Abstract
Background
Homeostasis is a crucial concept used to describe the condition of patients and the roles of herbs in traditional Chinese medicine. Qi-deficiency pattern is one of the conditions when loss of homeostasis and is usually characterized by symptoms including lassitude, spontaneous sweating, and a weak pulse, which are not easy to quantitate. Codonopsis pilosula and Astragalus membranaceus were usually prescribed for carriers with hepatitis and patients with metastatic colon cancer, because these patients tended to experience fatigue. However, crude drugs were prescribed based on the exterior symptoms of patients without controlling clinical setting, such as gender, age, and dietary habits. Limited molecular evidence of using gene expression as the guide for description is available. Therefore, the purpose of this study was to identify potential and objective biomarkers of these two qi-related drugs in a simplified cellular system.
Methods
Aqueous extracts of crude qi-tonifying herbs, C. pilosula and A. membranaceus, and that of a qi-consuming drug, Citrus reticulata, were prepared. Human liver cancer HepG2 cells were treated with the extracts of qi-tonifying herbs for 24 h. Differentially expressed genes were identified using microarrays and quantitative RT-PCR (qRT-PCR) and validated in two other hepatocellular cell lines, Huh7 and L-02.
Results
A total of 67 differentially expressed probes that responded to both herbs were identified. A pathway analysis revealed that these genes were involved in the development, growth, movement, and viability of the liver cells.
Conclusions
After qRT-PCR validation and examination of clinical data from public domains, our results showed that two genes, GDF15 and HMOX1, could serve as biomarkers in liver cells for identifying responses after treatment with C. pilosula and A. membranaceus.
Similar content being viewed by others
Background
Aqueous extract decoction is a widely used form of drug preparation in traditional Chinese medicine (TCM), and physical symptoms in patients are a crucial basis for prescribing herbal decoctions. Qi-deficiency symptoms include shortness of breath, lassitude, listlessness, spontaneous sweating, a pale tongue, and a weak pulse [1]. Many related single or complex formulas have been widely prescribed in TCM to ameliorate these symptoms [2, 3].
Qi-related crude drugs can be classified as qi-tonifying and qi-consuming drugs. The use of dried roots of Codonopsis pilosula and Astragalus membranaceus, qi-tonifying crude drugs, has been well documented in the ancient Chinese literatures, including in Ben cao gang mu [4], a few 100 years ago. C. pilosula is usually combined with other herbs to alleviate qi deficiency. For example, an animal study demonstrated that C. pilosula polysaccharide attenuated sepsis in a mouse model of cecal ligation [5]. In a randomized controlled trial, TCM formulas containing C. pilosula were used in patients with chronic obstructive pulmonary disease [6]. Extracts of A. membranaceus have been reported to accelerate wound healing, exhibit antitumor activity [7], inhibit inflammation [8], and inhibit melanogenesis through ERK pathway [9]. Because these two drugs, especially A. membranaceus, the most potent qi-tonifying drugs [10], were prescribed frequently in patients with liver cancer who tended to experience fatigue, they were selected for the present study. Citrus reticulata is a qi-consuming plant used in TCM [4]. The peel of C. reticulata is the major constituent of the drug. It contains many polyphenols [11], and its extract possesses antioxidant, anticancer, and antibacterial properties [12, 13].
A single drug, C. pilosula or A. membranaceus, was usually prescribed on the basis of patients’ external symptoms alone with little molecular-marker evidence. Furthermore, although studies have suggested some molecular pathways might be involved in the mechanisms underlying these drugs [5, 7, 8], the overall mechanisms remain elusive. Therefore, the purpose of this study was identify potential biomarkers for qi-related drug treatment and establish the possible underlying mechanisms through biological approaches.
We hypothesized that the gene expression levels could serve as biomarkers to assess the status of cell conditions. In addition, because patients with related symptoms usually exhibit large variations in biochemical examination, hepatocellular cell lines, HepG2, Huh7, and L-02, were treated with the herbs in this study. Therefore, to identify biological functions, a genome-wide approach, microarray technique, was first used to screen differentially expressed genes and their functions. Furthermore, PCR results and clinical data from an independent cohort revealed that the expression levels of GDF15 and HMOX1 could serve as biomarkers for identifying responses of these botanical drugs in liver cells.
Materials and methods
The Minimum Standards of Reporting Checklist contains details of the experimental design, and statistics, and resources used in this study (Additional file 1).
Sample preparation
Dried roots of C. pilosula and A. membranaceus were used as crude drugs, as reported in our previous papers [14,15,16]. The roots were soaked in water at a ratio of 1:3 (g:mL) and boiled at 100 °C for 4 h. The decoctions were dehydrated using a freeze dryer (Panchum FD DC-3000, Panchum, Kaohsiung, Taiwan), filtered using a 0.22-µm filter (Merck Millipore, Tullagreen, Carrigtwohill, IRL), and re-dissolved in ddH2O.
Quality control of crude drugs using high-performance liquid chromatography (HPLC) analysis and internal transcribed space 1 (ITS1) analysis
The details of the preparation steps and high-performance liquid chromatography (HPLC) equipment and results have been described previously [14]. Because the same herbs were used in the present study, we only showed the following key settings: The powdered roots of both plants (0.2 g) were suspended in 70% methanol and filtered through a 0.2-μm HPLC membrane. The HPLC conditions were as follows: gradient range, 10–90% methanol; flow time, 120 min; detector, 254 nm; flow rate, 1 mL/min; injection volume, 20 μL; and internal control, evodiamine.
The internal transcribed spacer (ITS) analysis protocols have been described previously but only ITS2 sequencing results have been published [14, 15]. The ITS1 for all herbs were sequenced using the following primers: (forward) 5′-GGAAGTAAAAGTCGTAACAAGG-3′; (reverse) 5′-TCCTCCTCCGCTTATTGATATGC-3′ (Table 1). The botanical origins of the plants were validated by comparing the ITS sequences with those in the National Center for Biotechnology Information (NCBI) nucleotide database.
Cell culture
The cancerous liver cell lines HepG2 and Huh7 cells were cultured in Dulbecco modified Eagle medium (GIBCO, Carlsbad, CA, USA) containing 1% streptomycin/puromycin (Biological Industries, Kibbutz Beit-Haemek, Israel) and 10% fetal bovine serum (Biological Industries). The human fetal hepatocyte line, L-02, a kind gift from Dr. Hui-Lin Wu from Hepatitis Research Center, National Taiwan University Hospital, was cultured in Roswell Park Memorial Institute medium 1640 (GIBCO). All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
Cell viability assay
The cells were seeded at a density of 3000 cells per well into a 96-well microtiter plate and treated with various concentrations (0–5 mg/mL) of each drug for 24 h. Cells were incubated with 0.46 mg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, BioTek, Winooski, VT, USA) at 37 °C and in an atmosphere containing 5% CO2 for 1.5 h. A fixed volume was sampled from each well and dissolved in 100 μL of dimethyl sulfoxide. Absorbance values were measured using a microtiter plate reader (BioTek, Winooski, VT, USA) at 570 nm.
The BrdU assay served as another detection method for cell viability. The BrdU proliferation analysis was performed with the BrdU cell proliferation ELISA kit according to the manufacturer’s protocol (Abcam). Cells were incubated with BrdU buffer for 3 h, and the wavelength was measured at 450 and 550 nm.
DNA ladder assay
The HepG2 cells were seeded at a density of 1 × 105 cells per well into a 6-well microtiter plate and treated with 3 mg/mL C. pilosula or A. membranaceus for 48 h. DNA was extracted according to the manufacturer’s protocol (Qiagen, Hilden, Germany). Three micrograms of DNA were loaded in 1.5% agarose gel (Seakenm, Lonza Rockland, ME, USA). DNA detection was done by safe DNA gel stain system (Invitrogen, USA) with BioSpectrum Imaging System (UVP, Upland, CA, USA). A newly synthesized compound 1-(9′-methyl-3′-carbazole)-3,4-dihydro-beta-carboline (MCDC) [17] was served as positive control.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using the TRIpure reagent (Roche Diagnostics, Branchburg, NJ, USA) according to the manufacturer’s protocol. One microgram of total RNA was reverse transcribed to cDNA by using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA). Ten percent of each cDNA sample was used as a template, and the mRNA levels of different genes were quantified through quantitative RT-PCR (qRT-PCR) analysis by using primers presented in Table 2. The intensity of SYBR Green was measured using StepOnePlus Real-Time PCR Systems (ThermoFisher, Waltham, MA USA) and OmicsGreen qPCR 5× Master Mix (OmicsBio, Taipei, Taiwan) according to the manufacturers’ instructions.
Human genome microarray analysis and Ingenuity Pathway Analysis (IPA)
An Illumina TotalPrep RNA Amplification Kit (Ambion Inc, Austin, TX, USA) and the T7 Oligo (dT) reagent were used to amplify total RNA. The cRNA was mixed with an equal volume of hybridization buffer, hybridized to Illumina Human HT-12 v4 BeadChips (Illumina) arrays at 58 °C for 16 h, and stained using the dye streptavidin-Cy3. An Illumina BeadArray Reader was used to read intensities, and the results were analyzed using BeadStudio v3.1 software. Background-adjusted signals were normalized using a quantile normalization algorithm, and the Partek Genomics Suite (Partek Incorporated, St. Louis, MO, USA) was used for principal components analysis (PCA). Cellular functions of the differentially expressed genes were examined using the Ingenuity Pathway Analysis (IPA) tool (Ingenuity Systems Inc. Redwood City, USA). Results of the microarray analysis are uploaded to the Gene Expression Omnibus (GEO) database with accession numbers GSE115506.
Cell cycle analysis
Cultured cells were harvested through trypsinization, treated with 0.5% Triton X-100 (Sigma, Saint Louis, MO, USA) in phosphate-buffered saline, and fixed with 100% methanol (Sigma). Cell pellets were then treated with 20 μg/mL propidium iodide (Life Technologies, Eugene, Oregon, USA), 0.1% Triton-X-100 (Sigma) and 100 μg/mL RNase A (Sigma) for 10 min. The suspension was analyzed using Beckman Coulter FC500 (Beckman, Brea, CA, USA) and ModFit LT analysis software.
Protein extraction and western blotting
Cells were lysed in radioimmunoprecipitation assay lysis buffer (Sigma). A total of 25 μg of protein lysate was loaded in each lane, separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories). We used the following primary antibodies: rabbit antihuman GCNT3, GDF15, HMOX1, and GAPDH polyclonal antibodies (GeneTex, Irvine, CA, USA). HRP-conjugated goat antirabbit IgG polyclonal antibody (GeneTex) was used as the secondary antibody. Visualization was performed using an enhanced chemiluminescence system (Millipore, Billerica, MA, USA). Images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/).
Results
Quality control of three crude drugs
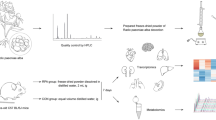
The workflow for identifying differentially expressed genes in HepG2 cells treated with C. pilosula or A. membranaceus was shown in Fig. 1. To examine the identity of crude drugs, high-performance liquid chromatography (HPLC) analysis and internal transcribed spacer (ITS) analysis were performed. The crude drugs used in this study are the same as that used in our previous study [14]. HPLC and ITS2 sequences data were published previously [14]. The results of HPLC confirmed the ingredients from same batch with same quality. However, ITS1 sequences have not yet been tested. Therefore, in this study, we sequenced ITS1 of C. pilosula, A. membranaceus, and C. reticulata (Table 1) and validated the botanical identity of these plants by comparing their sequences with those in the NCBI nucleotide database.
Growth inhibitory effects of crude drugs in HepG2 cells
To determine the optimal conditions for testing the effects of crude drugs (C. pilosula and A. membranaceus), dose–response studies were conducted using HepG2 cells (Fig. 2). Cytotoxic effects of the crude drugs were measured using the MTT (Fig. 2a) and BrdU (Fig. 2b) assays at 48 h. The dose close to a 50% inhibition rate was 3 mg/mL (Fig. 2a, b) for both C. pilosula and A. membranaceus; this dose was used for the following experiments.
Cytotoxic effects of C. pilosula and A. membranaceus on HepG2 cells. Cytotoxic effects of crude drugs were measured using the a MTT and b BrdU assay at 48 h. c Cell cycle analysis. HepG2 cells were stained with propidium iodide and subjected to flow cytometry analysis after 48 h of treatment with 3 mg/mL C. pilosula and A. membranaceus. The stacked bar chart summarizes three independent cell cycle experiments. d DNA ladder assay. DNA from HepG2 cells was extracted after 48 h of treatment with 3 mg/mL C. pilosula and A. membranaceus. 5 μM of 1-(9′-methyl-3′-carbazole)-3,4-dihydro-beta-carboline (MCDC) was served as positive control. All experiments were repeated at least three times and results are presented as mean ± SEM
Because 3 mg/mL of C. pilosula or A. membranaceus caused nearly 50% of growth inhibitory effects on HepG2 cells, we examined whether this phenomenon was caused by the triggering of apoptosis. Approximately only 2% of cells exhibited in the sub-G0/1 peaks of the cell cycle. Only 1-(9′-methyl-3′-carbazole)-3,4-dihydro-beta-carboline (MCDC), a known compound that induced strong cytotoxic and apoptosis effect against kinds cancer cells [17], triggered DNA ladder effect on HepG2 cells but C. pilosula and A. membranaceus did not (Fig. 2d). These two botanic drugs did not induce strong apoptosis on HepG2 cells with this dosage.
Downstream pathways of C. pilosula and A. membranaceus
Because the related pathways of these two drugs are unclear, we used microarrays to conduct the genomic profiling of differentially expressed genes in the HepG2 cells treated with two qi-tonifying plants, C. pilosula and A. membranaceus. HepG2 cells were treated with the drugs at a dosage of 3 mg/mL for 24 h, and total RNA was extracted afterwards. The selection criteria for the differentially expressed genes were a P value of < 0.05 and fold change of at least 1.5×. As shown in Fig. 3a and b, 124 probes were identified in the HepG2 cells treated with C. pilosula (Fig. 3a) and 454 probes were identified in HepG2 cells treated with A. membranaceus (Fig. 3b). To reduce false positives, 67 probes significantly responsive to both C. pilosula and A. membranaceus treatments were identified (Fig. 3c). To examine the drug effect at the transcriptional level, PCA was conducted using the common differentially expressed probes. As shown in Fig. 3d, the distribution of samples treated with C. pilosula (blue spots) and A. membranaceus (green spots) was closer to that of the samples treated with the control (black spots), indicating that the expression profiles of cells treated with C. pilosula and A. membranaceus were similar. The expression profiles of these differentially expressed probes were clustered and are shown in Fig. 3e.
Identification of differentially expressed genes in HepG2 cells treated with C. pilosula and A. membranaceus. a Volcano plots of HepG2 cells treated with C. pilosula for 24 h at a dose of 3 mg/mL. Total RNA was extracted and genomic profiling was performed using Illumina Human HT-12 v4 BeadChips. The red spots represent probes with fold changes ≥ 1.5× and P < 0.05, whereas the green spots represent probes with fold changes < 1.5× and P < 0.05. b Volcano plots of HepG2 cells treated with A. membranaceus for 24 h at a dose of 3 mg/mL. The orange–red spots represent probes with fold changes > 1.5× and P < 0.05, whereas the blue spots represent probes with fold changes < 1.5× and P < 0.05. c Venn diagram of differentially expressed probes. The blue circle represents probes identified from HepG2 cells treated with C. pilosula. The green circle represents probes identified from cells treated with A. membranaceus. d Principal component analysis of cells treated with C. pilosula or A. membranaceus with the common differentially expressed probes (n = 67). Blue spot: C. pilosula treatment; green spot: A. membranaceus treatment; black spot: non-treated control. e Heatmap and hierarchical cluster analysis of differentially expressed probes responsive to both C. pilosula and A. membranaceus treatment. Upregulated and downregulated probes are denoted in red and green, respectively
The 67 differentially expressed probes responsive to both C. pilosula and A. membranaceus were further examined. Of these, one probe exhibited an inconsistent pattern in both treatments and was hence removed from further analysis; of the remaining 66 probes corresponding to 66 genes, 20 genes were downregulated and 46 were upregulated. To explore the disorders and functions of these qi-tonifying related genes, IPA was performed. The results of the pathway analysis revealed that these genes are involved in organismal injury, reproductive system diseases, and cancer (Table 3). The top four molecular and cellular functions controlled by these genes were related to the development, growth, movement, and viability of cells (Table 4).
Validation of probes related to qi-tonifying herbs by using quantitative RT-PCR
To validate these drug-related genes as biomarkers for qi-tonifying herbs, we selected the top nine genes with maximal changes from the common genes responsive to qi-tonifying herbs and validated their expression levels through qRT-PCR. As shown in Fig. 4, the expression levels of GCNT3, GDF15, HMOX1, and IGFBP1 were significantly (P < 0.05) upregulated after A. membranaceus treatment and slightly upregulated after C. pilosula treatment. Conversely, the expression levels of HAMP, HEY1, HMGCS1, and MT1E were significantly (P < 0.05) downregulated after both A. membranaceus and C. pilosula treatment.
Validation of differentially expressed genes by using quantitative RT-PCR. Expression levels of nine genes were measured in HepG2 cells treated with C. pilosula or A. membranaceus (3 mg/mL) for 24 h by using qRT-PCR. Internal control: ACTB. The relative expression levels of genes were normalized to the non-treated group. All experiments were repeated at least three times and results are presented as mean ± SEM. *P < 0.05, Student t test
Effects of qi-consuming crude on drug-related gene markers
To reduce false positives, we expected qi-related genes to exhibit opposite responses to treatments with qi-tonifying and qi-consuming herbs. To identify the biomarkers that can accurately reflect the qi status, we examined the expression levels of the aforementioned nine genes in the HepG2 cells treated with the peel of C. reticulata, a qi-consuming crude drug. HepG2 cells were treated with 1.5 mg/mL of C. reticulata for 24 h (Fig. 5a), and total RNA was extracted to examine expression values. As shown in Fig. 5b, GCNT3, GDF15, and HMOX1 exhibited reciprocal responses to qi-tonifying (A. membranaceus and C. pilosula) (Fig. 4) and qi-consuming (C. reticulata) treatments, indicating that these three genes can be used as biomarkers to reflect the status after treatment with qi-related drugs.
Validation of drug-related biomarkers in HepG2 cells treated with C. reticulate. a Cytotoxic effects of C. reticulate on HepG2 cells were measured using the MTT at 48 h. b Relative expression levels of drug-related genes measured in HepG2 cells treated with C. reticulate at a dose of 1.5 mg/mL for 24 h
Expression patterns of GCNT3, GDF15, and HMOX1 in two other hepatocellular cell lines
To exclude the cell line–specific effect, we further examined the expression of these genes in a hepatocarcinoma cell line, Huh7, and normal hepatic L-02 cells. The doses that caused 50% of growth inhibition of C. pilosula and A. membranaceus were 4.5 and 4 mg/mL in Huh7 cells respectively (Fig. 6a, b). The same experiment revealed 50% growth inhibition doses of 5 and 4 mg/mL on L-02 cells (Fig. 6d, e). Expression patterns of GCNT3, GDF15, and HMOX1 in Huh7 and L-02 cells after C. pilosula or A. membranaceus treatments (Fig. 6c, f) were similar to those in hepatocellular carcinoma HepG2 cells (Fig. 4).
Expression levels of drug-related genes in Huh7 and L02 liver cells treated with C. pilosula and A. membranaceus. Cytotoxic effects of C. pilosula (a) and A. membranaceus (b) on Huh7 cells measured using MTT assay at 48 h. c Relative expression levels of drug-related genes in Huh7 cells treated with C. pilosula (4.5 mg/mL) or A. membranaceus (4 mg/mL) for 24 h. Cytotoxic effects of d C. pilosula and e A. membranaceus on L-02 cells measured by using the MTT assay at 48 h. f Relative expression levels of drug-related genes in L-02 cells treated with C. pilosula (5 mg/mL) or A. membranaceus (4 mg/mL) for 24 h
Expression of GDF15 and HMOX1 proteins was elevated in HepG2 cells
The protein expressions of GCNT3, GDF15 and HMOX1 were examined in HepG2 cells at the dose of 3 mg/mL of both C. pilosula or A. membranaceus for 48 h. Similar to the results of RNA (Fig. 6c, f), the GDF15 protein content drastically increased after treatments with C. pilosula or A. membranaceus, HMOX1 was increased after the treatment of A. membranaceus, and amount of GCNT3 did not change (Fig. 7a).
Validation of drug-related biomarkers at the protein level by using clinical data from external dataset. a Immunoblots of drug-related biomarker proteins. b Relative expression levels of drug-related genes in adjacent normal tissues compared with hepatocellular carcinoma were examined using the GEO data set, GSE60502 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60502)
Drug-related gene markers on clinical samples
After identifying the markers in the cell line model, we examined the relative expression levels of the three genes in clinical samples. The expression data were downloaded from the GEO data set GSE60502. As shown in Fig. 7b, the expression patterns of GDF15 and HMOX1, but not GCNT3, in adjacent normal tissues compared with hepatocellular carcinoma showed patterns similar to those in cells treated with qi-tonifying drugs (Figs. 4, 6), implicating the application of these two genes (GDF15 and HMOX1) as biomarkers after treatment with qi-related drugs.
Discussion
Although the definition and characteristics of qi-related botanical drugs, such as C. pilosula and A. membranaceus, have been debated for a long time, no decisive conclusion has been reached. The key reason for the controversial results is that these drugs are prescribed exclusively on the basis of apparent symptoms, which are hard to define neither qualitatively nor quantitatively. In this study, we qualitatively defined biological effects by measuring the expression levels of related genes in hepatocellular cells and hepatocellular carcinoma samples to identify indicators that reflect the qi status after treatment with these two herbs.
Astragalus membranaceus and other botanic drugs are prescribed frequently in patients with liver cancer [10]. However, identifying related biomarkers for each drug was difficult because numerous variables could not be controlled in a clinical setting, such as sex, age, and dietary habits. Therefore, we used a cell model in the current study to eliminate these variables. Furthermore, genomic approaches, such as microarray technique are ideal detection methods for surveying the mechanism of botanical herbs because one herbs may contain many ingredients and may trigger many downstream pathways.
To reduce the effects of false positives, we only selected differentially expressed genes that responded to both C. pilosula and A. membranaceus (Fig. 3c). These drug-related gene candidates were mainly involved in organismal injury, cellular development, and cellular growth. These results are consistent with those in previous studies [5, 7, 8]. For example, C. pilosula induced proliferation and migration in RSC96 Schwann cells [18] and exhibited a hepatoprotective effect in a rat model [19]. A. membranaceus exhibited cardioprotective activity [20] and wound-healing effects [8]. These studies have shed some light regarding the association between qi-related drugs and biological functions and could provide clues for additional explorative experiments in the future.
After microarray analysis, nine genes with relatively high fold changes were selected for validation through qRT-PCR. Except AKR1B15, the remaining eight genes had expression patterns similar to those in the microarray (Fig. 4). Because the drug-related genes were expected to exhibit opposite responses when the cells were treated with qi-tonifying and qi-consuming herbs, the genes were also examined in HepG2 cells treated with the qi-consuming drug C. reticulata. GCNT3, GDF15, and HMOX1 exhibited reciprocal responses to qi-tonifying (Fig. 4) and qi-consuming (Fig. 5) treatments.
The expression patterns of these three genes were examined in other hepatocellular cells. The expression patterns of GCNT3, GDF15, and HMOX1 in Huh7 and L-02 cells after treated with qi-tonifying herbs (Fig. 6) were similar to those in hepatocellular carcinoma HepG2 cells (Fig. 4). Other hepatoma cell lines, such as Hep3B and HA22T, were not selected because both C. pilosula and A. membranaceus cannot reach fifty percent of growth inhibitory effect at the maximum dose of 5 mg/mL (data not shown). Hep3B and HA22T belong to poorly differentiated malignant hepatocellular cell lines [21] and they might show more resistance against anticancer drugs.
Codonopsis pilosula and A. membranaceus are frequently prescribed according to patients’ external symptoms. Qi deficiency is characterized by physical symptoms such as lassitude, spontaneous sweating, pale complexion, and a weak pulse, which cannot be easily quantitated. The practical significance of the experiments was to identify the gene markers for these drugs so that these drugs can be prescribed by examining the gene marker expressions in patients. Therefore, the relative expressions levels of these three drug-related genes were examined in clinical samples from public repository, Gene Expression Omnibus (GEO). We hypothesized that the expression profiles of these drug-related genes in cells treated with qi-tonifying drugs, C. pilosula or A. membranaceus, are similar to those in adjacent normal tissues compared with cancerous tissues. GDF15 and HMOX1, but not GCNT3, were upregulated in adjacent normal tissues, which was similar to the pattern in cells treated with qi-tonifying drugs (Figs. 4, 6). Therefore, our results suggested that GDF15 and HMOX1 could be biomarkers for treatment with qi-related drugs. Although it remains unclear how these genes could reflect the cell status after treatment with qi-related drugs, previous studies have shown that GDF15 promoted angiogenesis through the p53/HIF-1 alpha pathway [22], and HMOX1 maintained bone mass by attenuating a redox imbalance [23]. However, additional experiments are required to explore the underlying mechanism by which these two genes reflect the cell status and to examine the expression of these genes in our collected clinical samples.
Conclusions
By using the genomic approach of microarrays in hepatocellular carcinoma cells treated with qi-tonifying botanic drugs, C. pilosula and A. membranaceus, followed by qRT-PCR validation of hepatic normal cells and clinical samples, we identified drug-related genes associated with qi and their functions. Additionally, our results suggested that GDF15 and HMOX1 are potential biomarkers in hepatic cells, which might serve as indicators of effectiveness while prescribing these qi-related drugs.
Abbreviations
- TCM:
-
traditional Chinese medicine
- ITS1:
-
internal transcribed space 1
- PCA:
-
principal components analysis
References
Hsu CH, Lee CJ, Chien TJ, Lin CP, Chen CH, Yuen MJ, Lai YL. The relationship between qi deficiency, cancer-related fatigue and quality of life in cancer patients. J Tradit Complement Med. 2012;2(2):129–35.
Zhu M, Liu Z, Gao M, Zhang Y, Li Y, Ling S, Zhang P, Zhao C, Jiang L, Liu Y, et al. The effect of Bu Zhong Yi Qi decoction on simulated weightlessness induced muscle atrophy and its mechanisms. Mol Med Rep. 2017;16(4):5165–74.
Kong L, Zhang H, Cao Y, Le J, Wu J, Liu B, Chen M, Du Y, Wang J, Wang G, et al. The anti-inflammatory effects of invigorating kidney and supplementing Qi Chinese herbal formulae in asthma patients. Evid Based Complement Alternat Med. 2017;2017:3754145.
Shizhen L. Ben-cao-gang-mu. 1596.
Zheng YS, Wu ZS, Ni HB, Ke L, Tong ZH, Li WQ, Li N, Li JS. Codonopsis pilosula polysaccharide attenuates cecal ligation and puncture sepsis via circuiting regulatory T cells in mice. Shock. 2014;41(3):250–5.
Shergis JL, Liu S, Chen X, Zhang AL, Guo X, Lu C, Xue CC. Dang shen [Codonopsis pilosula (Franch.) Nannf] herbal formulae for chronic obstructive pulmonary disease: a systematic review and meta-analysis. Phytother Res. 2015;29(2):167–86.
Liu AJ, Yu J, Ji HY, Zhang HC, Zhang Y, Liu HP. Extraction of a novel cold-water-soluble polysaccharide from astragalus membranaceus and its antitumor and immunological activities. Molecules. 2017;23(1):62.
Zhao B, Zhang X, Han W, Cheng J, Qin Y. Wound healing effect of an Astragalus membranaceus polysaccharide and its mechanism. Mol Med Rep. 2017;15(6):4077–83.
Tsao YT, Kuo CY, Kuan YD, Lin HC, Wu LH, Lee CH. The extracts of Astragalus membranaceus inhibit melanogenesis through the ERK signaling pathway. Int J Med Sci. 2017;14(11):1049–53.
Ting CT, Kuo CJ, Hu HY, Lee YL, Tsai TH. Prescription frequency and patterns of Chinese herbal medicine for liver cancer patients in Taiwan: a cross-sectional analysis of the National Health Insurance Research Database. BMC Complement Altern Med. 2017;17(1):118.
Safdar MN, Kausar T, Jabbar S, Mumtaz A, Ahad K, Saddozai AA. Extraction and quantification of polyphenols from kinnow (Citrus reticulate L.) peel using ultrasound and maceration techniques. J Food Drug Anal. 2017;25(3):488–500.
Wang Y, Qian J, Cao J, Wang D, Liu C, Yang R, Li X, Sun C. Antioxidant capacity, anticancer ability and flavonoids composition of 35 citrus (Citrus reticulata Blanco) varieties. Molecules. 2017;22:7.
Fomani M, Ngeufa Happi E, Nouga Bisoue A, Ndom JC, Kamdem Waffo AF, Sewald N, Wansi JD. Oxidative burst inhibition, cytotoxicity and antibacterial acriquinoline alkaloids from Citrus reticulate (Blanco). Bioorg Med Chem Lett. 2016;26(2):306–9.
Liu CY, Ko PH, Yen HR, Cheng CH, Li YH, Liao ZH, Hsu CH. The Chinese medicine Kuan-Sin-Yin improves liver function in patients with chronic hepatitis C: a randomised and placebo-controlled trial. Complement Ther Med. 2016;27:114–22.
Chien TJ, Liu CY, Ko PH, Hsu CH. A Chinese decoction, Kuan-Sin-Yin, improves autonomic function and cancer-related symptoms of metastatic colon cancer. Integr Cancer Ther. 2016;15(1):113–23.
Li TF, Lin CC, Tsai HP, Hsu CH, Fu SL. Effects of Kuan-Sin-Yin decoction on immunomodulation and tumorigenesis in mouse tumor models. BMC Complement Altern Med. 2014;14:488.
Ko PH, Shen YC, Murugan K, Huang CW, Sivakumar G, Pal P, Liao CC, Luo KS, Chuang EY, Tsai MH, et al. Macrophage migration inhibitory factor acts as the potential target of a newly synthesized compound, 1-(9′-methyl-3′-carbazole)-3, 4-dihydro-beta-carboline. Sci Rep. 2019;9(1):2147.
Chen HT, Tsai YL, Chen YS, Jong GP, Chen WK, Wang HL, Tsai FJ, Tsai CH, Lai TY, Tzang BS, et al. Dangshen (Codonopsis pilosula) activates IGF-I and FGF-2 pathways to induce proliferation and migration effects in RSC96 Schwann cells. Am J Chin Med. 2010;38(2):359–72.
Wu JP, Ho TJ, Tsai CC, Yeh YL, Lin CC, Lin KH, Hsieh DJ, Chen LM, Pan LF, Huang CY. Hepatoprotective effects of traditional chinese medicine on liver fibrosis from ethanol administration following partial hepatectomy. Chin J Physiol. 2015;58(6):393–403.
Liu D, Chen L, Zhao J, Cui K. Cardioprotection activity and mechanism of Astragalus polysaccharide in vivo and in vitro. Int J Biol Macromol. 2018;111:947–52.
Liu CJ, Hwang JM, Wu TT, Hsieh YH, Wu CC, Hsieh YS, Tsai CH, Wu HC, Huang CY, Liu JY. Anion exchanger inhibitor DIDS induces human poorly-differentiated malignant hepatocellular carcinoma HA22T cell apoptosis. Mol Cell Biochem. 2008;308(1–2):117–25.
Song H, Yin D, Liu Z. GDF-15 promotes angiogenesis through modulating p53/HIF-1alpha signaling pathway in hypoxic human umbilical vein endothelial cells. Mol Biol Rep. 2012;39(4):4017–22.
Ke K, Safder MA, Sul OJ, Kim WK, Suh JH, Joe Y, Chung HT, Choi HS. Hemeoxygenase-1 maintains bone mass via attenuating a redox imbalance in osteoclast. Mol Cell Endocrinol. 2015;409:11–20.
Authors’ contributions
PHK and LCL conceived and designed the experiments. PHK performed the experiments. PHK, CWH analyzed the data. HHC, EYC, MHT contributed reagents, materials, and/or analysis tools. PHK and LCL wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We would like to acknowledge the service provided by the DNA Sequencing Core of the First Core Laboratory, National Taiwan University College of Medicine.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The ITS and microarray data used to support the findings of this study have been deposited in the Genbank (A. membranaceus: MH445560; C. pilosula: MH445561) and Gene Expression Omnibus repository (Accession Number: GSE115506).
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
The work was funded by Taipei Chinese Medical Association (tpcma2012-Research01).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1.
Minimum standards of reporting checklist.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ko, PH., Huang, CW., Chang, HH. et al. Identifying the functions and biomarkers of Codonopsis pilosula and Astragalus membranaceus aqueous extracts in hepatic cells. Chin Med 14, 10 (2019). https://doi.org/10.1186/s13020-019-0233-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-019-0233-1