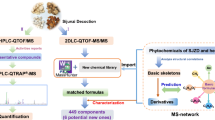
Abstract
Background
It is essential to identify the chemical components for the quality control methods establishment of Chinese Classical Formula (CCF). However, CCF are complex mixture of several herbal medicines with huge number of different compounds and they are not equal to the combination of chemical components from each herb due to particular formula ratio and preparation techniques. Therefore, it is time-consuming to identify compounds in a CCF by analyzing the LC–MS/MS data one by one, especially for unknown components.
Methods
An ultra-high pressure liquid chromatography-linear ion trap-orbitrap high resolution mass spectrometry (UHPLC-LTQ-Orbitrap-MS/MS) approach was developed to comprehensively profile and characterize multi-components in CCF with Erdong decoction composed of eight herbal medicines as an example. Then the MS data of Erdong decoction was analyzed by MS/MS-based molecular networking and these compounds with similar structures were connected to each other into a cluster in the network map. Then the unknown compounds connected to known compounds in a cluster of the network map were identified due to their similar structures.
Results
Based on the clusters of the molecular networking, 113 compounds were rapidly tentative identification from Erdong decoction for the first time in the negative mode, which including steroidal saponins, triterpenoid saponins, flavonoid O-glycosides and flavonoid C-glycosides. In addition, 10 alkaloids were tentatively identified in the positive mode from Nelumbinis folium by comparison with literatures.
Conclusion
MS/MS-based molecular networking technique is very useful for the rapid identification of components in CCF. In Erdong decoction, this method was very suitable for the identification of major steroidal saponins, triterpenoid saponins, and flavonoid C-glycosides.
Similar content being viewed by others
Background
The Chinese Classical Formula (CCF) are the essences of thousands of years of practical experience in the clinical application of traditional Chinese medicines (TCM). It is important and preferred direction of traditional Chinese medicine (TCM) to develop CCF into modern preparations to meet the needs of convenience. The chemical components analysis is of great significance for the study of pharmacologically active components and the establishment of quality control methods of CCF. The main chemical components of CCF are extremely complex and they are not equal to the combination of chemical components of each herb due to different formula proportions and preparation techniques. Therefore, how to quickly identify the main chemical components of a TCM formula is an important step for the modernization development of CCF.
Identification of chemical components of TCM formula have been facilitated by modern analytical techniques. In particularly, high-resolution mass spectrometry (HRMS) plays a critical role in characterizing structures of chemical compounds by providing precise molecular weight as well as fragmental structures with the advantages of high sensitivity and throughput in detecting versatile molecules [1]. Conventionally, liquid chromatography mass spectrometry (LC–MS) is one of the most widely used approaches to the preliminary characterization of chemical components of TCM formula extract. Nevertheless, it is time-consuming and difficult to analyze the MS data of a TCM formula due to its complex components, especially for unknown components.
Recently, the combination of LC-HRMS and molecular networking has facilitated the MS data analysis. Molecular networking (MN) is outstanding to dispose of complicated MS data. It is capable of gathering the molecules with similar structures together based on the similarity of their MS/MS fragments. Compounds that share similar MS/MS fragmentation patterns or molecular classes are likely to group together in MN. This improves the possibility of identification of unidentified nodes, if their spectra or the spectra of surrounding nodes are known by references [2,3,4]. Thus, the combination of LC-HRMS and molecular networking immensely enhances the efficiency and drastically reduces the time on data processing. In the last few decades, molecular networking was introduced in drug development and metabolomics, particularly for natural products containing hundreds of components.
As one example from the "Catalogue of Ancient Chinese Classic formula (First Batch)", Erdong decotion was record in yixuexinwu and used in nourishing Yin and quenching thirst. In modern clinical practice, Erdong decoction and its modified prescriptions have been mainly used to treat type 2 diabetes and its complications [5, 6]. It was composed of eight herbs including Asparagi Radix (the root of Asparagus cochinchinensis (Lour.)Merr.), Ophiopogonis Radix (the root of Ophiopogon japonicus.), Trichosanthis Radix (the root of Trichosanthes kirilowii Maxim.), Scutellariae Radix (the root of Scutellaria baicalensis Georgi.), Anemarrhenae Rhizoma (the rhizome of Anemarrhena asphodeloides Bunge.), Glycyrrhizae Radix Et Rhizoma (the root et rhizome of Glycyrrhiza uralensis Fisch.), Ginseng Radix Et Rhizoma (the root et rhizome of Panax ginseng C. A. Mey.) and Nelumbinis Folium (the leaf of Nelumbo nucifera Gaertn.). However, hitherto there is no report on systematic characterization of chemical components of Erdong decoction and its quality control methods.
In this study, the combination of LC-HRMS and molecular networking was applied to rapidly identify compounds in Erdong decoction as a case study to demonstrate the application of the combined techniques in TCM formula. An ultra-high pressure liquid chromatography-linear ion trap-orbitrap high resolution mass spectrometry (UHPLC-LTQ-Orbitrap-MS/MS) approach was developed to comprehensively profile and characterize multi-components in Erdong decoction. Then the MS data of Erdong decoction was analyzed by MS/MS-based molecular networking (Fig. 1). The results show that the combination of LC-HRMS and molecular networking greatly improves the efficiency of chemical components identification in CCF composed of many herbs.
Materials and methods
Materials and reagents
Asparagus cochinchinensis was purchased from Guizhou Province in July 2018. O. japonicus was purchased from Santai, Sichuan Province in July 2018. T. kirilowii was purchased from Feicheng, Shandong Province in July 2018. S. baicalensis was purchased from Lingchuan, Shanxi Province in July 2018. A. asphodeloides was purchased from Wanrong, Shanxi Province in July 2018. G. uralensis was purchased from Beitun Town, Xinjiang Province in July 2018. P. ginseng was purchased from Fushong, Jilin Province in July 2018. N. nucifera was purchased from Nanchang, Jiangxi Province in September 2018. Reference compounds, neomangiferin, oroxylin A-7-O-β-D-glucuronide and glycyrrhizin acid were purchased from Beijing Century Aoko Biotechnology Co. Ltd. (Beijing, China), mangiferin, baicalin and wogonoside were purchased from National Institutes for Food and Drug Control (Beijing, China), and quercetin-3-O-glucuronide and hyperoside were purchased from Chengdu Cloma Biological Technology Co. Ltd. (Sichuan, China). HPLC-grade acetonitrile and LC–MS-grade formic acid were purchased from Fisher Scientific (USA).
Sample preparation
The solutions of neomangiferin, mangiferin, hyperoside, quercetin-3-O-glucuronide, baicalin, oroxylin A-7-O-β-D-glucuronide, wogonoside and glycyrrhizic acid were prepared in methanol at appropriate concentrations. A mixture of 8 different slices consisting of 33.6 g of dried O. japonicus radixs, 22.5 g of dried A. cochinchinensis radixs, 11.1 g of dried T. kirilowii radixs, 11.1 g of dried S. baicalensis radixs, 11.1 g of dried A. asphodeloides naerhizomas, 11.1 g of dried N. nucifera foliums, 5.7 g of dried G. uralensis radix et rhizoma, and 5.7 g of dried P. ginseng radix et rhizome were subjected to decoction twice with 10-times amount of distilled water for 40 min and 6-times distilled water for 30 min, respectively. The extraction temperature is around 96–100 ℃, at which the decocting liquid keep boiling. All extraction solutions were concentrated to 560 mL at 60 ℃. One hundred microlitre of concentrated solution was dissolved in 900 μL of 10% acetonitrile and centrifuged at 13,000 r·min−1 for 5 min, then the supernatant solution was filtered through a 0.22 μm membrane filter prior to injection into the chromatographic system.
Data acquisition and molecular networking analysis
HPLC analysis was performed on DIONEX Ultimate 3000 UHPLC system (USA) with photodiode array (PDA) detector. Samples were separated on an Acquity UPLC HSS T3 column (100 × 2.1 mm i.d., 1.8 μm) at 40 ℃. The mobile phase consisted of acetonitrile (A) and water containing 0.1% formic acid (B). A gradient program was adopted as follows: 0–3 min, 10–13% A; 3–6 min, 13–14% A; 6–9 min, 14–17% A; 9–11 min, 17–25% A; 11–18 min, 25–30% A; 18–19 min, 30–48% A; 19–22 min, 48–48% A, with a flow rate of 0.4 mL/min. The PDA detector scanned at 254 nm.
The LTQ-Orbitrap XL mass spectrometer was purchased from Thermo Scientific equipped with electrospray ionization (EIS) and Xcalibur 2.1 workstation. The analysis was performed in both negative and positive mode with a mass range of m/z 100–1400. High-purity nitrogen (N2) was used as auxiliary gas (10 arb) and sheath gas (40 arb). The other parameters were as follows: capillary temperature, 350℃; capillary voltage, 3.3 kV (in the positive mode), 3.0 kV (in the negative mode).
The MS data of the targeted fraction was converted from the raw format to the mzXML format using the Proteo-Wizard 3.0.20014. Then, the mzXML file was uploaded by the suggested software of WinSCP (https://winscp.net/eng/download.php) to the GNPS platform (https://gnps.ucsd.edu). The resulting analysis and parameters for the network can be accessed via links http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4e68c1650ff24c9091a7a021d52531e0 (in the negative mode) and http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=bcd0018bf90d44c09353515f1ed7bdca (in the positive mode). The following settings were used for generation of the network: minimum pairs cos 0.6; parent mass tolerance, 2 Da; MS/MS fragment ion tolerance, 0.5 Da; network top, 10; minimum matched peaks, 5. The molecular networking data were analyzed and visualized using Cytoscape (ver. 3.7.2).
Results
Study on molecular networking of mass spectrometry of Erdong decoction
All the full-MS and MS/MS spectra were obtained in high-resolution FT-MS for robust identification. In order to quickly identify the main chemical components in Erdong decoction, LC–MS/MS based molecular networking was applied. The MS data was processed through GNPS online workflow and visualized by MS/MS molecular networking. Their spectral similarities were evaluated through cosine calculation (cos θ), the larger the cos θ value, the higher the similarity of the MS/MS fragments [7]. The results showed that the cluster of molecular networking in the negative mode (Fig. 2) was more obvious than that of the positive mode (Additional file 1: Figure S1). The MS data of steroids, triterpenes, and flavonoids in the LC–MS/MS molecular networking of Erdong decoction were split into different groups. Herein, a total of 430 nodes was incorporated into the MS/MS molecular networking of Erdong decoction in the negative mode, rendering 30 molecular clusters and 164 unconnected nodes (Fig. 2). Based on the clusters in the molecular networking, 113 compounds were rapidly tentative identification from Erdong decoction for the first time in the negative mode, which including steroidal saponins, triterpenoid saponins, flavonoid O-glycosides and flavonoid C-glycosides. The typical total ion chromatograms (TIC) of Erdong decoction in the positive mode and the negative mode are presented in Fig. 3. Details of the characterization of these compounds were further elaborated.
Rapid identification of steroidal saponins
Previous studies had reported that steroidal saponin was one of the main compounds of Asparagi radix [8]. Taking aspacochioside A at m/z 903.495 as an example, its MS/MS spectrum showed three characteristic fragments of m/z 757.432, m/z 595.383, and m/z 433.330, which in turn lost rhamnosyl, glucosyl and glucosyl, the fragment of m/z 433.330 corresponding to the aglycone of aspacochioside A (Additional file 1: Figure S2). The fragmentation scheme of aspacochioside A was further elaborated in Additional file 1: Figure S2. In comparison to aspacochioside A, its adjacent node of m/z 919.491 gave a MS/MS spectrum showing identical aglycone and three identical characteristic fragments, with different [M−H]− ion (Fig. 4a). The node of m/z 919.491 was preliminarily deduced as aspacochioside A analogue with one more hydroxyl group to the rhamnose of aspacochioside A, finally annotated as 3-O-β-d-glucopyranosyl (1 → 2)-β-d-glucopyranosyl-26-O-β-d-glucopyranosyl-(25S)-5β-furostane-3β,22α,26-triol according literature [8]. According to the clusters, the structures of these compounds could be rapidly identified. Sixteen steroidal saponins were tentatively identified from Asparagi radix and 14 steroidal saponins were tentatively identified from Anemarrhenae rhizoma by comparison with reported literatures [8,9,10] (Table 1), and they were annotated in red and light green in Fig. 2, respectively.
Steroidal saponins in Erdong Decoction are partly from Asparagi radix and Anemarrhenae rhizoma, and partly from Ophiopogonis radix. But only two steroidal saponins from Ophiopogonis radix were tentatively identified by comparison with literature [11] (Table 1) and no saponins from Trichosanthis radix were identified in Erdong Decoction.
Rapid identification of triterpenoid saponins
Triterpenoid saponins in Erdong decoction were derived from Glycyrrhizae radix and Ginseng radix. Glycyrrhizin acid as the mainly active compound in Glycyrrhizae radix [12], its MS/MS fragments mainly showed the fragment of disaccharides chain at m/z 351.057 and the weak signal of aglycone fragment at m/z 469.332. The fragmentation scheme of glycyrrhizin acid was further elaborated in Fig. 5a. In comparison to glycyrrhizin acid, its adjacent node of m/z 837.392 gave a MS/MS spectra of an identical disaccharides chain fragment, with different fragment of aglycone at m/z 485.330 (Fig. 4b). The node of m/z 837.392 was preliminarily deduced as glycyrrhizin acid analogue with one more hydroxyl group in the aglycone moiety of glycyrrhizin acid, finally annotated as macedonoside A by comparison with literature [12]. Based on the cluster, twenty-four triterpenoid saponins were rapidly tentative identification from Glycyrrhizae radix by comparison with literatures [12, 13], including 3 groups of isomers (Table 1), they were annotated in dark green in Fig. 2.
Ginsenosides could not be quickly identified by LC–MS/MS molecular networking under the condition of negative mode. Only 8 triterpenoid saponins from ginseng were tentatively identified by comparison with literatures [14, 15] (Table 1), they were annotated in purple in Fig. 2.
Rapid identification of flavonoids
The flavonoids in Erdong decoction were derived from four herbs, Anemarrhenae rhizoma, Nelumbinis folium, Glycyrrhizae radix and Scutellariae radix. According to the difference of glycoside bond atoms, flavonoids in Erdong decoction were divided into two types. Identified flavonoids were annotated in blue for flavonoid O-glycosides and light blue for flavonoid C-glycosides (Fig. 2).
Flavonoid O-glycosides
The flavonoid O-glycosides in the Erdong decoction are mainly from Scutellariae radix and Glycyrrhizae radix. The types of aglycone are mainly flavone and flavanone. It was well known that baicalin and wogonoside were mainly active components in Scutellariae radix [16, 17]. Peak 72 was identified as wogonoside by comparison with its standard compound, and its MS/MS spectra showed three characteristic fragments of m/z 283.061, m/z 268.038, and m/z 240.042, which in turn lost C6H8O6, CH3 and CO, the fragment of m/z 283.061 corresponding to the aglycone moiety of wogonoside by the loss of Da 176 (C6H8O6) from the [M−H]− [18] (Additional file 1: Figure S3). The fragmentation scheme of wogonoside was further elaborated in Additional file 1: Figure S3. In comparison to wogonoside, its adjacent node of m/z 475.088 gave a MS/MS spectrum of different aglycone fragment at m/z 299.056 by the loss of Da 176 (C6H8O6), with one more hydroxyl group to the aglycone of wogonoside. The node of m/z 475.088 was annotated as the isomer of hydroxyl wogonoside according to literatures [16, 19] (Fig. 4c). Notably, another adjacent node of m/z 445.078 was connected to wogonoside in the molecular networking with a relatively low similarity (Fig. 4c). Comparing with wogonoside, the node of m/z 445.078 gave a MS/MS spectrum showing a different aglycone fragment at m/z 269.045 by the loss of Da 176 (C6H8O6), with one less methyl group to the aglycone of wogonoside. The node of m/z 445.078 was annotated as baicalin by comparison with standard compound. Basing on the cluster, forty-one flavonoid O-glycosides were tentatively identified from Scutellariae radix and Glycyrrhizae radix by comparison with literatures [12, 16, 17].
Some studies have shown that liquiritin and isoliquiritin are the active compounds in Glycyrrhizae radix [12]. It is noteworthy that some of isomers could not be distinguished by MS/MS and MN, but these isomers could be separated by retention time during LC–MS/MS analysis. Therefore, two groups of flavonoid isomers (peaks 9, 11, 44, 48, 14, 38, and 46) from Glycyrrhizae radix were tentatively identified by comparison with literatures [12, 13] (Table 1).
Flavonoid C-glycosides
The flavonoid C-glycosides in Erdong decoction were mainly from Scutellariae radix and Anemarrhenae rhizoma. Taking peak 19 at m/z 547.146 as an example, at m/z 487.125, m/z 457.114, m/z 427.123 involved serial losses of 60 Da, 90 Da, 120 Da, revealed that these compounds were flavonoid C-glycosides with two attached saccharides: glucose and arabinose [16]. So peak 19 was identified as Chrysin 6-C-arabinoside-8-C-glucoside. The fragmentation scheme of Chrysin 6-C-arabinoside-8-C-glucoside was further elaborated in Fig. 5b and it shows special cleavage rule in the glucosyl part. In comparison to Chrysin 6-C-arabinoside-8-C-glucoside, its adjacent node of m/z 561.161 gave a MS/MS spectrum showing two characteristic fragments at m/z 471.130 and at m/z 441.118 by the loss of 90 Da, 120 Da, and so one more methyl group should be connected to the aglycone of Chrysin 6-C-arabinoside-8-C-glucoside. The node of m/z 561.161 was annotated as 5-hydroxy-7-methoxyflavone 6-C-arabinoside-8-C-glucoside or 7-hydroxy-5-methoxyflavone 6-C-arabinoside-8-C-glucoside [16] (Fig. 4d). Basing on the cluster, six flavonoid C-glycosides were tentatively identified from Scutellariae radix by comparison with literature [16].
Previous studies showed that the flavonoids from Anemarrhenae rhizoma were main xanthones, which was a special structure type of flavonoids, so it was not clustered with most of flavonoids in the molecular networking. Finally, 3 flavonoid C-glycosides were tentatively identified from Anemarrhenae rhizoma by comparison with literature [10] (Table 1).
Identification of alkaloids
A total of 169 nodes were incorporated into the MS/MS molecular network (in the positive mode) of the Erdong decoction, rendering 15 molecular clusters and 88 unconnected nodes (Additional file 1: Figure S1). Besides the above three types of main compounds detected in Erdong decoction in negative mode, there are alkaloids from Nelumbinis folium mainly detected in positive mode. The mass spectrum of nuciferine at m/z 296.164 was detected and its MS/MS spectrum showed four characteristic fragments of m/z 265.123, m/z 250.098, m/z 234.103 and m/z 235.075 (Additional file 1: Figure S4). The fragmentation scheme of nuciferine was further elaborated in Additional file 1: Figure S4. It was well known that alkaloids were the major active compound of Nelumbinis folium [20], however, it was not shown in molecular networking and alkaloids could not be rapidly identified through the clusters in the LC–MS/MS molecular networking due to its various structural types. Finally, a total of 10 alkaloids were tentatively identified from Nelumbinis folium by comparison with literatures [20, 21] (Table 2).
Discussion
In this study, the cluster of molecular networking in the negative mode (Fig. 2) was more obvious than that in the positive mode (Additional file 1: Figure S1). And more flavonoids, steroidal saponins, and triterpenoid saponins were tentatively identified in the negative mode than in positive mode. So, in this study, the flavonoids, steroidal saponins, and triterpenoid saponins in Table 1 were tentatively identified in the negative mode. The alkaloids were the major active compound of Nelumbinis folium, which were mainly detected in positive mode. And no cluster were observed in the molecular networking of the alkaloids, that might be due to the various types of structural framework of alkaloids, and it leads to the MS/MS fragments of alkaloids doesn’t have a certain similarity. Therefore, 10 alkaloids were tentatively identified in the positive mode from Nelumbinis folium by comparison with literatures.
According to the above results, LC–MS/MS molecular networking is suitable for the rapid identification of steroidal saponins, glycyrrhizin saponins, and flavonoids. Because of the stable structure of steroidal saponins and glycyrrhizin saponins, and special cleavage rule of flavonoid C-glycosides, their analogues in the LC–MS/MS molecular networking were obviously clustered with a high similarity. Based on the clusters, the structures of these compounds could be rapidly tentative identification by MN. In addition, the flavonoid O-glycosides obviously clustered in LC–MS/MS molecular networking, but the similarity between nodes was low, which might be due to different substituents sites on aglycones. Therefore, the identification of flavonoid O-glycosides could be facilitated by the combination of LC–MS/MS and molecular networking, but standard compounds are needed for the finally identification of isomers.
Notably, MS/MS-based molecular networking technique is not suitable for the rapid identification of compounds without cluster in MN. Steroidal saponins from Ophiopogonis radix and triterpenoid saponins from Ginseng radix in Erdong decoction couldn’t be rapidly identified, which might be due to their low content caused by both low formula ratio in Erdong decoction and low content in each herb itself. According to the unpublished quantification data by our laboratory, the content of saponins from Glycyrrhizae Radix Et Rhizoma, Anemarrhenae Rhizoma, Asparagi Radix are very high, whereas the content of saponins from Ophiopogonis Radix and Ginseng Radix Et Rhizoma are very low. The content of those compounds might be too low to generate fragment of aglycones in this study, so the MS/MS fragments of these compounds were not clustered in this study. The second type of compounds without cluster in the molecular networking is the alkaloids from Nelumbinis folium.
Conclusions
In this study, the combination of LC-HRMS and molecular networking was applied to rapidly identify compounds in Erdong decoction as a case study to demonstrate the application of this technique in complex TCM formula. MS/MS-based molecular networking technique is very useful for the rapid identification of major components in CCF. Finally, 113 compounds were rapidly tentative identification in the negative mode by the MS/MS-based molecular networking, the types of these compounds mainly include steroidal saponin, triterpenoid saponins and flavonoids in Erdong decoction. MS/MS-based molecular networking greatly improves the efficiency of chemical components identification in CCF. In addition, 10 alkaloids were tentatively identified in the positive mode of Nelumbinis folium by comparison with literatures.
Availability of data and materials
All data included in this article are available from the corresponding author upon request.
Abbreviations
- CCF:
-
Chinese Classical Formula
- UHPLC-LTQ-Orbitrap-MS/MS:
-
Ultra-high pressure liquid chromatography-linear ion trap-orbitrap high resolution mass spectrometry
- TCM:
-
Traditional Chinese medicine
- HRMS:
-
High-resolution mass spectrometry
- LC–MS:
-
Liquid chromatography mass spectrometry
- MN:
-
Molecular networking
References
Yao C, Yang W, Si W, Pan H, Qiu S, Wu J, et al. A strategy for establishment of practical identification methods for chinese patent medicine from systematic multi-component characterization to selective ion monitoring of chemical markers: Shuxiong tablet as a case study. RSC Adv. 2016;6(69):65055–66.
Wei W, Hou J, Yao C, et al. A high-efficiency strategy integrating offline two-dimensional separation and data post-processing with dereplication: caracterization of bufadienolides in Venenum Bufonis as a case study. J Chromatogr A. 2019;1603:179–89.
Wang T, Lu Q, Sun C, et al. Hetiamacin E and F, new amicoumacin antibiotics from Bacillus subtilis PJS using MS/MS-based molecular networking. Molecules. 2020;25(19):4446.
Han YK, Kim H, Shin H, et al. Characterization of anti-inflammatory and antioxidant constituents from Scutellaria baicalensis using LC-MS coupled with a bioassay method. Molecules. 2020;25(16):3617.
Zou XH, Zhang JX, Liu SZ, et al. Clinical Study on Treatment of Diabetic Peripheral Neuropathy with Erdong Decoction and Zhufeng Tongbi Decoction Combined with Western Medicine. Chinese Archives of Traditional Chinese Medicine. 2012. (In Chinese)
Yuan X, Xie M, Yang Y, et al. Effect of erdong decoction and aerobic exercise on glucose metabolism and lipid metabolism in type 2 diabetes rats. Chongqing Medicine. 2018. (In Chinese)
Kuo TH, Huang HC, Hsu CC. Mass spectrometry imaging guided molecular networking to expedite discovery and structural analysis of agarwood natural products. Anal Chim Acta. 2019;1080:95–103.
Ling Y, He X, Jiang R, et al. Rapid detection and characterization of steroidal Saponins in the root of Asparagus cochinchinensis by high-performance liquid chromatography coupled to electrospray Ionization and quadrupole time-of-flight mass spectrometry. J Chromatogr Sci. 2020;5:5.
Jaiswal Y, Liang Z, Ho A, et al. A comparative tissue-specific metabolite analysis and determination of protodioscin content in Asparagus species used in traditional Chinese medicine and Ayurveda by use of laser microdissection, UHPLC-QTOF/MS and LC-MS/MS. Phytochem Anal. 2014;25(6):514–28.
Sun Y, Du Y, Yong L, et al. Simultaneous determination of nine components in Anemarrhena asphodeloides by liquid chromatography-tandem mass spectrometry combined with chemometric techniques. J Sep Sci. 2015;35(14):1796–807.
Wu Y. Studies on chemical composition analysis and pharmacokinetics of Radix Ophiopogonis based on LC-MS/MS technology. Hebei Medicine University. 2015. (In Chinese)
Zhang Q, Ye M. Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice). J Chromatogr A. 2009;1216(11):1954–69.
Ji S, Li Z, Song W, et al. Bioactive constituents of Glycyrrhiza uralensis (Licorice): discovery of the effective components of a traditional herbal medicine. J Nat Prod. 2017;79(2):281.
Hu C, Kong H, Zhu C, et al. An ultra performance liquid chromatography-time-of-flight-mass spectrometric method for fast analysis of ginsenosides in Panax ginseng root. Chin J Chromatogr. 2011;29(06):488–94.
Meng Q, Zhong YM, Guo XL, Feng YF. Rapid identification and preliminary study of fragmentation regularity of saponins components of ginseng. Zhong Yao Cai. 2013;36(2):240–5 (In Chinese).
Zhang F, Li Z, Li M, et al. An integrated strategy for profiling the chemical components of Scutellariae Radix and their exogenous substances in rats by ultra-high-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2020;34(18):e8823.
Huang S, Fu Y, Xu B, et al. Wogonoside alleviates colitis by improving intestinal epithelial barrier function via the MLCK/pMLC2 pathway. Phytomedicine. 2020;68:153179.
Tong L, Wan M, Zhang L, Zhu Y, Sun H, Bi K. Simultaneous determination of baicalin, wogonoside, baicalein, wogonin, oroxylin A and chrysin of Radix scutellariae extract in rat plasma by liquid chromatography tandem mass spectrometry. J Pharm Biomed Anal. 2012;70:6–12.
Zhang W, Saif M, Dutschman GE, et al. Identification of chemicals and their metabolites from PHY906, a Chinese medicine formulation, in the plasma of a patient treated with irinotecan and PHY906 using liquid chromatography/tandem mass spectrometry (LC/MS/MS). J Chromatogr A. 2010;1217(37):5785–93.
Menéndez-Perdomo IM, Facchini PJ. Isolation and characterization of two O-methyltransferases involved in benzylisoquinoline alkaloid biosynthesis in sacred lotus (Nelumbo nucifera). J Biol Chem. 2020;295(6):1598–612.
Ma W, Lu Y, Hu R, Chen J, Zhang Z, Pan Y. Application of ionic liquids based microwave-assisted extraction of three alkaloids N-nornuciferine, O-nornuciferine, and nuciferine from lotus leaf. Talanta. 2010;80(3):1292–7.
Acknowledgements
The authors greatly appreciate the financial support from Sunflower Pharmaceutical Group (Xiangyang) Longzhong Co. Ltd.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
YD, SC, and ZH designed the experiment. RJ, XW, PL and SS carried out the experiment. QJ contributed analysis tools. XX contributed to the data analysis. XX, QJ, SC, and YD. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
MS/MS molecular networking of Erdong decoction in the positive mode. Figure S2. The proposed fragmentation pathways and the MS/MS spectra for aspacochioside A in the negative mode. Figure S3. The proposed fragmentation pathways and the MS/MS spectra for wogonoside in the negative mode. Figure S4. The proposed fragmentation pathways and the MS/MS spectra for nuciferine in the positive mode.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xue, X., Jiao, Q., Jin, R. et al. The combination of UHPLC-HRMS and molecular networking improving discovery efficiency of chemical components in Chinese Classical Formula. Chin Med 16, 50 (2021). https://doi.org/10.1186/s13020-021-00459-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-021-00459-6