Abstract
Background
Baduanjin, a traditional Chinese exercise therapy, has been widely used in China to treat type 2 diabetes (T2DM) with depression (DD). However, the underlying mechanism of Baduanjin in anti-DD is unclear. This study was focused on investigating the effects of Baduanjin on symptoms of depression and blood glucose in patients with DD and the underlying mechanism.
Methods
We performed a 12-week Baduanjin intervention on patients with DD and longitudinally compared the differential expressions of lncRNAs, circRNAs, and mRNAs between pre- (BDD) and post- (ADD) Baduanjin intervention in the same group. Subsequently, Gene Ontology (GO) and pathway analysis was performed to investigate the function of differentially expressed mRNAs. Finally, Reverse Transcription-Polymerase Chain Reaction (RT-PCR) was used to verify the sequencing result and the mRNA-lncRNA regulatory network was constructed.
Results
The blood glucose level, depression index scores, and PHQ9 scores of the patients with DD were significantly decreased (P < 0.05) after Baduanjin intervention. Compared to BDD, 207 lncRNAs, 266 circRNAs, and 610 differentially expressed mRNAs were identified in ADD. Kyoto Encyclopedia of Genes and Genomes (KEGG) and GO showed that the significantly dysregulated mRNAs were mainly involved in immune function and inflammatory response pathways, and various signaling pathways including IL-17 and TNF. In addition, we selected five differentially expressed lncRNAs to construct an lncRNA-mRNA regulatory network, and found a total of 1045 mRNAs associated with them.
Conclusions
Our research is the first systematic profiling of mRNA, lncRNA, and circRNA in patients of ADD and BDD, and provides valuable insights in the potential mechanism of Baduanjin in anti-DD. Further, it was confirmed that Baduanjin is a safe and effective intervention for patients with DD because it can effectively ameliorate the symptoms of depression and blood glucose levels in patients with DD by regulating the dysregulated expression of lncRNA, mRNA, and circRNA.
Similar content being viewed by others
Background
Baduanjin (Eight-Section Brocades) is a traditional form of exercise that has been extensively practiced in China for many centuries [1]. Baduanjin contains eight simple actions based on Traditional Chinese Medicine (TCM) theory. The main focus is to assimilate energy in the body, which causes a variety of health benefits [2]. Baduanjin makes the posture and movement of the body symmetrical, and the movement of the mind and breathing become harmonious [3]. Due to the unique physical and mental relaxation, improved concentration, and better respiratory control it helps attain, it is suggested to improve Type 2 diabetes (T2DM) with symptoms of depression [4, 5]. Previous studies have shown that Baduanjin can relieve musculoskeletal pain, reduce anxiety and depression, and regulate glucose and lipid metabolism [6,7,8,9]. These studies provide a foundation for us to study Baduanjin as a potential non-drug anti-DD intervention.
People with T2DM have a 24% higher risk of depression than healthy people, and people developing depression are more likely to develop T2DM [10, 11]. Patients with depression often have poor compliance, which in turn worsens glycemic control and life quality, further aggravating the progression of T2DM [12]. Therefore, it is crucial to provide an effective treatment to such patients with symptoms of depression.
Abnormal expression of RNA during transcription is involved in the pathogenesis of DD [13, 14]. Long non-coding RNA (lncRNA) is an endogenous RNA molecule with a length of over 200 bp and limited protein-coding ability [15]. Circular RNAs (circRNAs) are a group of ring RNA molecules which are generally composed of more than one exon [16]. Recent studies have shown that lncRNAs and circRNAs are closely related to DD and suggest that may be promising targets for its treatment [15,16,17]. However, whether Baduanjin plays a role as an anti-DD intervention in association with lncRNA and circRNA remains unclear. Therefore, in this study, we aimed to investigate the hypoglycemic effect and the reduction in depression index due to Baduanjin, as well as its effects on the expression of lncRNAs and circRNAs, in patients with DD.
Materials and methods
The Minimum Standards of Reporting Checklist contains details of the experimental design, and statistics, and resources used in this study (Additional file 1).
Experimental objects and grouping
This study was approved by the Ethics Committee of Beijing University of Chinese Medicine (BUCM) (2016BZHYLL0105) and conducted in accordance with the principles of the Declaration of Helsinki. All enrolled patients were from hospitalized patients at the BUCM affiliated hospital (2016.1–2016.4) and were confirmed to have signed informed consent after enrollment. Cutoff points of 44 (total coarse points) and 55 (standard score) were used to define depression for the Self-Rating Depression Scale (SDS), whereas a score of 5 or more was used as a cutoff to define depression for patient health questionnaire 9 (PHQ9). We performed a 12-week Baduanjin intervention on DD patients enrolled in the study (starting at 8:30 am every day for approximately 30 min). Fasting blood samples of patients in the DD group were collected before and after the intervention of Baduanjin for further experiments. More specifically speaking, at 8:00 am on the 0th and 12th week of the intervention, the patient was in a state of peace and fasting, and blood was collected by nurse.
RNA extraction and library construction
Total RNA was extracted from whole blood according to a previously reported method [18]. Briefly, purification was performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and with the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturers protocol. Total RNA samples were then subjected to agarose electrophoresis and Nanodrop quality control and quantification. Enrich mRNA with oligo (dT) beads (rRNA removal kit for RNA degradation or prokaryotic); RNA sequencing library are completed by the KAPA Stranded RNA-Seq Library Prep Kit (Illumina). Finally, the constructed library was checked with the Agilent 2100 (NanoDrop ND-1000) and quantified by qPCR.
Illumina sequencing
Sequencing libraries of well-pooled different samples were denatured by 0.1 M NaOH to generate single-stranded DNA, which was in situ expanded on TruSeq SR Cluster Kit v3-cBot-HS (#GD-401-3001, Illumina) after dilution to 8 pM concentration The resulting fragment ends are sequenced using a Sequencer such as the Illumina HiSeq 4000 for 150 cycles.
Quantitative analysis of genes and circRNAs expression
Transcript abundance calculations were performed by software StringTie by aligning the results to known transcriptomes and then calculated by Ballgown. The units of expression were expressed as FPKM (Fragments Per Kilobase of gene/transcript model per Million mapped fragments). Genes were expressed at a threshold of FPKM ≥ 0.5 and the average number of genes with FPKM exceeding 0.5 in each group was considered to be expressed in the subgroup and statistically analyzed.
The expression of circRNA (Backsplice junction reads count) was quantified by the software STAR and then compared to the known transcriptome for junction locus detection and then calculated by CIRCexplorer2 for Backsplice junction reads. Unlike genes, the circRNA expression threshold is that CircRNAs with mean CPM exceeding 100 in each group are considered as expressed in the group and statistically analyzed.
qRT-PCR
qRT-PCR was performed to verify the expressions of lncRNAs and mRNAs related to the occurrence and development of DD. Purified total RNAs isolated from 14 samples were reverse-transcribed into cDNA according to the manufacturer’s instructions. Data were analyzed using the 2−△△CT relative quantification method and normalized to β-actin to calculate relative lncRNA and mRNA concentrations. All primers are shown in Table 1.
Gene Ontology and KEGG pathway analysis
We used software topGO for Gene Ontology (GO) analysis to obtain significantly enriched GO articles and corresponding genes to deduce important biological functions involved in differentially expressed genes. Meanwhile, based on the newest KEGG database (Kyoto Encyclopedia of Genes and Genomes), find out the pathways which are most associated with differentially expressed genes.
Gene set enrichment analysis (GSEA)
GSEA is a computational method used to determine whether a given gene set has significant differences among different groups [19]. Genes in these sets have some degree of correlation. Therefore, enrichment analysis of gene sets can make up for the shortcomings of single gene in the analysis.
Correlation and co-expression analysis
An lncRNA-mRNA regulatory network was constructed based on the inter-regulatory association between differentially expressed genes in the blood of DD patients following the Baduanjin intervention.
Statistical analysis
The results are expressed as mean ± standard error (SEM). GO and pathway enrichment analysis was done using Fisher’s test, the smaller the P value the greater the contact between the GO and pathway terms and the input differentially expressed genes. Two-sided P values < 0.05 were considered statistically significant.
Results
Hypoglycemic effect and reduction in depression index due to Baduanjin
We compared fasting blood glucose levels (GLU), HbA1c %, SDS, and PHQ9 in patients with DD before and after the Baduanjin intervention. The results showed that after the intervention of Baduanjin (ADD), HbA1c % did not significantly differ than that before Baduanjin intervention (BDD). However, the levels of fasting blood glucose, PHQ9, total coarse points, and standard score of ADD were significantly lower than those of BDD (Fig. 1 and Table 2).
Effect of Baduanjin on mRNA and circRNA expression in patients with DD
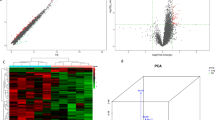
Compared to BDD, 610 (99 up-regulated and 511 down-regulated) and 266 (170 down-regulated and 96 up-regulated) differentially expressed mRNAs and circRNAs were identified in ADD, respectively (P < 0.05 and fold change > 1.5; Additional file 2: Table S1 and Additional file 3: Table S2). The scatterplot and the volcano plot of the differentially expressed mRNAs and circRNAs are shown in Fig. 2a–h. In addition, the fragment distribution of each sample is balanced and concentrated within 15 to 80 bp, indicating that the file construction quality is good (Fig. 2d).
Volcano-Plot, Scatter plot and Hierarchical clustering of differentially expressed mRNAs (a, b, c), circRNAs (e, f, g) and lncRNAs (i, j, k). Volcano-Plot above the green parallel line (P < 0.05) and outside the two longitudinal green lines indicated differentially expressed genes between the two compared samples. Scatter plot x-axis and y-axis represent FPKM mean values (log2 transitions) of two groups of gene. Analogous, green and red dots represent down-and up-regulated genes respectively, and the gray dots represent non-significantly different genes. d The transcriptional coverage profile is used to assess the 5 ‘or 3’ preference of the sample library fragment. h circRNA length distribution chart. X-axis and y-axis represent the number and the longth of circRNAs, respectively. i Types and counts of differentially regulated lncRNAs detected by Illumina sequencing (log2FC ≥ 0.585 and P < 0.05). The amount of each lncRNA type was accumulated in a bar graph, showing up-regulated and down-regulated lncRNAs, respectively
Effect of Baduanjin on the expression of lncRNAs in patients with DD
We detected 33 up-regulated and 174 down-regulated lncRNAs in ADD as compared to BDD (P ≤ 0.05, log2FC ≥ 0.585; Additional file 4: Table S3). According to the relative positions of lncRNAs and protein-coding genes in the genome, we classified these differentially expressed lncRNAs into 6 categories (Fig. 2i–l). In our data, the number of antisense lncRNAs was the most, in which 12 were up-regulated and 54 were down-regulated.
Effect of Baduanjin on DD-related antisense lncRNAs and their corresponding mRNA
Antisense lncRNA regulates the expression of the corresponding sense mRNA by inducing epigenetic changes in DNA [20]. Therefore, in the current study, we integrated differentially expressed antisense-lncRNAs and their differentially expressed mRNAs to deduce the function of the lncRNAs. We found a total of 153 corresponding sense mRNAs, 33 of which were up-regulated and 120 were down-regulated. In addition, 41 of these antisense lncRNAs were intronic antisense and 112 were natural antisense. As shown in Table 3, we listed the top 20 differentially expressed antisense LncRNAs based on their log2FC values. Furthermore, we also defined highly conserved, tissue-specific, disease-related, and biological processes-related lncRNAs.
Enrichment analysis of differentially expressed genes
GO analysis revealed the functional effects of Baduanjin on differentially expressed mRNAs in blood samples of patients with DD. Our results showed that up-regulated mRNAs were associated with biological processes such as inflammatory reactions, while the down-regulated mRNAs were associated with immune-related processes. Interestingly, the inflammatory response is closely related to depression, and it can play an important role in controlling the symptoms of depression (Fig. 3a, b).
Circle drawing represented the mainly involved BP of up-regulated (a) and down-regulated (b) differentially expressed genes. Bar chart of the top ten entries KEGG pathway analysis up (c) and down (d). Sorts from low to high according to P-values and the abscissa indicates enrichment score (− log10 (P value)). Difference lncRNAs GSEA Cluster Heat Map. (e) Biological Process and (f) KEGG Pathway, each row represents a functional entry, and each column represents an lncRNA
Pathway analysis of differentially expressed genes is devised to comprehend pathways and molecular interactions of the related mRNAs. The results showed that a total of 22 pathways were associated with the up-regulated mRNAs and 15 to the down-regulated mRNAs. Of the up-regulated protein-coding mRNAs, the highest number of mRNAs coded proteins involved in the IL-17 and TNF signaling pathways, whereas, the highest number of down-regulated mRNAs were related to the ribosome and spliceosome machineries (Fig. 3c, d). Hence, it is suggestive that these pathways might play a role in this intervention.
GSEA analysis of differentially expressed lncRNAs
In order to more intuitively understand the pathways and biological processes the differentially expressed lncRNAs are involved in, we used a powerful analytical method called GSEA to interpret lncRNA expression data (Fig. 3e, f). Compared with BDD, we found that the differentially expressed lncRNAs in the blood of ADD were mainly associated with biological processes such as response to cytokines, cellular response to hormonal stimuli, response to CAMP, positive regulation of NF-ƙB, and pathways related to ribosome machinery, cytosolic DNA-sensing pathway, and steroid biosynthesis. Therefore, the lncRNA-GSEA makes up for the deficiencies of the single lncRNA in the analysis, providing insights for the next in-depth study of the role of Baduanjin in decreasing the symptoms of DD.
Effect of Baduanjin on lncRNA-mRNA regulatory network
We screened 5 up-regulated lncRNAs (RP13-516M14.10, NEAT1, CTD-2530H12.2, CTD-3014M21.1, and AC068580.5) to construct the lncRNA-mRNA regulatory network map. A total of 1045 related mRNA were identified, of which 688 were negatively correlated with the corresponding lncRNAs and 357 mRNAs were positively correlated with the corresponding lncRNAs (Fig. 4). Five of our differentially expressed mRNAs (CXCL8, DUSP2, OSM, CXCR4, and NR4A2) were identified in the above lncRNA-mRNA regulatory network and defined as the key nodes of the lncRNA-mRNA regulatory network.
Discussion
In this study, we found that Baduanjin intervention significantly reduced blood glucose levels, and lowered the depression coefficient in patients with DD, showing an anti-DD effect. Results of the current study are in line with a previous review indicating that physical activity has a positive effect on regulating mood and reducing the symptoms of depression [21]. 610 and 207 differentially expressed mRNAs and lncRNAs, respectively, were screened before and after Baduanjin intervention. Subsequently, we selected six differentially expressed mRNAs and 1 differentially expressed lncRNA for RT-PCR validation, confirming the reliability of our sequencing results (Fig. 5). Our results revealed a 1.69 fold up-regulation in lncRNA- nuclear enriched abundant transcript 1 (NEAT1) in BDD group, which is consistent with previous studies [22, 23]. It has been known that NEAT1 is a competitive endogenous RNA, and it was closely related to T2DM, neurodegenerative diseases, epilepsy, and other mental illnesses [24,25,26].
The disks large-associated protein (DLGAP) family serves as a scaffold in postsynaptic density, which can link the glutamate receptors in the postsynaptic membrane with other glutamate receptors, signaling proteins, and cytoskeletal components [27]. With the central localization in the postsynapse, the DLGAP family plays a significant role in synaptic scaling by adjusting the turnover of both ionotropic and metabotropic glutamate receptors in response to synaptic activity [28]. Therefore, the DLGAP family is involved in the development of a variety of psychological and neurological diseases. As a member of the DLGAP family, DLGAP1-AS1 is expected to become a molecular target for the treatment of DD. Compared with BDD, we found that lncRNA-DLGAP1-AS1 was down-regulated in ADD. This shows that Baduanjin may play a role by down-regulating DLGAP1-AS1.
Since most of the differentially expressed lncRNAs have not been studied, we have constructed mRNA expression profiles to help study the effect of Baduanjin on DD. Depression can be perceived as a psycho-neuro-immunological disorder and the chemokine system plays a crucial role in its pathogenesis [29,30,31,32]. In this study, we also found increased expression of two chemokines (CXCL8 and CXCR4) after Baduanjin intervention. OSM are associated with depression [33]. NR4A2 (Nur-related receptor 2), is an orphan nuclear receptor that can be constitutively active as a transcription factor and can influence the expression of genes important for human brain development and regulation [34]. In our results, compared with BDD, the expression level of NR4A2 was significantly up-regulated in ADD (FD = 3.448, P < 0.05), suggesting that the anti-DD effect of the Baduanjin may be exerted through the regulation of the expression of NR4A2.
KEGG pathway analysis revealed that Baduanjin may play a pivotal role in DD progression through the significantly enriched 20 pathways including IL-17 and TNF. Our results support previous discovery [35, 36]. Studies have shown that IL-17 pathway is closely related to severe depression and depression can inhibit the activity of the IL-17 pathway [37, 38]. Our study found that the activity of the IL-17 pathway was significantly increased after Baduanjin intervention in patients with DD, suggesting that Baduanjin exerts anti-DD effects by up-regulating the IL-17 pathway.
The improvement of the depression index and blood glucose levels in patients with DD by Baduanjin intervention was significantly associated with the significant enrichment of GO processes in the blood of these patients. According to the annotation results, the most significant GO processes are cellular response to lipopolysaccharide, nuclear-transcribed mRNA catabolic process nonsense-mediated decay, and inflammatory response, indicating that the related coding genes contributed to development of DD. Interestingly, the top up-regulated biological processes belong to the immune pathway, which is closely related to depression [39]. Therefore, it is further confirmed that the Baduanjin intervention can improve the immune system function which is related to the anti-DD effect.
In addition, five significant differentially expression mRNAs (CXCL8, DUSP2, OSM, CXCR4, and NR4A2) were identified in the lncRNA-mRNA regulatory network and defined as the key nodes of the network. Among them, differentially expressed mRNA-CXCL8 was positively correlated with AC068580.5, DUSP2 was positively correlated with RP13-516M14.10, mRNA-OSM was positively correlated with RP13-516M14.10, AC068580.5, and CTD-2530H12.2, mRNA-CXCR4 was positively correlated with RP13-516M14.10, CTD-3014M21.1, AC068580.5, and CTD-2530H12.2, and mRNA-NR4A2 was positively correlated with RP13-516M14.10. The lncRNA-mRNA regulatory network analysis confirmed the crosslinking and complex regulation patterns of lncRNA and mRNA expression and verified the authenticity of the regulatory network. DD-related lncRNA-mRNA regulatory network and the exploration of their regulatory relationships have provided a molecular basis for the study for the treatment of DD with Baduanjin.
Conclusions
To conclude, our research is the first systematic profiling of mRNA, lncRNA, and circRNA in patients with DD undergoing Baduanjin intervention. We demonstrated that a 12-week Baduanjin intervention could significantly reduce blood glucose, SDS, and PHQ9 scale scores in patients with DD. These effects may be achieved by regulating the expression of the non-coding RNAs. This study hints towards an epigenetic role of Baduanjin intervention in DD and provides valuable insights in its potential mechanism.
Abbreviations
- T2DM:
-
type 2 diabetes
- DD:
-
T2DM with depression
- ADD:
-
after Baduanjin intervention group
- BDD:
-
before Baduanjin intervention
- GO:
-
Gene Ontology
- RT-PCR:
-
reverse transcription-polymerase chain reaction
- lncRNAs:
-
long non-coding RNAs
- circRNAs:
-
circular RNAs
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GSEA:
-
gene set enrichment analysis
- GLU:
-
glucose
- SDS:
-
Self-Rating Depression Scale
- PHQ9:
-
patient health questionnaire 9
- NEAT1:
-
nuclear enriched abundant transcript 1
- DLGAP:
-
discs large associated protein
- NR4A2:
-
Nur-related receptor 2
References
Li M, Fang Q, Li J, et al. The effect of chinese traditional exercise-baduanjin on physical and psychological well-being of college students: a randomized controlled trial. PLoS ONE. 2015;10(7):e0130544.
Liye Z, Eidi SJ, Huiru W, et al. A systematic review and meta-analysis of Baduanjin Qigong for health benefits: randomized controlled trials. Evi Based Complement Altern Med. 2017;2017:1–17.
Zou L, Pan Z, Yeung A, et al. A review study on the beneficial effects of Baduanjin. J Altern Complement Med. 2018;24(4):324.
Zheng G, Zheng Y, Xiong Z, et al. Effect of Baduanjin exercise on cognitive function in patients with post-stroke cognitive impairment: study protocol for a randomised controlled trial. BMJ Open. 2018;8(6):e020954.
Liu SJ, Ren Z, Wang L, et al. Mind body (Baduanjin) exercise prescription for chronic obstructive pulmonary disease: a systematic review with meta-analysis. Int J Environ Res Public Health. 2018;15(9):1830.
Chan JS, Li A, Ng SM, et al. Adiponectin potentially contributes to the antidepressive effects of Baduanjin Qigong exercise in women with chronic fatigue syndrome-like illness. Cell Transplant. 2017;26(3):493–501.
Chan JS, Ho RT, Chung KF, et al. Qigong exercise alleviates fatigue, anxiety, and depressive symptoms, improves sleep quality, and shortens sleep latency in persons with chronic fatigue syndrome-like illness. Evid Based Complement Altern Med. 2014, (2014-12-24), 2014, 2014:106048.
Zou L, Yeung A, Quan X, Boyden SD, Wang H. A systematic review and meta-analysis of mindfulness-based (Baduanjin) exercise for alleviating musculoskeletal pain and improving sleep quality in people with chronic diseases. Int J Environ Res Public Health. 2018;15:206.
Ma Y, Li X, Zhao D, et al. Association between cognitive vulnerability to depression—dysfunctional attitudes and glycaemic control among in-patients with type 2 diabetes in a hospital in Beijing: a multivariate regression analysis. Psychol Health Med. 2017;23:189–97.
Roy T, Lloyd CE. Epidemiology of depression and diabetes: a systematic review. J Affect Disord. 2012;142(Suppl):S8.
Moulton CD, Pickup JC, Ismail K. The link between depression and diabetes: the search for shared mechanisms. Lancet Diabetes Endocrinol. 2015;3(6):461–71.
Associations between psychological factors and health-related quality of life and global quality of life in patients with ALS: a systematic review. Health and Quality of Life Outcomes. 2016; 14: 107.
Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359.
Cui X, Niu W, Kong L, et al. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in major depressive disorder. Biomark Med. 2016;10(9):943–52.
Huang X, Luo YL, Mao YS, et al. The link between long noncoding RNAs and depression. Prog Neuropsychopharmacol Biol Psychiatry. 2016;73:73.
Jiang G, Ma Y, An T, et al. Relationships of circular RNA with diabetes and depression. Sci Rep. 2017;7(1):7285.
Xiao X, Zheng F, Chang H, et al. The Gene encoding protocadherin 9 (PCDH9), a novel risk factor for major depressive disorder. Neuropsychopharmacology Official Publication of the American College of Neuropsychopharmacology, 2017.
An T, Zhang T, Teng F, et al. Long non-coding RNAs could act as vectors for paternal heredity of high fat diet-induced obesity. Oncotarget. 2017;8(29):47876–89.
Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50.
Ørom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;27(4):359.
Liye Z, Albert Y, Xinfeng Q, et al. Mindfulness-based Baduanjin exercise for depression and anxiety in people with physical or mental illnesses: a systematic review and meta-analysis. Int J Environ Res Public Health. 2018;15(2):321.
Lin Z, Li X, Zhan X, et al. Construction of competitive endogenous RNA network reveals regulatory role of long non-coding RNAs in type 2 diabetes mellitus. J Cell Mol Med. 2017;21:3204–13.
Blüthgen N, Bentum MV, Merz B, et al. Profiling the MAPK/ERK dependent and independent activity regulated transcriptional programs in the murine hippocampus in vivo. Sci Rep. 2017;7:45101.
Barry G, Briggs JA, Hwang DW, et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci Rep. 2017;7:40127.
Riva P, Ratti A, Venturin M. The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res. 2016;13(11):1219–31.
Yan W, Chen ZY, Chen JQ, et al. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem Biophys Res Commun. 2017;496:1019–24.
Mathias SR, Knowles EEM, Kent JW, et al. Recurrent major depression and right hippocampal volume: a bivariate linkage and association study. Hum Brain Mapp. 2016;37(1):191–202.
Rasmussen AH, Rasmussen HB, Silahtaroglu A. The DLGAP family: neuronal expression, function and role in brain disorders. Mol Brain. 2017;10(1):43.
Leighton SP, Nerurkar L, Krishnadas R, et al. Chemokines in depression in health and in inflammatory illness: a systematic review and meta-analysis. Mol Psychiatry. 2017;23:48–58.
Eyre HA, Air T, Pradhan A, et al. A meta-analysis of chemokines in major depression. Prog Neuropsychopharmacol Biol Psychiatry. 2016;68:1–8.
Trojan E, Ślusarczyk J, Chamera K, et al. The modulatory properties of chronic antidepressant drugs treatment on the brain chemokine—chemokine receptor network: a molecular study in an animal model of depression. Front Pharmacol. 2017;8:779.
Grabowska P, Targowski T, Rozyńska R, et al. Alexithymia and depression: relationship to cigarette smoking, nicotine dependence and motivation to quit smoking. Przegla̧d Lekarski. 2005;62(10):1004.
Rodríguez-Morales A J, Mejia-Bernal Y V, Meneses-Quintero O M, et al. Chronic depression and post-chikungunya rheumatological diseases: is the IL-8/CXCL8 another associated mediator? Travel Medicine & Infectious Disease, 2017: article in press.
Ruano D, Macedo A, Dourado A, et al. NR4A2 and schizophrenia: lack of association in a Portuguese/Brazilian study. Am J Med Genet B Neuropsychiatr Genet. 2004;128B(1):41.
Beurel E, Lowell JA. Th17 cells in depression. Brain Behav Immunity. 2017;69:28–34.
Wang Y, Jiang H, Meng H, et al. Antidepressant mechanism research of acupuncture: insights from a genome-wide transcriptome analysis of frontal cortex in rats with chronic restraint stress. Evid Based Complement Alternat Med. 2017;2017(1):1676808.
Strober BE, Langley RGB, Menter A, et al. No elevated risk for depression, anxiety, or suicidality with secukinumab in a pooled analysis of data from 10 clinical studies in moderate to severe plaque psoriasis. Br J Dermatol. 2017;178:e105–7.
Lebwohl MG, Papp KA, Marangell LB, et al. Psychiatric adverse events during treatment with brodalumab: analysis of psoriasis clinical trials. J Am Acad Dermatol. 2017;78:81–9.
Guloksuz S, Rutten BP, Arts B, et al. The immune system and electroconvulsive therapy for depression. J Ect. 2014;30(2):132–7.
Authors’ contributions
SHG and GJJ designed the experiments; TA wrote the manuscript; ZCH, XQZ, JL, ALC and FT performed the experiments; HDC, JL and BHL analyzed the data. All authors read and approved the final manuscript.
Acknowledgements
We thank Yue Ma for data collection.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data and materials can be found from the first author and corresponding author.
Consent for publication
Consent to publish from the participant to report individual patient data. All enrolled patients were confirmed to have signed informed consent after enrollment.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Beijing University of Chinese Medicine (BUCM) (2016BZHYLL0105). All enrolled patients were from hospitalized patients at the BUCM affiliated hospital and were confirmed to have signed informed consent after enrollment.
Funding
This study was support by the grants from the National Natural Science Foundation of China (81774171), the Tangshan Science and Technology Innovation Team Training Program (18130219A), the Beijing University of Chinese Medicine university research funding (2180072220002), the Beijing Municipal Education Commission scientific research and graduate training to build projects (1000062520025) and Collaborative innovation project of Chaoyang District in Beijing (CYXC1719).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1.
Minimum Standards of Reporting Checklist.
Additional file 2.
Differentially expressed mRNAs.
Additional file 3.
Differentially expressed circRNAs.
Additional file 4.
Differentially expressed lncRNAs.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
An, T., He, ZC., Zhang, XQ. et al. Baduanjin exerts anti-diabetic and anti-depression effects by regulating the expression of mRNA, lncRNA, and circRNA. Chin Med 14, 3 (2019). https://doi.org/10.1186/s13020-019-0225-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-019-0225-1