Abstract
Background
Tuberous sclerosis complex (TSC) is a genetic disorder that cause tumors to form in many organs. These lesions may lead to epilepsy, autism, developmental delay, renal, and pulmonary failure. Loss of function mutations in TSC1 and TSC2 genes by aberrant activation of the mechanistic target of rapamycin (mTORC1) signaling pathway are the known causes of TSC. Therefore, targeting mTORC1 becomes a most available therapeutic strategy for TSC. Although mTORC1 inhibitor rapamycin and Rapalogs have demonstrated exciting results in the recent clinical trials, however, tumors rebound and upon the discontinuation of the mTORC1 inhibition. Thus, understanding the underlying molecular mechanisms responsible for rapamycin-induced cell survival becomes an urgent need. Identification of additional molecular targets and development more effective remission-inducing therapeutic strategies are necessary for TSC patients.
Results
We have discovered an Mitogen-activated protein kinase (MAPK)-evoked positive feedback loop that dampens the efficacy of mTORC1 inhibition. Mechanistically, mTORC1 inhibition increased MEK1-dependent activation of MAPK in TSC-deficient cells. Pharmacological inhibition of MAPK abrogated this feedback loop activation. Importantly, the combinatorial inhibition of mTORC1 and MAPK induces the death of TSC2-deficient cells.
Conclusions
Our results provide a rationale for dual targeting of mTORC1 and MAPK pathways in TSC and other mTORC1 hyperactive neoplasm.
Similar content being viewed by others
Background
Tuberous sclerosis complex (TSC) is a genetic disorder that is associated with tumors to form in many organs, primarily in the brain, eyes, heart, kidney, skin and lungs [1]. These lesions cause morbidity and mortality in patients with TSC, as they may lead to epilepsy, autism, developmental delay, renal, and pulmonary failure [2]. Loss of function mutations in TSC1 and TSC2 genes are the known causes of TSC. The TSC1 and TSC2 gene products combine with TBC1D7 to form a ternary complex which have GTPase activating protein (GAP) activity for the GTPase Ras homologue enriched in brain (Rheb), therefore inhibiting mTOR complex 1 (mTORC1) kinase activity [3, 4]. Therefore, Targeting mTORC1 becomes a most available therapeutic strategy for TSC.
The mechanistic target of rapamycin (mTOR) is a serine/threonine protein kinase that regulates cell growth, proliferation, cell motility, cell survival, protein synthesis, autophagy, and transcription [5]. The mTOR functions as a catalytic subunit in two distinct multiprotein complexes, mTORC1 and mTORC2 [6]. mTORC1, a complex including regulatory-associated protein of mTOR (RAPTOR), phosphorylates and controls, at least, two regulators of protein synthesis, the 40S ribosomal protein subunit S6 kinase (S6K) and the translational repressor 4E-binding protein 1, referred as 4E-BP1. mTORC2, characterized by rapamycin-insensitive companion of mTOR (RICTOR), phosphorylates several AGC protein kinases, including AKT at Ser473. Deregulation of mTORC1 has been observed with various human diseases [7]. Thus, this renders mTORC1 as an attractive drug target for cancer therapy. Although mTORC1 inhibitors showed very convincing results in some TSC clinical studies, tumors or lung function returned to their original states when drugs were discontinued, addressing the cytostatic instead of cytotoxic effects of mTORC1 inhibition [8,9,10]. Thus, there is an urgent need to identify additional molecular targets and develop novel combinatorial therapies with mTORC1 inhibitors that could render tumor cell death.
To explore the possibility of selectively killing tumor cells with high mTORC1 activity, we performed bioinformatic analysis and identified signaling pathways that were activated in response to rapamycin treatment, including focal adhesion, adherent junction, Jak-Stat, and MAPK signaling pathways. Recently, the FAK inhibitor and JAK-STAT inhibitor have shown benefits in mTORC1 inhibitor-resistant pancreatic cancer and breast cancer, respectively [11, 12]. MAPK inhibitors have been studied with a synergistic effect with mTOR inhibitors in several cancers [13, 14]. However, the mechanism of MAPK inhibitor-attenuated resistance to mTORC1 inhibition in cancers and especially in TSC have not been extensively explored.
Here we report that mTORC1 inhibition results in a compensatory activation of MAPK signaling pathway in TSC-deficient cells in vitro. This enhanced MPAK signaling pathway was associated with enhanced survival of TSC-deficient cells. Pharmacological suppression of MEK1/2-MAPK sensitized TSC-deficient cells to cell death. Taken together, our study reveals a novel approach of combined suppression of pro-survival signaling pathways that informs future preclinical studies and potential clinical application of remission-inducing therapies for TSC and other mTOR1 hyperactive neoplasms.
Results
MAPK signaling pathway is activated in response to rapamycin treatment
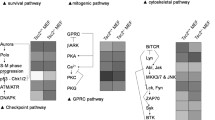
To explore the possibility of selectively killing tumor cells with high mTORC1 activity, we performed bioinformatic analysis using various tumor cells including TSC1 and TSC2-deficient cells (GEO accession number GSE16944 [15], GSE21755 [16], GSE5332 [17], GSE27982 [18], GSE28021 [18], GSE67529, GSE28992 [19], GSE18571 [20], GSE7344 [21], GSE37129 [22] and GSE17662 [23]) (Fig. 1a). Gene set enrichment analysis identified top 10 up-regulated signaling pathways in resposne to rapamycin treatment that were conserved in all cell types analysed (Fig. 1b). MAPK signaling pathway is one of the upregulated pathways induced by rapamycin treatment. Other rapamycin-upregulated pathways include axon guidance, notch signaling pathway, small cell lung cancer, adherent junction, B cell receptor signaling pathway, chemokine signaling pathway, ECM receptor interaction, focal adhesion, and JAK/STAT signaling pathway.
Bioinfomatic analysis of rapamycin enhanced signaling pathway in TSC deficient cells. a Publically available gene expression datasets were re-analyzed. b Gene set enrichment analysis was performed. Top 10 upregulated signaling pathways in response to rapamycin treatment relative to vehicle-treatment were indicated
Tsc2-deficient xenograft tumors become refractory to rapamycin treatment
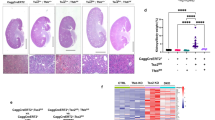
To determine the in vivo efficacy of rapamycin on tumor growth, we first generated xenograft tumors of Tsc2-deficient Eker Rat uterine leiomyoma-derived luciferase-tagged cells [24,25,26]. The tumor growth was recorded by non-invasive imaging. Rapamycin treatment for one-week resulted in drastic decrease of tumor volume dramatically due to one-week rapamycin treatment. However, tumor rebounded rapidly despite rapamycin treatment was continued for 1 week (Fig. 2a). The tumor growth was monitored for 5 weeks during rapamycin treatment. Interestingly, xenograft tumors persistently progressed from week 2 of the treatment (Fig. 2b). By week 5 of rapamycin treatment, tumors became ulcerated and reached the study endpoints. We performed immunohistochemistry using cell proliferative marker proliferating cell nuclear antigen (PCNA) and found that rapamycin-treated xenograft tumors exhibited high levels of nuclear PCNA staining, comparable to those detected in control tumors (Fig. 2c), indicating that rapamycin does not affect cell proliferative status. To determine the effect of rapamycin on tumor cell death, TUNEL staining was performed in the same set xenograft tumor specimens used for PCNA staining. We did not observe positive TUNEL staining in xenograft tumors of control vehicle-treated or rapamycin-treated mice (Fig. 2c), indicating that rapamyicn does not induce the death of tumor cells. To assess the effect of rapamycin on mTORC1 inhibition in xenograft tumors, we performed immunoblotting analysis and found that S6 phosphorylation was markedly decreased in response to rapamycin treatment in xenograft tumors relative to vehicle control (Fig. 2d). Collectively, our data show that long-term effective inhibition of mTORC1 by rapamycin promotes tumor refractory growth in TSC.
Tsc2-deficient xenograft tumors become refractory to rapamycin treatment. a Female CB17-scid mice were inoculated with ELT3-luciferase cells subcutaneously. Mice were treated with either vehicle or rapamycin for 5 weeks. b Bioluminescent intensity in xenograft tumors was recorded and quantified weekly. The left Y-axis indicated the relative tumor growth versus the baseline quantification before drug treatment. c Immunohistochemistry staining of PCNA and TUNEL. d Immunoblotting analysis of phospho-S6 (S235/236) of xenograft tumors
Rapamycin is cytostatic but not cytotoxic in TSC2-deficient cells in vitro
To determine whether rapamycin affects cell proliferation and death in vitro, we use TSC2-deficient patient-derived cells [27, 28], Tsc2-deficient rat uterine leiomyoma-derived cells [24, 25], and Tsc2−/−p53−/− mouse embryonic fibroblasts (MEF) and their TSC2-exressing counterpart controls [29]. Crystal Violet staining showed that rapamycin treatment up to 96 h (1 nM to 100 nM) significantly decreased cell proliferation relative to vehicle control (Fig. 3a). Phase-contrast microscopy showed rapamycin slowed the growth of TSC2-deficient cells without inducing the death of TSC2-deficient patient-derived cells, rat uterine-leiomyoma-derived cells, or Tsc2−/− MEFs (Fig. 3b). Furthermore, Propidium iodide exclusion assay showed that rapamycin treatment (1 nM – 100 nM) did not induce the death of TSC2-deficient patient-derived 621–101 cells (Fig. 3c). Collectively, our data demonstrate that rapamycin exhibits cytostatic effect but not cytocidal effect on TSC2-deficient cells in vitro.
Rapamycin is cytostatic but not cytotoxic in vitro. a Cell proliferation of TSC2-deficient patient-derived cells treated with escalating concentrations of rapamycin. b Phase contrast microscopy of TSC2-deficient patient-derived, rat uterine leiomyoma-derived, Tsc2−/− MEFs and Tsc1−/− MEFs treated with rapamycin. c Propidium iodide exclusion assay of TSC2-deficient patient-derived cells treated with escalating concentrations of rapamycin as indicated. ** P < 0.01, the Student’s t-test
Rapamycin promotes MAPK activation in mTORC1-hyperactive tumor cells in vitro
Our bioinformatic analysis identified activation of MAPK signaling pathway genes in a panel of TSC-deficient cells (Fig. 1). To further assess the MAPK activation, we performed immunoblotting analysis of MAPK phosphorylation in TSC2-deficient patient-derived cells, Tsc2-deficient rat uterine-leiomyoma-derived cells, Tsc2−/− mouse embryonic fibroblast (MEFs), and Tsc1−/− MEFs, and their TSC2- or TSC1-reexpressing counterparts cultured in nutrient-rich medium containing 10% FBS or nutrient-deprived FBS-free medium, repersenting two basal levels of MAPK phosphorylation. We found that rapamycin selectively promoted MAPK phosphorylation in TSC1- or TSC2-deficient cells but not in TSC1- or TSC2-reexpressing cells (Fig. 4a-d). We also observed that rapamycin treatment decreased S6 phosphorylation as expected.
Rapamycin promotes MAPK signaling pathway in TSC2 deficient cells. Immunoblots of TSC2-deficient (a) patient-derived cells, b rat uterine leiomyoma-derived cell, c Tsc2−/− MEFs, and d Tsc1−/− MEFs, treated with rapamycin at indicated time points. Immunoblotting analyses of TSC2, phospho-MAPK (T202/Y204), and phospho-S6 (S235/236) were shown
Dual inhibition of mTORC1 and MAPK induces the death of TSC2-deficient patient-derived cells in vitro
To test whether dual inhibition of mTORC1 and MAPK synergistically affects cell survival, we first examined cell viability using crystal violet staining. Rapamycin single treatment decreased the viability of TSC2-deficient patient-derived cells (Fig. 5a), but not TSC2-reexpressing cells (Fig. 5b). Importantly, dual treatment of rapamycin and CI-1040, an MEK1/2 inhibitor, significantly decreased the viability of TSC2-deficient cells, and moderately reduced the viability of TSC2-reexpressing patient-derived cells, relative to rapamycin treatment alone (Fig. 5a, b). However, rapamycin plus AZD6244, an MEK1 inhibitor, did not affect the viability of TSC2-deficient or TSC2-reexpressing patient-derived cells (Fig. 5a, b), indicative of a differential effect of MEK1/2 and MEK1 on cell viability in TSC2-deficient patient-derived cells.
Combinational suppression of mTORC1 and MAPK induces cell death in vitro. TSC2-deficient or TSC2 re-expressing patient-derived cells were treated with vehicle control, rapamycin, or rapamycin combined with AZD6244 or CI-1040. a-b Cell viability was exmained using crystal violet staining assay (n = 8). c-d Cell death was quantified using Propidum iodine excursion assay (n = 8). * P < 0.05; ** P < 0.01, the Student’s t-test
To determine the combinatorial effect of rapamycin and MEK1/2 inhibitor on cell survival, we preformed Propidium iodide exclusion assay and found that CI-1040 in combination with rapamycin substantially induced cell death relative to rapamycin treatment in TSC2-deficient and TSC2-reexpressing patient-derived cells (Fig. 5c, d). AZD6244 in combination with rapamycin moderately induced the death of TSC2-deficient and TSC2-reexpresing patient-derived cells (Fig. 5c, d), further indicating the differentially effect of MEK1/2 and MEK1 on the survival of TSC2-deficient patient-derived cells.
Discussion
The mTORC1 is a serine/threonine protein kinase and plays crucial roles in transcriptional regulation, initiation of protein synthesis, ribosome biogenesis, metabolism, and apoptosis. The deregulation of mTORC1 signaling pathway is frequently observed in cancers and other diseases due to aberrant expression of numerous oncogenes and tumor suppressors [5, 30]. mTORC1 signaling pathway has been the key targets for cancer treatment [31,32,33]. Although mTORC1 inhibitors have activity in some cancer types, only small population of patients treated with these agents exhibited substantial clinical benefit [34].
mTOR1 pathway is the main therapeutic target for TSC and LAM patients. mTORC1 inhibitors, sirolimus (rapamycin) and everolimus (RAD001), have been approved by FDA for the treatment of TSC-associated subependymal Giant cell astrocytoma in brain (everolimus) [2, 35], renal angiomyolipoma (everolimus) [36], and pulmonary lymphangioleiomyomatosis (sirolimus) [10]. Everolimus has also been approved for the treatment of TSC-associated SEGA and renal angiomyolipoma [37]. Rapamycin (sirolimus) acts by forming complex with the intracellular binding protein FK506-binding protein (FKBP121), such complex in turn binds to the FKBP12-rapamycin binding (FRB) domain of the mTORC1 molecule to inhibit mTORC1 activity [38, 39]. mTORC2 function is intact under acute inhibition, however, it has been noted that long-term rapamycin treatment decreases mTORC2 signaling in primary human dermal microvascular endothelial cells [40] and several cell lines [41]. Everolimus, known as RAD001, is a derivative of sirolimus that acts via similar mechanism [42]. It shares the central macrolide chemical structure with sirolimus, which allows for interaction with FKBP12 [43, 44]. The tissue selectivity of everolimus has also been noted, preferably accumulated in brain mitochondria relative to sirolimus [44].
Currently, there is no single study that directly compares the therapeutic effect of sirolimus and everolimus in TSC management [38, 44, 45]. Clinical decisions are based on clinical trial experiences in the setting of certain TSC manifestations. Sirolimus is generally used to manage TSC-LAM [10], while Everolimus is favored over sirolimus in treating SEGA [44, 46]. Although our current studies focus on the impact of rapamycin on pro-survival of TSC mutant cells, it will be interesting to examine the effect of everolimus on the survival of TSC mutant cells.
Recently, the therapeutic benefit of cannabidiol has been proposed in TSC associated epilepsy [47]. Cannabidiol is a marijuana plant extract that has been studied as an anticonvulsant medicine for treatment-resistant epilepsy with acceptable tolerance [48, 49]. Hess and colleague observed decreased weekly seizure frequency in TSC patients with refractory epilepsy under cannabidiol treatment. In addition to the fact that cannabidiol is yet to be FDA approved, there is no conclusive evidence supporting the effect of cannabidiol exceeding traditional anti-seizure therapy such as the benzodiazepine, GABA analog vigabatrin and ketogenic diet in the management of TSC associated epilepsy [50,51,52,53]. Moreover, the specificity of cannabidiol to target the unique mechanism of TSC pathogenesis has not been elucidated.
Preclinical studies including ours have demonstrated the effectiveness of sirolimus, an mTORC1 inhibitor, in multiple animal models of TSC [26, 54,55,56,57,58]. The effect of mTOR inhibitors on TSC tumors in these experiments has been consistently cytostatic rather than cytotoxic, and is variable in efficacy; tumors typically regrow upon the cessation of treatment [59, 60]. Therefore, these preclinical models have become powerful tools in the assessment of potential therapies for TSC. However, the molecular mechanism of the sirolimus-induced cytostatic effect on TSC tumors is not totally elucidated. Our recent study reported that xenograft tumors of Tsc2-deficient rat uterine leiomyoma-derived ELT3 cells became resistant to rapamycin treatment [61]. In this study, we observed that xenograft tumors of ELT3 cells potently responded to rapamycin within 1 week of treatment, however, tumors became refractory from week 2 of rapamycin treatment. This rapamycin resistant growth is consistent with the study by Valianou et al. [61]. In our xenograft tumor study, we used bioluminescent imaging approach to quantify the tumor growth in response to rapamycin treatment, enabling quantification of viable tumor cells in vivo.
Treatment with sirolimus alone has a suppressive rather than remission-inducing effect in majority of tumor models with dysregulated mTORC1 [62, 63]. mTORC1 inhibition leads to upregulation of pro-survival mediators including autophagy and paradoxically increases the growth of Tsc2-null cells [58, 64,65,66]. Specifically, inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer [67]. Using bioinformatic approach and immunoblotting analyses, we identified activation of MPAK signaling pathway among other pro-survival pathways in a panel of TSC-deficient cells, and rapid and sustained activation of MAPK in TSC-deficient cells, in agreement with other findings in prostate cancer cells [67].
High-throughput chemical screens in mTORC1-hyperactive patient renal angiomyolipoma-derived and Tsc2−/− MEFs cells identified compounds that selectively induce cell death through oxidative stress-dependent mechanisms within 72 h of drug treatment [68, 69]. Thus, there is an unmet need for identifying agents that act with chronic sirolimus treatment to kill mTORC1-hyperactive cells. Our identification of rapamycin-induced MAPK activation prompted us to perform studies of dual inhibition of MAPK and mTORC1 in TSC-deficient cells. We found that MAPK inhibition attenuated rapamycin-induced cytostasis and promoted the death of TSC-deficient cells in vitro.
A potential mechanism by which active-site mTOR or dual inhibitors of PI3K/mTOR promotes MEK1/2-MAPK signaling pathway activation is via enhanced EGFR activity. A recent RNAseq analysis by Valianou et al. identified rapamycin-induced upregulation of EGFR signaling pathway in rapamycin-resistant ELT3 cells [61]. The EGFR tyrosine kinase activity and affinity for its ligands are negatively regulated by protein kinase C (PKCα) via phosphorylation at Thr654 [70]. Studies indicate that mTORC2 mediates PKCα phosphorylation [71, 72]. Interestingly, the mTORC2-dependent phosphorylation of PKCα plays an important role in its maturation, stability, and signaling [71, 72]. It is plausible, therefore, that suppression of mTORC2-mediated post-translational processing of PKCα interferes with negative feedback of PKCα on EGFR, thereby leading to hyperactivation of EGFR and activation of MAPK signaling in response to EGFR agonists or GPCR transactivation [73]. Future studies of the impact of EGFR-mediated MAPK activation on the survival of mTROC1 hyperactive cells will provide novel mechanistic targets for therapeutic application for TSC.
Conclusions
In the past decade, remarkable progress has been made in demonstrating the efficacy of sirolimus and everolimus in management of TSC and LAM patients. Rapamycin and Rapalogs that target mTOR activity offer an additional value which would help in the treatment of TSC and LAM. However, the effect of sirolimus and everolimus on reducing tumor size or improving symptoms has been consistently cytostatic rather than cytotoxic; tumors typically regrow and symptoms resume upon the cessation of treatment. In this study, we have revealed that mTORC1 inhibition using rapamycin results in a compensatory activation of MAPK in TSC1- and TSC2-deficient cells. This enhanced MAPK signaling pathway was associated with enhanced survival of TSC-deficient cells in vitro. Dual inhibition of mTORC1 and MAPK triggers the death of TSC2-deficient cells. Taken together, our study reveals a novel approach of dual targeting of mTORC1 and MAPK pathways to induce tumor remission in TSC and other mTORC1 hyperactive neoplasms.
Materials and methods
Gene set enrichment analysis
Re-analysis of publicly available expression array data (GEO accession number GSE16944 [15], GSE21755 [16], GSE5332 [17], GSE27982 [18], GSE28021 [18], GSE67529, GSE28992 [19], GSE18571 [20], GSE7344 [21], GSE37129 [22] and GSE17662 [23]) was performed using the online tool Gene Pattern (Broad Institute).
Cell culture and reagents
Cell culture media and supplements were from GIBCO (Frederick, MD). Tsc2−/−p53−/− and Tsc2+/+p53−/− mouse embryonic fibroblasts (MEFs) were developed previously [29]. Mouse expression arrays of Tsc2−/−p53−/− and Tsc2+/+p53−/− MEFs were preformed [16, 18]. An immortalized TSC2-deficient human cell line derived from angiomyolipoma of a LAM patient [28], and its corresponding TSC2-rescued control cell line has been described previously [15]. In brief, patient-derived cells were transfected with pcDNA3.1zeo-hTSC2 or its corresponding empty vector control pcDNA3.1zeo. Stable clones expressing TSC2 were selected using Zeocin for 2 weeks as described previously [74]. Eker rat uterine leiomyoma-derived Tsc2-deficient cells (ELT3) were developed by Howe et al. [24, 25]. ELT3 cells were transduced with a retroviral plasmid pMSCVneo-hTSC2 or its corresponding empty vector pMSCVneo, and then selected with neomycin for 2 weeks. Stable clones were characterized for TSC2 expression [75]. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin-amphotericin B (PSA). Experiments were performed in triplicate for biochemical analyses. Cells were seeded at a density of 2 × 105 cells/ml in 6-well plates in regular growth media for 24 h. Six or 24 h later, cell lysates were prepared using RIPA buffer supplemented with protease inhibitor cocktail (Sigma) and phosphatase inhibitor cocktail (Sigma). Protein concentration was determined using Bradford assay (BioRad Laboratories Inc. Hercules, CA).
Cell viability assay
Cells were seeded at a density of 5 × 104/ml in a 96-well plate for 24 h and then treated with inhibitors or vehicle control for 24 h. Cell numbers were quantified using CyQuant (Invitrogen, Carlsbad, CA) or crystal violet staining assay. Values are expressed as mean ± SEM; n = 8/group.
Animal studies
The University of Cincinnati Institutional Animal Care and Use Committee approved all procedures described according to standards as outlined in The Guide for the Care and Use of Laboratory Animals. For xenograft tumor study, 2 × 106 ERL4-luciferase-tagged (TSC2-null) cells were inoculated bilaterally into the posterior back region of female intact CB17-SCID mice (Taconic) as previously described [26, 76]. For the current study, 9–10 week-old CB-17 SCID mice were treated with vehicle control or 2 mg/kg rapamycin (dissolve in 0.25% Tween 80, 0.25% polyethylene glycol 400, i.p.) every day for 3 weeks. The tumors were harvested 3 weeks post cell inoculation. Tumor growth were monitored weekly using a non-invasive imaging by IVIS (Perkin Elmer). All efforts were made to reduce suffering of the animals and minimize the number of animals used in the study.
Bioluminescent reporter imaging
Ten minutes before imaging, animals were injected with luciferin (Xenogen) (120 mg/kg, i.p.). Bioluminescent signals were recorded using the Xenogen IVIS System. The total photon flux of tumors was analyzed [26].
Immunohistochemistry
Immunohistochemistry (IHC) was performed on paraffin-embedded 10 μm-sections. Slides were deparaffinized, and antigen retrieval was performed using Dako Target Retrieval Solution pH 6 (Dako, Carpinteria, CA). Sections were stained by the immunoperoxidase technique using DAB substrate (Dako EnVision System HRP) and counterstaining with hematoxylin. After staining, slides were viewed on a Nikon Eclipse E400 microscope, and images captured using Spot Insight digital camera with Spot software (Diagnostic Instruments, Sterling Heights, MI).
Western blotting
Protein samples were analyzed by SDS-PAGE using 4–12% NuPAGE Gel (Invitrogen, Carlsbad, CA), and transferred to a nitrocellulose membrane. Immunoblotting was performed by standard methods using HRP-conjugated secondary antibodies, and chemiluminescence using Supersignal West Pico Chemiluminescent substrate (Thermo Scientific) and exposure using Syngene G:Box. All antibodies were purchased from Cell Signaling (Danvers, MA).
Statistical analyses
All data are shown as the mean ± S.E.M. Measurements at single time points were analyzed by ANOVA and then using a two-tailed t-test (Student’s t test). Time courses were analyzed by repeated measurements (mixed model) ANOVA and Bonferroni post-t-tests. All statistical tests were performed using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA) and p < 0.05 indicated statistical significance.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ELT3:
-
Eker rat uterine leiomyoma-derived cells
- ERL4:
-
ELT3 cells expressing luciferase
- LAM:
-
Lymphangioleiomyomatosis
- MAPK:
-
Mitogen-activated protein kinase
- mTOR:
-
Mechanistic target of rapamycin
- PCNA:
-
Proliferating cell nuclear antigen
- PS6:
-
Phospho-ribosomal protein S6 (Ser235/236)
- RAPTOR:
-
Regulatory-associated protein of mTOR
- Rheb:
-
Ras homologue enriched in brain
- RICTOR:
-
Rapamycin-insensitive companion of mTOR
- S6K:
-
Ribosomal protein S6 kinase
- TSC:
-
Tuberous Sclerosis Complex
- 4E-BP1:
-
Translational repressor 4E-binding protein 1
References
Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–56.
Krueger DA, Care MM, Holland K, Agricola K, Tudor C, Mangeshkar P, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med. 2010;363(19):1801–11.
Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47(4):535–46.
Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10(1):151–62.
Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–45.
Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35.
Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–93.
Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373–9.
Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358(2):140–51.
McCormack FX, Inoue Y, Moss J, Singer LG, Strange C, Nakata K, et al. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med. 2011;364(17):1595–606.
Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Muller U, et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell. 2012;22(6):796–811.
Francois RA, Maeng K, Nawab A, Kaye FJ, Hochwald SN, Zajac-Kaye M. Targeting focal adhesion kinase and resistance to mTOR inhibition in pancreatic neuroendocrine tumors. J Natl Cancer Inst. 2015;107(8):djv123.
Posch C, Moslehi H, Feeney L, Green GA, Ebaee A, Feichtenschlager V, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci U S A. 2013;110(10):4015–20.
Zhao H, Cui K, Nie F, Wang L, Brandl MB, Jin G, et al. The effect of mTOR inhibition alone or combined with MEK inhibitors on brain metastasis: an in vivo analysis in triple-negative breast cancer models. Breast Cancer Res Treat. 2012;131(2):425–36.
Lee PS, Tsang SW, Moses MA, Trayes-Gibson Z, Hsiao LL, Jensen R, et al. Rapamycin-insensitive up-regulation of MMP2 and other genes in tuberous sclerosis complex 2-deficient lymphangioleiomyomatosis-like cells. Am J Respir Cell Mol Biol. 2010;42(2):227–34.
Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–83.
Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–40.
Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30(16):3242–58.
Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21(2):227–39.
Akcakanat A, Zhang L, Tsavachidis S, Meric-Bernstam F. The rapamycin-regulated gene expression signature determines prognosis for breast cancer. Mol Cancer. 2009;8:75.
Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, et al. Antitumor activity of rapamycin in a phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5(1):e8.
Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, et al. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med. 2012;4(139):139ra84.
Jimenez RH, Boylan JM, Lee JS, Francesconi M, Castellani G, Sanders JA, et al. Rapamycin response in tumorigenic and non-tumorigenic hepatic cell lines. PLoS One. 2009;4(10):e7373.
Howe SR, Gottardis MM, Everitt JI, Goldsworthy TL, Wolf DC, Walker C. Rodent model of reproductive tract leiomyomata. Establishment and characterization of tumor-derived cell lines. Am J Pathol. 1995;146(6):1568–79.
Howe SR, Gottardis MM, Everitt JI, Walker C. Estrogen stimulation and tamoxifen inhibition of leiomyoma cell growth in vitro and in vivo. Endocrinology. 1995;136(11):4996–5003.
Yu JJ, Robb VA, Morrison TA, Ariazi EA, Karbowniczek M, Astrinidis A, et al. Estrogen promotes the survival and pulmonary metastasis of tuberin-null cells. Proc Natl Acad Sci U S A. 2009;106(8):2635–40.
Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28(12):4104–15.
Yu J, Astrinidis A, Howard S, Henske EP. Estradiol and tamoxifen stimulate LAM-associated angiomyolipoma cell growth and activate both genomic and nongenomic signaling pathways. Am J Physiol Lung Cell Mol Physiol. 2004;286(4):L694–700.
Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003;112(8):1223–33.
Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103(2):253–62.
Baldo P, Cecco S, Giacomin E, Lazzarini R, Ros B, Marastoni S. mTOR pathway and mTOR inhibitors as agents for cancer therapy. Curr Cancer Drug Targets. 2008;8(8):647–65.
Li A, Hardy R, Stoner S, Tuckermann J, Seibel M, Zhou H. Deletion of mesenchymal glucocorticoid receptor attenuates embryonic lung development and abdominal wall closure. PLoS One. 2013;8(5):e63578.
Lim HJ, Crowe P, Yang JL. Current clinical regulation of PI3K/PTEN/Akt/mTOR signalling in treatment of human cancer. J Cancer Res Clin Oncol. 2015;141(4):671–89.
Carew JS, Kelly KR, Nawrocki ST. Mechanisms of mTOR inhibitor resistance in cancer therapy. Target Oncol. 2011;6(1):17–27.
Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, Kuperman R, et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2013;381(9861):125–32.
Bissler JJ, Kingswood JC, Radzikowska E, Zonnenberg BA, Frost M, Belousova E, et al. Everolimus for angiomyolipoma associated with tuberous sclerosis complex or sporadic lymphangioleiomyomatosis (EXIST-2): a multicentre, randomised, double-blind, placebo-controlled trial. Lancet. 2013;381(9869):817–24.
Summary of the risk management plan for Afinitor and Votubia (everolimus). European Medicines Agency. 2011:Online.
Curatolo P, Moavero R. mTOR inhibitors in tuberous sclerosis complex. Curr Neuropharmacol. 2012;10(4):404–15.
Xie J, Wang X, Proud CG. mTOR inhibitors in cancer therapy. F1000Res. 2016;5:F1000 Faculty Rev-2078.
Phung TL, Ziv K, Dabydeen D, Eyiah-Mensah G, Riveros M, Perruzzi C, et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. 2006;10(2):159–70.
Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–68.
Hasskarl J. Everolimus. Recent Results Cancer Res. 2018;211:101–23.
Granata S, Dalla Gassa A, Carraro A, Brunelli M, Stallone G, Lupo A, et al. Sirolimus and everolimus pathway: reviewing candidate genes influencing their intracellular effects. Int J Mol Sci. 2016;17(5):735.
MacKeigan JP, Krueger DA. Differentiating the mTOR inhibitors everolimus and sirolimus in the treatment of tuberous sclerosis complex. Neuro-Oncology. 2015;17(12):1550–9.
Henske EP, Jóźwiak S, Kingswood JC, Sampson JR, Thiele EA. Tuberous sclerosis complex. Nat Rev Dis Primers. 2016;2:16035.
Palavra F, Robalo C, Reis F. Recent advances and challenges of mTOR inhibitors use in the treatment of patients with tuberous sclerosis complex. Oxidative Med Cell Longev. 2017;2017:9820181.
Hess EJ, Moody KA, Geffrey AL, Pollack SF, Skirvin LA, Bruno PL, et al. Cannabidiol as a new treatment for drug-resistant epilepsy in tuberous sclerosis complex. Epilepsia. 2016;57(10):1617–24.
Devinsky O, Marsh E, Friedman D, Thiele E, Laux L, Sullivan J, et al. Cannabidiol in patients with treatment-resistant epilepsy: an open-label interventional trial. Lancet Neurol. 2016;15(3):270–8.
Gloss D, Vickrey B. Cannabinoids for epilepsy. Cochrane Database Syst Rev. 2014(3):CD009270.
Jennesson M, van Eeghen AM, Caruso PA, Paolini JL, Thiele EA. Clobazam therapy of refractory epilepsy in tuberous sclerosis complex. Epilepsy Res. 2013;104(3):269–74.
Kossoff EH, Thiele EA, Pfeifer HH, McGrogan JR, Freeman JM. Tuberous sclerosis complex and the ketogenic diet. Epilepsia. 2005;46(10):1684–6.
Krueger DA, Wilfong AA, Mays M, Talley CM, Agricola K, Tudor C, et al. Long-term treatment of epilepsy with everolimus in tuberous sclerosis. Neurology. 2016;87(23):2408–15.
Nickels K. Cannabidiol in patients with intractable epilepsy due to TSC: a possible medication but not a miracle. Epilepsy Curr. 2017;17(2):91–2.
El-Hashemite N, Walker V, Zhang H, Kwiatkowski DJ. Loss of Tsc1 or Tsc2 induces vascular endothelial growth factor production through mammalian target of rapamycin. Cancer Res. 2003;63(17):5173–7.
Karbowniczek M, Zitserman D, Khabibullin D, Hartman T, Yu J, Morrison T, et al. The evolutionarily conserved TSC/Rheb pathway activates Notch in tuberous sclerosis complex and Drosophila external sensory organ development. J Clin Invest. 2010;120(1):93–102.
Kenerson HL, Aicher LD, True LD, Yeung RS. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 2002;62(20):5645–50.
Lee L, Sudentas P, Donohue B, Asrican K, Worku A, Walker V, et al. Efficacy of a rapamycin analog (CCI-779) and IFN-gamma in tuberous sclerosis mouse models. Genes Chromosom Cancer. 2005;42(3):213–27.
Parkhitko A, Myachina F, Morrison TA, Hindi KM, Auricchio N, Karbowniczek M, et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci U S A. 2011;108(30):12455–60.
Kwiatkowski DJ. Animal models of lymphangioleiomyomatosis (LAM) and tuberous sclerosis complex (TSC). Lymphat Res Biol. 2010;8(1):51–7.
Yu J, Henske EP. mTOR activation, lymphangiogenesis, and estrogen-mediated cell survival: the “perfect storm” of pro-metastatic factors in LAM pathogenesis. Lymphat Res Biol. 2010;8(1):43–9.
Valianou M, Filippidou N, Johnson DL, Vogel P, Zhang EY, Liu X, et al. Rapalog resistance is associated with mesenchymal-type changes in Tsc2-null cells. Sci Rep. 2019;9(1):3015.
Ilagan E, Manning BD. Emerging role of mTOR in the response to cancer therapeutics. Trends Cancer. 2016;2(5):241–51.
Sahin M, Henske EP, Manning BD, Ess KC, Bissler JJ, Klann E, et al. Advances and future directions for tuberous sclerosis complex research: recommendations from the 2015 strategic planning conference. Pediatr Neurol. 2016;60:1–12.
Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22(2):169–76.
Parkhitko AA, Priolo C, Coloff JL, Yun J, Wu JJ, Mizumura K, et al. Autophagy-dependent metabolic reprogramming sensitizes TSC2-deficient cells to the antimetabolite 6-Aminonicotinamide. Mol Cancer Res. 2013;12(1):48–57.
Xu J, Pham CG, Albanese SK, Dong Y, Oyama T, Lee CH, et al. Mechanistically distinct cancer-associated mTOR activation clusters predict sensitivity to rapamycin. J Clin Invest. 2016;126(9):3526–40.
Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–74.
Li J, Shin S, Sun Y, Yoon SO, Li C, Zhang E, et al. mTORC1-driven tumor cells are highly sensitive to therapeutic targeting by antagonists of oxidative stress. Cancer Res. 2016;76(16):4816–27.
Medvetz D, Sun Y, Li C, Khabibullin D, Balan M, Parkhitko A, et al. High-throughput drug screen identifies chelerythrine as a selective inducer of death in a TSC2-null setting. Mol Cancer Res. 2015;13(1):50–62.
Santiskulvong C, Rozengurt E. Protein kinase Calpha mediates feedback inhibition of EGF receptor transactivation induced by Gq-coupled receptor agonists. Cell Signal. 2007;19(6):1348–57.
Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27(14):1932–43.
Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27(14):1919–31.
Rozengurt E. Mitogenic signaling pathways induced by G protein-coupled receptors. J Cell Physiol. 2007;213(3):589–602.
Hong F, Larrea MD, Doughty C, Kwiatkowski DJ, Squillace R, Slingerland JM. mTOR-raptor binds and activates SGK1 to regulate p27 phosphorylation. Mol Cell. 2008;30(6):701–11.
Astrinidis A, Cash TP, Hunter DS, Walker CL, Chernoff J, Henske EP. Tuberin, the tuberous sclerosis complex 2 tumor suppressor gene product, regulates Rho activation, cell adhesion and migration. Oncogene. 2002;21(55):8470–6.
Li C, Lee PS, Sun Y, Gu X, Zhang E, Guo Y, et al. Estradiol and mTORC2 cooperate to enhance prostaglandin biosynthesis and tumorigenesis in TSC2-deficient LAM cells. J Exp Med. 2014;211(1):15–28.
Acknowledgements
We thank members of the Yu Laboratory for critical discussions and technical assistance. We are grateful to Dr. C. Walker (Texas A&M Health Science Center) for providing ELT3 cells, Dr. E.P. Henske (Brigham and Women’s Hospital) for providing 621-101 and 621-103 cells, and Dr. D.K.J. Kwiatkowski to provide Tsc1 and Tsc2 MEFs (Brigham and Women’s Hospital).
Funding
NIHRO1HL138481, NIHRO1HL098216, NIHRO1DK098331, ARMY-W81XWH-19-1-0474, and The LAM Foundation Bridge Fund 0124B07–17 to JJY.
Author information
Authors and Affiliations
Contributions
YL and JJY designed all in vitro and in vivo studies. YL and EYZ performed in vitro and in vivo studies. YL, EYZ and JJY analyzed all data. EYZ performed bioinformatics data analysis. JL provided therapeutic perspectives of mTOR inhibitors in LAM and TSC management. YL and JJY wrote the manuscript. All authors had full access to the data, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The University of Cincinnati Institutional Animal Care and Use Committee approved all procedures described according to standards as outlined in The Guide for the Care and Use of Laboratory Animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lu, Y., Zhang, E.Y., Liu, J. et al. Inhibition of the mechanistic target of rapamycin induces cell survival via MAPK in tuberous sclerosis complex. Orphanet J Rare Dis 15, 209 (2020). https://doi.org/10.1186/s13023-020-01490-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13023-020-01490-w