu Z, Gao Y, Mao J, Wang X, Hao M, et al. Novel NR5A1 mutations found in Chinese patients with 46, XY disorders of sex development. Clin Endocrinol (Oxf). 2018;89:613–20.
Acknowledgements
We thank the subject for participation in this study and Prof. Xiuli Zhao for a gift of the pCAS2 vector.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81971375 and 81771576), National Key Research and Development Program of China (2016YFC0905100), CAMS Innovation Fund for Medical Sciences (2016-I2M-1-002), and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2017PT32020, No. 2018PT32001).
Author information
Authors and Affiliations
Contributions
X.Y. Wu and M. Nie conceived and designed the experiments. X.Y. Wu, J.F. Mao, and X. Wang collected blood samples and clinical data. B.Q. Yu and Y.J. Gao performed the experiments. B.Q. Yu wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Each patient gave the informed written consent for the study. The study protocol was reviewed and approved by the Peking Union Medical College Hospital ethics committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yu, B., Gao, Y., Mao, J. et al. Mutation of c.244G>T in NR5A1 gene causing 46, XY DSD by affecting RNA splicing. Orphanet J Rare Dis 16, 370 (2021). https://doi.org/10.1186/s13023-021-02002-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13023-021-02002-0