Abstract
Background
Leonuri Herba (Yimucao) is a very common Chinese herbs for treating menstrual and maternal diseases for thousands of years in China. However, the herb collected in different origins was easily found in the markets which induce the unstable quality for clinic use. In this study, a comprehensive strategy of using multiple chromatographic analysis and chemometric analysis was firstly investigated for chemical discrimination of Leonuri Herba from different geographical origins.
Methods
UHPLC-QTOF-MS/MS was applied to identify the peaks of Leonuri Herba and chemical fingerprints were established in 30 batches from different geographical origins. Meanwhile, dissimilarities of chemical compositions among different origins were further investigated by principal component analysis and cluster analysis. And a quantitative UHPLC-QTOF-MS/MS approach were established to investigate the potential marker for quality control of Leonuri Herba.
Results
A total of 49 chromatographic peaks of Leonuri Herba were identified by UHPLC-QTOF-MS/MS. Leonuri Herba were classified into four categories, and eight major compounds detected could be used as chemical markers for discrimination. Also, the eight components, including leonurine, 4',5-dihydroxy-7-methoxyflavone, rutin, hyperoside, apigenin, quercetin, kaempferol and salicylic acid, were simultaneously quantified using the extracting ion mode of UHPLC-QTOF-MS/MS.
Conclusion
The current strategy not only clearly expounded the correlation between quality and geographical origins of Leonuri Herba, but also provided a fast, accurate and comprehensive qualitative and quantitative method for assessing the quality of Leonuri Herba.
Similar content being viewed by others
Background
Leonuri Herba (Yimucao) is the aerial part of Leonurus japonicus Houtt., which is a very common Chinese herb for treating menstrual and maternal diseases. It has been used for thousands of years in Chinese Medicine, and it is recognized to be a non-toxic herb [1]. Leonurus japonicus Houtt. grows on wastelands of mountains and plains, rand, grasslands and streamside. It was distributed or cultivated in all parts of China. The medicinal materials are mainly produced in Henan (Songxian and Luanzhou), Anhui (Liuan and Bengbu), Sichuan (Wenjiang), Jiangsu (Nanjing), Zhejiang (Fenghua) [2]. Besides, it is also native to other countries such as Europe, North America, Japan and Malaysia [3]. According to Chinese Pharmacopoeia [4], the dried herb is collected in summer before flowering and dried under the sun to obtain Herba Leonuri. However, the herb collected in different stages with the varied proportion of leaves and stems was easily found in the markets which induce the unstable quality for clinic use. Thus, the quality of this traditional Chinese Materia Medica has already attracted public concern. Leonuri Herba has plenty of pharmacological effects for treating human diseases [5], especially in gynecopathy [6], as well as other functions with its low toxic property [7]. Thus, it has been using for a long time as traditional Chinese medicine (TCM). A substantial number of studies have been conducted to investigate its active components and related mechanisms [5].
Leonuri Herba is not a geo-authentic herb (daodi yaochai). It is difficult to determine the quality of Leonuri Herba in different geographical origins. Furthermore, different harvesting period of Leonuri Herba is also one of the major factors affecting the quality. Nowadays, Leonuri Herba at seedling stage is commonly used in Hong Kong market while the mature one is widely used in mainland China. These situations show the necessity and importance to clarify which growing stage and geographical origins that should be chosen. There are around 140 compounds discovered in Leonuri Herba, alkaloids, diterpenes and flavones are its main chemicals [1]. Apart from leonurine and stachydrine, the studies of other components such as diterpenes and flavones that monitoring the quality of Leonuri Herba is still deficient [3]. This leads to the significance of analyzing the quality of Leonuri Herba by other chemical components.
In this study, we investigated whether and how the quality of Leonuri Herba correlates with its geographical origins by combining UHPLC-QTOF-MS/MS-based qualitative fingerprint profiling and quantitative determination of potential markers. The data obtained were processed by multivariate statistical analysis, including hierarchical cluster analysis (HCA), principal component analysis (PCA) and supervised orthogonal partial least squared discriminant analysis (OPLS-DA), to evaluate the differences in quality of these samples. Finally, in order to better understand the differences from quantitative levels, potential markers that played key roles in differentiating Leonuri Herba from different locations, were simultaneously determined in different samples.
Materials and methods
Herbal materials
Thirty batches of Leonuri Herba were acquired for this study. Of these, ten batches were obtained from the Hong Kong TCM market, other batches were purchased directly from certified production regions in China, as specified in Table 1. Thirty batches of Leonuri Herba were all authenticated by Prof. Zhongzhen Zhao from the School of Chinese Medicine (SCM), Hong Kong Baptist University (HKBU). Samples were dried, ground, then sifted through a 24-mesh sieve. Voucher specimens were deposited in SCM of HKBU (Additional file 1).
Instrument, chemicals and reagents
Chemical markers of leonurine, rutin, 4',5-dihydroxy-7-methoxyflavone, hyperoside, salicylic acid, kaempferol were obtained from Shanghai Tauto Biotech Co. Ltd. (Shanghai, China). Chemical marker of apigenin was purchased from Chengdu Mansite Pharmacetical Co. Ltd. (Sichuan, China). Chemical marker of quercetin was obtained from the National Institute of Control of Pharmaceutical and Biological Products (Beijing, China). The purity of each chemical marker was above 95%. Methanol and acetonitrile for UHPLC-QTOF-MS/MS analysis were obtained from Merck (Darmstadt, Germany). Ultrapure water was prepared by a Milli-Q water purification system (Millipore, Bedford, MA, USA).
The analyses were performed on UHPLC (Agilent Technologies Inc., Palo Alto, CA, USA), Agilent 6540 ultra-high-definition accurate mass quadrupole time-of-flight spectrometer (Agilent Technologies Inc., Wilmington, DE, USA).
Sample extraction
Powdered sample (0.5 g) accurately weighed, was extracted with 5 mL of methanol for 60 min at 60 °C in an ultrasonic water bath (300 W), and then cooled to room temperature. After compensating the lost weight of methanol, the extracted solution was filtered through a 0.22 μm PTFE syringe filter UHPLC fingerprint and identification analysis by UHPLC-PDA-QTOF-MS/MS [8].
UHPLC-QTOF-MS/MS identification
According to the research reports on the chemical components of Leonuri Herba, 121 chemical compounds are collected from Leonuri Herba. Agilent’s “Mass Hunter PCDL Manager” software is used to calculate the relatively molecular mass accurately and establish a database of known chemical components of Leonuri Herba.
Chromatographic conditions
Waters ACQUITY UPLC® BEH-C18 analytical column (2.1 mm × 100 mm, I.D. 1.7 µm); VanGuardTM BEH-C18 guard column (2.1 mm × 5 mm, I.D. 1.7 µm); Mobile phase: 0.1% FA (A) and acetonitrile (B); Linear gradient elution program: 0–8 min, 25% B; 8–16 min, 25–75% B; 16–18 min, 75–100% B; 18–21 min, 100% B; 21 min, 2% B; 21–24 min, 2% B; Flow rate: 0.35 mL/min; Column temperature: 40 ℃; Injection volume: 2 µL.
Mass spectrometry conditions
Mode: positive and negative(scanning from 100 to 900 m/z); Dry gas temperature 300 ℃; Dry nitrogen gas flow rate 8 L/min; Nebulizer pressure 40 psi; Vcap 3500; Nozzle voltage 500 V; Fragmentor voltage 120 V. The mass spectrum was calibrated by Tune mix every time (positive mode, 118–1521 m/z; negative mode, 112–1634 m/z).
Fingerprint similarity evaluation
According to the UHPLC-MS/MS spectra of 30 samples, the common chromatographic fingerprint peaks and internal reference peaks contained in each batch of samples were confirmed. The Similarity evaluation system for chromatographic fingerprint of TCM was used to establish the fingerprint of 30 batches of Leonuri Herba. The similarity was calculated by the angle cosine method.
Hierarchical clustering analysis and principal component analysis
For HCA, the common fingerprint peak area of Leonuri Herba from different origins and batches are used as the source data of hierarchical clustering analysis. The hierarchical clustering algorithm in microarray analysis software (MeV 4.7.4) is used. For PCA, the common fingerprint peak area of Leonuri Herba fingerprints from different origins and batches are used as the source data of PCA by using factor analysis in SPSS 20.0(SPSS Corporation, Armonk, NY, USA).
Quantitative analysis
The accurately weighed powdered samples of 30 batches (0.5 g) were respectively extracted with methanol (2.5 mL) for 60 min, and then cooled to 25 ℃. After centrifugation at 3500g for 10 min, the supernatant was transferred to a 10 mL volumetric flask. This process was repeated twice and then the volume was adjusted to the calibration mark with methanol. The extracted solution was filtered through a 0.22 μm PTFE syringe filter before analysis. The standards were accurately weighed and dissolved into methanol. A series of standard solutions for constructing working standard curves was prepared by diluting the mixed stock standard solution with methanol, and a calibration curve was established by plotting peak areas (y axis) versus concentrations (x axis).
Statistical analysis
Peaks above the S/N ratio of the chromatogram were labeled and manually integrated using version 7.2 of the Chromeleon Chromatography Data System software (Thermo Fisher Scientific) to distinguish between the different batches. PCA and OPLS-DA were performed using the software SIMCA-P Version 13.0 (Umetrics). Summary data were expressed as the mean ± standard deviation (SD) for n = 3–7.
Results and discussion
Component authentication
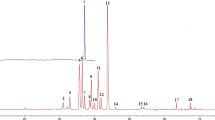
Figure 1A shows the Base peak chromatogram (BPC) of Leonuri Herba in negative mode. According to TR, Mass, (−)-ESI–MS/MS Fragment Ions and compared with the reference and standards, 49 components were identified or preliminarily identified. There were 2 alkaloids, 18 flavonoids, 7 Terpenoids, 8 aromatic acids and 14 other classes (Table 2).
Fingerprint analysis
According to the UHPLC-MS/MS spectrum of 30 batches of samples, the chromatographic peaks was showed within 24 min. Thirty-three of them were the common peaks to each batch of samples (Table 3). The total peak area of each batch of samples accounted for more than 85% of the total peak area and the reproducibility was good, which fulfilled the requirements of fingerprints. Thus, they were determined as common fingerprint peaks. The common fingerprint peak was determined by the standard retention time. When the retention time was 6.53 min, it was leonurine (Fig. 1B). Since the peak area of leonurine was relatively big, the peak time was moderate and the shape was good, it was selected as the reference peak. Using Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine (2004A edition). Thirty batches of Leonuri Herba from different origins were introduced (Fig. 2) and the time width was 0.1 min, gaining the control fingerprint by median generation method. The similarity of 30 batches of samples was calculated by the angle cosine method. The result showed the similarity between each fingerprint and the control fingerprint was less than 0.90, indicating there was a big difference in each origin including Hubei, Guangdong, Henan, Anhui, Yunnan, Zhejiang and Sichuan. Thus, principal component and hierarchical cluster analysis were further analysed to clarify the relationship between origins and quality.
Results of hierarchical clustering analysis (HCA)
The analysis showed the samples could be roughly clustered into two big groups. Five batches of Leonuri Herba from Henan, Anhui and Sichuan could be classified as one group, other samples could be classified as another group by the first chemical component clustering analysis. Based on the second to fifth chemical component clustering analysis, they were further clustered into small group: Anhui as a group; Yunnan as a group; Guangdong and Hubei as a group; Henan as a group. However, many factors such as harvest time, proportion of medical parts, soil, environment, and water were different, resulting in significant differences in chemical components of Leonuri Herba. Thus, it is necessary to find quality markers in different origins to clarify the relationship between habitat and quality. From the chemical composition clustering analysis, as shown in Fig. 3, the first component was leonurine. The second and third classes were quercetin-3-O-robinosideand 4',5-dihydroxy-7-methoxyflavone. The fourth class was hyperoside, quercetin-3-O-β-d-glucopyranoside and tiliroside. The fifth class was rutin, lavandulifolioside, syringic acid, salicylic acid and 2'''-syringylrutin. The sixth to eighth classes were apigenin. Other classes also included kaempferol, quercetin, tryptophan, 7α(H)-eudesmane-4,11(12)-diene-3-one-2β-hydroxy-13-β-d-glucopyranoside, etc. These different classes of components could be used to distinguish the quality of Leonuri Herba and as a basis for quantitative analysis.
Leonurine (C1), 4',5-dihydroxy-7-methoxyflavone (C3), rutin (C4), hyperoside (C11), apigenin (C15), quercetin (C16), kaempferol (C17) and salicylic acid (C30) were selected for further quantitative analysis of the indicator components.
Results of multivariate statistical analysis
The common fingerprint peak area of Leonuri Herba fingerprints from different origins and batches were used as the source data of PCA by using factor analysis in SPSS 20.0. The results of SPSS analysis indicated that the first principal component should integrate the information of C1, C2, C3, C18, C21, C28, C29, C30 compounds. Thus, the chemical composition differences of different origins and batches of Leonuri Herba were mainly reflected to these chemical components. The results showed that the content of C1 (leonurine), C3 (4',5-dihydroxy-7-methoxyflavone) and C30 (salicylic acid) in Henan samples is higher, which was consistent with the cluster analysis results. The matrix coefficient of main components 1–8 in Leonuri Herba are shown in Fig. 4. In Fig. 4A, PCA divided Leonuri Herba from different origins into four parts: Henan, Yunnan as a large category; Zhejiang, Sichuan as a large category; Anhui as a large category; Guangdong, Hubei (at seedling and flowering stage) as a large category; Hubei (at mature stage) as a small category, which is different from the one at seedling and flowering stage. In order to further characterize the differences in chemical profiles among different Leonuri Herba samples, OPLS-DA, a supervised latent structures-discriminant analysis technique, which utilizes class information to maximize the separation between classes and minimize the discrimination between intra-groups, was performed to achieve better separation among different samples. The score plot of OPLS-DA indicated that all 30 samples were unambiguously classified into seven groups (Fig. 4B). Interestingly, these seven groups were highly consistent with the collection locations of these samples (Fig. 4C), which demonstrated that the chemical components of different samples are heavily influenced by growing area.
Comprehensive evaluation analysis
The relationship formula between the principal component load matrix U, the factor load matrix A and the eigenvalue λ was Ui = A/ SQRT (λi). By calculating the variables, eight eigenvectors U1-8 were obtained, and the expressions of the 8 principal components could be obtained, as follows.
Normalizing the original variables and using SPSS to compute variables and calculate the principal componentsY1, Y2, Y3, Y4, Y5, Y6, Y7 and Y8. Taking the variance contribution rate corresponding to each principal component as the weight, the principal component scores and the corresponding weights were linearly weighted to construct a comprehensive evaluation function of different habitats and batches of Leonuri Herba:
The comprehensive evaluation scores of Leonuri Herba from different origins and batches were calculated from the above formula are shown in Table 4. The higher of the score, the higher content of active ingredient in the sample. The comprehensive score was greater than 0. The results showed that batch S8 (from Hubei at seedling stage) had the highest comprehensive score, followed by batch S4 (from Guangdong at seedling stage), S15 (Henan) S10 (from Hubei at seedling stage) and S13 (Henan). The above results showed that the quality of three batches at seedling stage was better than the others. The chemical components of Leonuri Herba produced in Hubei and Henan are relatively higher than other origins.
Result of quantitative analysis
The LOD was calculated according to the signal-to-noise ratio of 3:1; the LOQ was calculated according to the signal-to-noise ratio of 10:1, and the results were shown in (Additional file 2: Table S1). The precision test results showed that the precision of the instrument was good; the stability test results showed that the test solution had a good stability within 24 h after preparation; the repeatability test results showed that the method had a good repeatability; the results of the sample recovery rate showed that the accuracy was good (Additional file 3: Table S2). The results of 30 batches of Leonuri Herba from different origins are shown in Table 5. The detailed content trends of eight analytes in the 30 samples from different origins are exhibited in Fig. 5. In Fig. 5A, the results showed that Leonuri Herba from Henan had the highest content in hyperoside (C11) and salicylic acid (C30). The content of leonurine (C1) in Anhui was the highest while the content of rutin (C4) in Zhejiang was the highest. For apigenin (C15), quercetin(C16) and kaempferol (C17), Hubei, Henan and Sichuan had similar result (Fig. 5B). By adding all the eight analytes together, the total content of Henan is the highest (Fig. 5C).
The contents of 8 components in the 30 samples from different regions. A The content of Leonurine (C1), 4',5-dihydroxy-7-methoxyflavone (C3), rutin (C4) and hyperoside (C11). B The content of apigenin(C15), quercetin(C16), kaempferol (C17) and salicylic acid (C30). C The content of combination of 8 components in 30 samples
In summary, the quality of Leonuri Herba at seedling stage is better than the mature ones. Apart from the quality of medicinal part, Leonuri Herba from different origins are mainly clustered into four big categories: Henan and Yunnan are grouped together; Zhejiang and Sichuan are grouped together; Anhui is grouped as one group; Guangdong and Hubei (at seedlings and flowering stage) are grouped together while Hubei (at mature stage) is in different group due to growing stage difference. The comprehensive evaluation analysis showed that the quality of Leonuri Herba at seedling stage was good. And the quality of Leonuri Herba in Henan was relatively good when compared to other origins. However, the quality difference between stems and leaves from different origins and different growing stage is not very clear. Further studies should be conducted to address this issue. Laser microdissection combined with chromatographic analysis could be one of the powerful tools to investigate the chemical composition distribution and change in different growing stages of Leonuri Herba. This may also help to unify the standard of Leonuri Herba so as to decrease the confusion in markets.
Conclusion
The current study revealed a clear correlation between quality and geographical origin of Leonuri Herba. According to the correlation, the raw materials of Leonuri Herba from different regions can be formulated in a certain proportion and used for its traditional Chinese medicine preparations, thereby ensuring the safety, stability and effectiveness of clinical application; however, extensive pharmacological research is still required to address this issue. In a word, the proposed fingerprint will be important for authentication; the developed quantitative method will be useful for quality control of Leonuri Herba and its preparations; the revealed correlation could be significant for conservation and utilization of Leonuri Herba as a natural resource.
Availability of data and materials
The data used to support the current study are available from the corresponding author on reasonable request.
Abbreviations
- BPC:
-
Base peak chromatogram
- LOD:
-
Limit of detection
- LOQ:
-
Limit of quantification
- MF:
-
Molecular formula
- MS:
-
Mass spectrometry
- PCA:
-
Principal component analysis
- TCM:
-
Traditional Chinese medicine
- UHPLC:
-
Ultra-high performance liquid chromatography
- OPLS-DA:
-
Orthogonal partial least squares discriminant analysis
References
Shang X, Pan H, Wang X, He H, Li M. Leonurus japonicus Houtt.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J Ethnopharmacol. 2014;152(1):14–32.
Dong S, He J, Hou H, Shuai Y, Wang Q, Yang W, Liu R. Quality assessment of Herba Leonuri based on the analysis of multiple components using normal-and reversed-phase chromatographic methods. J Sep Sci. 2017;40(23):4482–94.
Zhu YZ, Wu W, Zhu Q, Liu X. Discovery of Leonuri and therapeutical applications: from bench to bedside. Pharmacol Ther. 2018;188:26–35.
Chinese Pharmacopoeia Commission. The Pharmacopoeia of the Peopleʼs Republic of China. 2020th ed. Beijing: China Medical Science Press; 2020.
Wang C, Lv X, Liu W, Liu S, Sun Z. Uncovering the pharmacological mechanism of motherwort (Leonurus japonicus Houtt.) for treating menstrual disorders: a systems pharmacology approach. Comput Biol Chem. 2020;89:107384.
Liu J, Peng C, Zhou QM, Guo L, Liu ZH, Xiong L. Alkaloids and flavonoid glycosides from the aerial parts of Leonurus japonicus and their opposite effects on uterine smooth muscle. Phytochemistry. 2018;145:128–36.
He YL, Shi JY, Peng C, Hu LJ, Liu J, Zhou QM, Xiong L. Angiogenic effect of motherwort (Leonurus japonicus) alkaloids and toxicity of motherwort essential oil on zebrafish embryos. Fitoterapia. 2018;128:36–42.
Zhu LX, Xu J, Wang RJ, Li HX, Tan YZ, Chen HB, Zhao ZZ. Correlation between quality and geographical origins of Poria cocos revealed by qualitative fingerprint profiling and quantitative determination of triterpenoid acids. Molecules. 2018;23(9):2200.
Wojtyniak K, Szymański M, Matławska I. Leonurus cardiaca L. (motherwort): a review of its phytochemistry and pharmacology. Phytother Res. 2013;27(8):1115–20.
Xiong L, Peng C, Zhou QM, Wan F, Xie XF, Guo L, Dai O. Chemical composition and antibacterial activity of essential oils from different parts of Leonurus japonicus Houtt. Molecules. 2013;18(1):963–73.
Kuchta K, Ortwein J, Savtschenko A, Briel D, Volk RB, Rauwald HW. Leonurus cardiaca, L. Japonicus, Leonotis leonurus: quantitative HPLC and instrumental HPTLC determination of fourteen phenolics. Planta Med. 2012;78(11):PJ50.
Li Y, Chen Z, Feng Z, Yang Y, Jiang J, Zhang P. Hepatoprotective glycosides from Leonurus japonicus Houtt. Carbohyd Res. 2012;348:42–6.
Tan YJ, Zhou GS, Guo S, Yan H, Zhang J, Zhu ZH, Duan JA. Simultaneous optimization of ultrasonic-assisted extraction of antioxidant and anticoagulation activities of compounds from Leonurus japonicus Houtt. by response surface methodology. RSC Adv. 2018;8(71):40748–59.
Wagner H, Bauer R, Melchart D, Xiao PG, Staudinger A. Herba Leonuri—Yimucao. In: Chromatographic fingerprint analysis of herbal medicines; 2011, p. 707–717; Vienna: Springer.
Chen Z, Wu JB, Liao XJ, Yang W, Song K. Development and validation of an UPLC-DAD-MS method for the determination of leonurine in Chinese motherwort (Leonurus japonicus). J Chromatogr Sci. 2010;48(10):802–6.
Deng S, Wang T, Wu CH, Qu L, Han LF, Zhang Y. Isolation and identification of constituents from Leonurus japonicus. Chin J Med Chem. 2013;23:209–12.
Xiong L, Zhou QM, Peng C, Xie XF, Guo L, Li XH, Dai O. Sesquiterpenoids from the herb of Leonurus japonicus. Molecules. 2013;18(5):5051–8.
Seo HK, Kim JS, Kang SS. Labdane diterpenes and flavonoids from Leonurus japonicus. Helv Chim Acta. 2010;93(10):2045–51.
Chang JM, Shen CC, Huang YL, Shieh BJ, Chen CC. Two new glycosides from Leonurus japonicus. J Asian Nat Prod Res. 2010;12(9):740–4.
Garran TA, Ji R, Chen JL, Xie D, Guo L, Huang LQ. Elucidation of metabolite isomers of Leonurus japonicus and Leonurus cardiaca using discriminating metabolite isomerism strategy based on ultra-high performance liquid chromatography tandem quadrupole time-of-flight mass spectrometry. J Chromatogr A. 2019;1598:141–53.
Miao LL, Zhou QM, Peng C, Liu ZH, Xiong L. Leonurus japonicus (Chinese motherwort), an excellent traditional medicine for obstetrical and gynecological diseases: a comprehensive overview. Biomed Pharmacother. 2019;117:109060.
Acknowledgements
We thank Alan Ho from the School of Chinese Medicine, Hong Kong Baptist University, for his technical assistance.
Funding
This work was supported by the Research Grants Council (Project No. 12102217 and 12103919).
Author information
Authors and Affiliations
Contributions
ZZ and KYL were in charge of idea and concept of the paper; KYL, YW and TL performed the experiment; KYL, YW processed the data and wrote the paper. CK and WY were in charge of sample collection; ZZ acquired funding for the research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Photograph of Leonuri Herba at seedling stage (upper) and mature stage (lower).
Additional file 2: Table S1.
Calibration curves, correlation factors, linear ranges, LOD and LOQ for the eight compounds.
Additional file 3: Table S2.
The RSD values of precisions, reproducibility, stability and recovery for the eight compounds.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lam, K.Y., Wang, Y., Lam, T. et al. Correlation between quality and geographical origins of Leonuri Herba revealed by the qualitative fingerprint profiling and quantitative determination of chemical components. Chin Med 17, 46 (2022). https://doi.org/10.1186/s13020-022-00592-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-022-00592-w