Abstract
Background
Osteoporosis results from decreased bone mass and disturbed bone structure. Human bone marrow mesenchymal stem cells (hBMSCs) demonstrate robust osteogenic differentiation, a critical process for bone formation. This research was designed to examine the functions of LINC01133 in osteogenic differentiation.
Methods
Differentially expressed lncRNAs affecting osteogenic differentiation in hBMSCs were identified from the GEO database. A total of 74 osteoporosis patients and 70 controls were enrolled. hBMSCs were stimulated to undergo osteogenic differentiation using an osteogenic differentiation medium (OM). RT-qPCR was performed to evaluate LINC01133 levels and osteogenesis-related genes such as osteocalcin, osteopontin, and RUNX2. An alkaline phosphates (ALP) activity assay was conducted to assess osteogenic differentiation. Cell apoptosis was detected using flow cytometry. Dual luciferase reporter assay and RIP assay were employed to investigate the association between miR-214-3p and LINC01133 or CTNNB1. Loss or gain of function assays were conducted to elucidate the impact of LINC01133 and miR-214-3p on osteogenic differentiation of hBMSCs.
Results
LINC01133 and CTNNB1 expression decreased in osteoporotic patients but increased in OM-cultured hBMSCs, whereas miR-214-3p showed an opposite trend. Depletion of LINC01133 suppressed the expression of genes associated with bone formation and ALP activity triggered by OM in hBMSCs, leading to increased cell apoptosis. Nevertheless, this suppression was partially counteracted by the reduced miR-214-3p levels. Mechanistically, LINC01133 and CTNNB1 were identified as direct targets of miR-214-3p.
Conclusions
Our study highlights the role of LINC01133 in positively regulating CTNNB1 expression by inhibiting miR-214-3p, thereby promoting osteogenic differentiation of BMSCs. These findings may provide valuable insights into bone regeneration in osteoporosis.
Similar content being viewed by others
Background
Osteoporosis (OP) is a common disease in the elderly, characterized by an imbalance in bone formation, which can increase fracture risk and decrease bone strength [1]. It is estimated that more than 200 million people worldwide suffer from OP, and the incidence increases with age [2]. Marrow mesenchymal stem cell (MSC) dysfunction is associated with the pathogenesis of OP. MSCs are widely distributed in various organs and tissues and are the preferred cell type for bone regeneration. Among adult stem cells, bone marrow mesenchymal stem cells (BMSCs) are considered the most suitable option for bone tissue engineering due to their ability to self-renew, high differentiation potential, and low immunogenicity [3]. The clinical use of BMSCs depends on a comprehensive analysis of the molecular mechanisms that govern their regulation of osteogenic differentiation. BMSCs can differentiate into osteoblasts, chondrocytes, and adipocytes, and any impairment in their osteogenic differentiation process leads to conditions such as OP [4]. A deeper understanding of the osteogenic differentiation mechanisms of BMSCs could present new approaches for managing and preventing metabolic bone disorders like OP.
Long non-coding RNAs (LncRNAs) are a subclass of RNAs with limited protein-coding potential. Prior investigations have indicated that disrupted lncRNAs are linked to significant illnesses like OP and musculoskeletal aging [5]. Additionally, various differentially expressed LncRNAs (DELncRNAs) are involved in osteogenesis, such as USP2-AS1 [6], H19 [7], HOTAIR [8], and LINC00638 [9]. Long intergenic non-protein coding RNA 1133 (LINC01133) is located on human chromosome 1q23.0 and contains 3 exons. LINC01133 enhances the characteristics of cancer stem cells and serves as a direct effector of the signaling pathways activated by MSCs in triple-negative breast cancer [10]. LINC01133 also promotes the osteogenic differentiation of periodontal ligament stem cells [11]. Moreover, hydrostatic pressure influences the osteogenic differentiation of MSCs and regulates bone dynamic homeostasis by increasing LINC01133 expression [12]. We identified differentially expressed LncRNAs on day 10 post hBMSCs induction of osteogenic differentiation using the GEO database. Among them, LINC01133 showed a significant 2.3-fold increase. Nevertheless, the precise roles and molecular mechanisms remain largely unknown.
We proposed that LINC01133 might regulate the osteogenic differentiation of BMSCs. To validate this hypothesis, our ongoing research examined the expression of LINC01133 and elucidated the molecular mechanisms in patients with OP, along with osteogenic differentiation induction by BMSCs in vitro. Our discoveries could present new perspectives and offer fresh possibilities for bone repair therapy.
Methods
Data source
Differentially expressed LncRNAs (DElncRNAs) in the osteogenic differentiation of hBMSCs were retrieved from the Gene Expression Omnibus (GEO) database. Download the original data from the GSE113359 dataset, which includes DElncRNAs in non-osteogenic differentiated hBMSCs and 10-day induced osteogenic differentiated hBMSCs. Use GEO’s GEO2R to analyze the data and adjust the adj.P values of potential false positive results using the Benjamin-Hochberg method.
Participants
Prior to this study, the informed consent of the subjects was obtained and the guidelines of the Declaration of Helsinki were followed with the approval of the Zhucheng People’s Hospital Ethics Committee. Seventy-four patients with osteoporotic fractures (51 females, age 68 ± 10 years) who underwent hip arthroplasty at Zhucheng People’s Hospital from June 2020 to July 2022, and 70 patients with traumatic fractures without OP symptoms were included as controls (43 females, age 66 ± 9 years). All participants had no history of bone metabolism disorders, cancer, diabetes, hyperthyroidism, or renal disease, and were not on any medication. Blood samples and femoral trabecular bone tissue were collected from the participants. Blood samples were centrifuged at 3000 rpm for 15 min before serum preservation.
Cell lines and culture
hBMSCs were purchased from BioBw (#bio-1342278). Flow cytometry showed that hBMSCs positively expressed CD29, CD44, CD73, and CD105, and negatively expressed CD48 (Supplemental Fig. 1). hBMSCs were cultured in Dulbecco’s modified Eagle’s proliferation medium (PM) with 10% FBS. The incubator was set to 37℃ with 5% CO2 and appropriate humidity.
Osteogenic differentiation of hBMSCs
To promote bone formation, the third-generation hBMSCs were suspended in osteogenic differentiation DMEM medium (OM) containing 15% FBS, 1% penicillin/streptomycin, 50 µg/mL ascorbic acid, 10 mmol/L β-glycerophosphate, and 0.1 mM dexamethasone. The cells were inoculated in a 6-well plate at a density of 5 × 105 cells per well. The hBMSCs were cultured in the OM medium for 0, 1, 3, 7, 14, and 21 days, with the medium changed every 3 days.
Plasmid construction and transfection
Third-generation hBMSCs were plated in 6 well dishes are cultured until reaching 80% confluency before transfection. The specific small interfering RNA (siRNA) targeting LINC01133 (si-LINC01133) or catenin beta 1 (CTNNB1) (si-CTNNB1), negative control (si-NC) and miR-214-3p inhibitor or inhibitor NC (RioBio, Guangzhou, China) were mixed with Lipofectamine 3000 transfection reagent. After a 0.5 h incubation, the transfection medium was replenished to the hBMSCs.
RNA extraction and real-time quantitative reverse transcription PCR (RT-qPCR)
Total RNA from hBMSCs and serum or bone tissues of subjects was extracted using TRIzol reagent. The RNA concentration and purity were assessed using the A26/A280 ratio. Afterward, 1 µg of RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit or miRcute miRNA cDNA First strand synthesis kit. RT-qPCR was conducted with the CFX Connect Real-Time Fluorescent PCR System and the SYBR Green Preix Pro kit (for mRNA) or miRcute miRNA RT-qPCR kit (for miRNA). Each sample was analyzed in triplicate. For lncRNA and CTNNB1, GAPDH served as the internal reference, whereas for miRNA, U6 was utilized. The relative levels of LncRNA or miRNA were determined using the 2−ΔΔCT methodology.
Cell apoptosis assay
An Annexin V-FITC/Propidium iodide (PI) cell apoptosis assay kit was for apoptosis detection. The hBMSCs were washed with pre-cooled PBS and the cells were digested with EDTA-free trypsin. Subsequently, cells were centrifuged at 1000 g for 5 min to remove the supernatant. The cell pellet was resuspended in 200 µL of binding buffer. Next, 5 µL of PI and 5 µL of Annexin V-FITC were added and incubated in a dark room for 15 min. Early apoptotic and late apoptotic cells were then analyzed by flow cytometric to count the total number of apoptotic cells.
Alkaline phosphatase (ALP) activity assay
hBMSCs were cultured in an osteogenic medium. After 7 days, the hBMSCs were covered with a lysis buffer without inhibitors, centrifuged to obtain the supernatant, and the ALP activity was measured using an ALP assay kit (Beyotime, Shanghai) in the enzyme-linked immunoassay (ELX88, BioTek) at 405 nm optical density (OD). Each sample had at least three replications.
Bioinformatics prediction
The target miRNA of LINC01133 was predicated by the LncBook and LncRNASNP2 online databases, the binding site of miR-214-3p. Targetscan, miRDB, ENCORI, miRWalk, and miRTarBase respectively predicted the targets of miR-214-3p, and overlapping mRNAs were searched for using the Bioinformatics Website. Additionally, STRING constructed a protein-protein interaction (PPI) network of these overlapping mRNAs and analyzed the connectivity degree of proteins, as well as the top 10 hub nodes.
Nuclear-cytoplasmic separation assay
Subcellular localization of LINC01133 was examined by the nuclear-cytoplasmic separation assay. According to the instructions for the PARIS reagent kit, hBMSCs were washed in PBS digested with trypsin, and suspended in lysis buffer. The cytoplasmic fraction was collected from the supernatant by centrifugation at 400 g for 15 min at 4 ℃. Subsequently, PBS, nuclear division buffer, and 0.3 mL of RNAase-free H2O were added to the centrifuged cell precipitate for resuspension, followed by an ice bath for 20 min and centrifugation to obtain the cytosolic fragment. The levels of LINC01133 in different cell components were assessed by RT-qPCR using GAPDH and U6 as controls for cytoplasmic and nuclear fractions, respectively.
RNA immunoprecipitation (RIP) assay
The RIP assay was conducted using Magna RIP assay kits. hBMSCs were lysed and incubated with magnetic beads linked to either an anti-Ago2 antibody or an anti-IgG antibody for RNA compound capture. The bound RNA compounds were then eluted and separated for RT-qPCR.
Dual-luciferase reporter (DLR) assay
Segments of LINC01133 and CTNNB1 3’UTR containing the predicted miR-214-3p complementary sequence or mismatched seed sequences were subcloned into the pMIR-REPORT vector. This was done to construct luciferase reporter gene wild type LINC01133 (LINC01133-WT) or CTNNB1-WT, and mutant LINC01133 (LINC01133-MT) or CTNNB1-MT recombined plasmids. hBMSCs were inoculated into 48-well plates and co-transfected with miR-214-3p mimic, inhibitor, mimic NC, and inhibitor NC along with recombined plasmids. The transfection reagent used was lipofectamine 3000. Luciferase activity was monitored 48 h later using the dual luciferase reporter assay kit.
Statistical analysis
The data acquired from three experiments were gathered and displayed as mean ± standard deviation. Pearson coefficient analysis was used to investigate the correlation between the variables. Comparisons between groups were performed using unpaired Student’s t-tests or ANOVA. Statistical analysis and data visualization were conducted using GraphPad Prism version 9.0 software. The significance levels were set at P < 0.05.
Results
The expression of LINC01133 was detected in patients with OP
The GEO database (GSE113359) detected various differently expressed LncRNAs in hBMSCs on days 0 and 10 of osteogenic differentiation (Fig. 1A and B), with a log2 (Fold change) of 2.33 for LINC01133. Based on the research background of LINC01133, we chose LINC01133 as the research object. OP patients and controls were then enrolled. As shown in Fig. 1C-D, the levels of serum LINC01133 were typically lower in OP patients compared to the control group, and the levels of bone tissue LINC01133 exhibited similar findings (P < 0.001). In addition, the serum LINC01133 demonstrated 89.33% sensitivity and 75.71% specificity in significantly distinguishing OP patients from controls (AUC = 0.837, Fig. 1E).
The levels of LINC01133 in patients with OP. The volcano plot (A) and heatmap (B) from the GEO database (GESE113359 dataset) were used to detect differentially expressed LncRNAs in human bone marrow mesenchymal stem cells (hBMSCs) on days 0 and 10 of osteogenic differentiation. C-D. Patients with OP had significantly lower levels of LINC01133 in serum and bone tissue compared to controls. E. Diagnostic value of ROC curves for analyzing serum LINC01133 in patients with OP. *** P < 0.001
LINC01133 is highly expressed during the osteogenic differentiation of hBMSCs
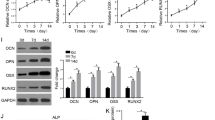
To investigate the impact of LINC01133 in OP and the bone formation process, we established cellular models in vitro by culturing hBMSCs using PM and OM. As illustrated in Fig. 2A-C, OM greatly promoted the mRNA levels of the osteogenic markers OCN, OPN, and RUNX2 in hBMSCs at days 3, 7, 14, and 21 compared to PM (P < 0.001). In addition, apoptosis of hBMSCs on days 3, 7, 14, and 21 was notably diminished as OM-induced osteogenic differentiation of hBMSCs (P < 0.05, Fig. 2D). Concurrently, mRNA levels of LINC01133 were significantly enhanced by OM at days 3, 7, 14, and 21 of induction of osteogenic differentiation of hBMSCs compared to PM culture (P < 0.001, Fig. 2E).
The expression of LINC01133 and osteogenesis-relative genes in hBMSCs cultured in PM and OM. A-C, the mRNA levels of OCN, OPN, and RUNX2 in hBMSCs were analyzed in both OM and PM at the same time points (0, 3, 7, 14, and 21 days). D. Flow cytometry was conducted to examine the apoptosis of hBMSCs during the OM-induced osteogenic differentiation. E. RT-qPCR was employed to analyze the mRNA levels of LINC01133 in hBMSCs cultured for the same time (0, 3, 7, 14, and 21 days) in OM and PM. PM, proliferation medium; OM, osteogenic differentiation medium. * P < 0.05, *** P < 0.001
Effect of silencing LINC01133 on apoptosis and osteogenic differentiation of hBMSCs cells
To evaluate the functions of LINC01133 on the apoptosis, and osteogenic differentiation of hBMSCs, we silenced the expression of LINC01133 in hBMSCs and verified its inhibitory effect by RT-qPCR. Since si-LINC01133#1 showed the greatest disruption efficacy among the three siRNA, it was selected for further study (P < 0.001, Fig. 3A). Furthermore, si-LINC01133 significantly reduced the OM-induced upregulation of LINC01133 levels in hBMSCs (P < 0.001, Fig. 3B). Additionally, OM induction enhanced ALP activity in hBMSCs, but this was partially reduced by downregulated LINC01133 (P < 0.001, Fig. 3C). The deletion of LINC01133 also led to a notable decrease in the levels of osteogenic differentiation markers OCN, OPN, and RUNX2 (P < 0.001, Fig. 3D). Compared to PM, OM notably suppressed cell apoptosis, but this effect was effectively reversed by si-LINC01133 (P < 0.001, Fig. 3E).
Effect of silencing LINC01133 on cell viability, apoptosis, and osteogenic differentiation of hBMSCs cells. A. LINC01133 levels were detected by the RT-qPCR after siRNA transfection. B. The expression of LINC01133 was detected after transfection of si-LINC01133 in OM-cultured hBMSCs. C. si-LINC01133 regulated ALP activity in OM-induced hBMSCs. D. RT-qPCR analysis of the effect of si-LINC01133 on the levels of osteogenic differentiation markers OCN, OPN, and RUNX2. E. Flow cytometry was used to detect the effects of si-LINC01133 on OM-induced cell apoptosis of hBMSCs. ** P < 0.01, *** P < 0.001
LINC01133 interacts with miR-214-3p
Cytoplasmic LncRNA can function as molecular sponges for miRNAs, so subcellular isolation methods were initially assessed to examine the localization of LINC01133 in hBMSCs. As depicted in Fig. 4A, LINC01133 in hBMSCs was predominantly observed in the cytoplasm. Subsequently, the miRNAs interacting with LINC01133 were identified through database prediction, with their binding sites found in miR-214-3p (Fig. 4B). Additionally, the upregulation of miR-214-3p resulted in a significant reduction in the luciferase activity of LINC01133-WT (P < 0.001, Fig. 4C), while it had no impact on the luciferase activity of LINC01133-MT (P > 0.05). Furthermore, both LINC01133 and miR-214-3p were enriched in the anti-Ago2 group more than the anti-IgG (P < 0.001, Fig. 4D). Conversely, serum miR-214-3p was notably promoted in patients with OP (P < 0.001, Fig. 4E), and the same results were observed in bone tissues of OP patients (P < 0.001, Supplemental Fig. 2). Furthermore, there was a marked negative association between serum LINC01133 expression and serum miR-214-3p in patients with OP (r = -0.735, Fig. 4F). However, miR-214-3p levels were diminished at days 3, 7, 14, and 21 of OM-induced osteogenic differentiation of hBMSCs compared to PM (P < 0.001, Fig. 4G). Finally, the suppression of miR-214-3p levels by OM was significantly restored by si-LINC01133 (P < 0.001, Fig. 4H).
LINC01133 directly targets miR-214-3p. A. Subcellular fractionation assay detects the distribution of LINC01133 in hBMSCs. B. The binding sequences between miR-214-3p and LINC01133. Dual luciferase reporter assay (C) and RIP assay (D) were conducted to verify the correlation between miR-214-3p and LINC01133. E. The serum miR-214-3p levels in the subjects. F. The Pearson coefficient correlation was employed to examine the relationship between miR-214-3p and LINC01133 in patients with OP. G. RT-qPCR was employed to analyze the mRNA levels of miR-214-3p in hBMSCs cultured for the same time (0, 3, 7, 14, and 21 days) in OM and PM. H. Effect of silencing LINC01133 in OM-induced hBMSCs on miR-214-3p levels. *** P < 0.001
Effects of miR-214-3p and LINC01133 on proliferation, apoptosis, and osteogenic differentiation of hBMSCs
The co-regulatory effects of LINC01133 and miR-214-3p were analyzed in OM-induced hBMSCs. As shown in Fig. 5A, compared to the inhibitor NC, the miR-214-3p inhibitor could diminished miR-214-3p levels in hBMSCs (P < 0.05). Furthermore, the promotion of miR-214-3p levels by silencing LINC01133 in OM-induced hBMSCs was suppressed by miR-214-3p inhibitor (P < 0.05, Fig. 5B). Silencing LINC01133 markedly suppressed ALP activity, but this was restored by the miR-214-3p inhibitor (P < 0.05, Fig. 5C). In OM-induced hBMSCs, reduced LINC01133 led to decreased expression of osteogenic markers, which was partially restored by the downregulating miR-214-3p (P < 0.05, Fig. 5D). Additionally, the apoptosis-promoting effect of silencing LINC01133 in OM-induced hBMSCs was partially eliminated by downregulating miR-214-3p (P < 0.05, Fig. 5E).
Effects of miR-214-3p and LINC01133 on proliferation, apoptosis, and osteogenic differentiation of hBMSCs. A. Inhibition of miR-214-3p levels in hBMSCs by transfection with miR-214-3p inhibitor. B. miR-214-3p levels in the hBMSCs under OM induced downregulation of LINC01133. C. Co-regulation of miR-214-3p and LINC01133 on ALP activity in hBMSCs. D. Effects of co-regulation of miR-214-3p and LINC01133 on mRNA levels of osteogenic differentiation markers were analyzed by RT-qPCR. E. Flow cytometry used to detect the effects of miR-214-3p inhibitors on OM-induced cell apoptosis of hBMSCs. ** P < 0.01, *** P < 0.001
CTNNB1 is the target of miR-214-3p
Targetscan, miRDB, ENCORI, miRWalk, and miRTarBase online software were employed to predict the targets of miR-214-3p. As shown in Fig. 6A, a total of 49 overlapping targets were identified. The PPI network of these overlapping genes was constructed, containing 49 nodes and 45 edges, with an average node degree of 1.84 and a PPI-enriched P value of 0.00017 (Fig. 6B). The top 10 highest-degree nodes in the network are also shown in Fig. 6B, which includes CTNNB1. The binding sequences of miR-214-3p and CTNNB1 are shown in Fig. 6C. The miR-214-3p mimic reduced the luciferase activity of CTNNB1-WT, and the miR-214-3p inhibitor promoted the luciferase activity of CTNNB1-WT, while they had no functions in the CTNNB1-MT (P > 0.05, Fig. 6D). Additionally, LINC01133, miR-214-3p, and CTNNB1 were all enriched on anti-Ago2 (P < 0.001, Fig. 6E).
CTNNB1 is the target of miR-214-3p. A. The overlapping targets of miR-214-3p were predicted by the online software Targetscan, miRDB, ENCORI, microT, miRWalk, and EVmiRNA. B. The PPI network was constructed through the overlapping targets and the top 10 hub genes in the PPI network were also displayed according to node degree. C. The binding sequences between miR-214-3p and CTNNB1. Dual luciferase reporter assay (D) and RIP assay (E) were conducted to verify the correlation between miR-214-3p and CTNNB1. F. The serum CTNNB1 levels in the subjects. The Pearson coefficient correlation was employed to examine the relationship between CTNNB1 with LINC01133 (G) and miR-214-3p (H). I. RT-qPCR was employed to analyze the mRNA levels of CTNNB1 in hBMSCs cultured for the same time (0, 3, 7, 14, and 21 days) in OM and PM. J. LINC01133 and miR-214-3p regulated CTNNB1 levels in hBMSCs. *** P < 0.001
Furthermore, compared with the controls, the serum CTNNB1 level was significantly lower in the patients with OP (P < 0.05, Fig. 6F), and the levels of bone tissue CTNNB1 exhibited similar findings (P < 0.001, Supplemental Fig. 3). Serum CTNNB1 was positively correlated with serum LINC01133 and negatively correlated with serum miR-214-3p in OP patients (P < 0.05, Fig. 6G-H). Furthermore, OM greatly promoted the expression of CTNNB1 in hBMSCs after days 3, 7, 14, and 21 compared to PM (Fig. 6I). Silencing of LINC01133 increased the expression of CTNNB1, while miR-214-3p suppressed its expression (P < 0.05, Fig. 6J).
Discussion
OP is a condition that affects the skeletal system, leading to decreased bone density and quality, and a higher risk of fractures [13, 14]. Current research indicates that LncRNAs play a role in the progression of OP and hold promise as clinical biomarkers or treatment targets. In our research, we identified a highly preserved lncRNA in OP that impacts the osteogenic differentiation process. Initially, we screened for differentially expressed LncRNAs influencing osteogenic differentiation of BMSCs from the GEO database and discovered an irregularly high level of LINC01133. through studying OP patients, we observed that LINC01133 expression was typically reduced in OP individuals, making it a distinguishing factor between OP patients and controls. We also identified that LINC01133 serves as a positive regulator of osteogenic differentiation in BMSCs and suppresses apoptosis. This effect may be mediated by modulating the miR-214-3p/CTNNB1 axis. In summary, our findings indicate that LINC01133 could enhance osteogenic differentiation and prevent apoptosis in hBMSCs, offering the potential for OP treatment.
Regeneration following bone defects, whether congenital or caused by trauma or disease, continues to pose significant challenges in clinical settings. Bones have an inherent capacity for regeneration, with the response to damage being integral to the repair process [15]. BMSCs play a pivotal role in bone formation and reconstruction in patients with bone metabolic disorders, showing great promise in medical research and being extensively utilized in numerous clinical trials. Many clinical and basic research efforts aim to repair and regenerate bone using BMSCs. Emerging evidence suggests that LncRNA contributes to the process of osteogenic differentiation in BMSCs, which may be critical to restoring bone reconstruction in patients with OP. For example, LncRNA KCNQ1OT1 was downregulated in OP patients, and it inhibited osteogenic differentiation of BMSCs by suppressing miR-205-5p to promote RICTOR expression [16]. Li et al. reported that peripheral blood LncRNA MALAT1 was abnormally reduced in elderly OP patients but elevated in BMSCs after osteogenic differentiation, and was involved in disease onset by regulating the Wnt/β-catenin signaling pathway [17]. Despite the growing body of literature on lncRNA, the precise mechanism by which lncRNA intricately regulates the osteogenic differentiation of hBMSCs remains a significant enigma that warrants further in-depth investigation.
LINC01133, a novel non-coding RNA, has been identified in pancreatic cancer [18], lung squamous cell carcinoma [19], and colorectal cancer [20]. Additionally, significant involvement of LINC01133 has been observed in orthopedic-related conditions. It exhibits dysregulated expression in osteosarcoma [21] and is implicated in regulating osteogenic differentiation. For instance, LINC01133 is generally downregulated in periodontitis and enhances the osteogenic differentiation of periodontal ligament stem cells through the miR-30 [11]. Moreover, LINC01133 was reported to participate in periodontal bone differentiation by regulating the mitochondrial function of periodontal ligament stem cells [22]. In our research, we identified DElncRNAs after osteogenic differentiation of BMSCs in the GEO database and found that LINC01133 was typically increased. Previous studies have reported defective osteogenic differentiation of BMSCs during OP progression [23]. In our cohort of OP patients, LINC01133 demonstrated low expression levels. Conversely, following osteogenic differentiation induction in hBMSCs, LINC01133 exhibited high expression. Earlier research has shown that targeting the molecular foundation of OP is feasible using siRNA, allowing for the development of specific therapies aimed at disease prevention [24]. Numerous authors have unitized siRNAs to pinpoint target molecules or as therapeutic agents [25]. In our in vitro study, we suppressed LINC01133 expression by introducing a siRNA specifically designed for it. We found that inhibition of LINC01133 could lead to decreased osteogenic differentiation and a notable increase in apoptosis. These findings indicate that LINC01133 might impede the progression of osteoporosis by enhancing osteogenic differentiation.
LncRNA function is dependent on subcellular localization. It was found that the cytoplasmic LncRNAs can act as molecular sponges for miRNAs, inhibiting their expression and restoring mRNA levels by competitively binding miRNAs. miRNAs have been widely reported for their therapeutic potential in musculoskeletal disorders [26,27,28,29]. In our study, we found that LINC01133 predominantly localized in the cytoplasm based on subcellular localization analysis, and the prediction of its target miRNAs identified miR-214-3p. miRNA sequencing displayed a generalized elevation of miR-214-3p in exosomes from bone loss in osseous de-ovulated mice [30]. miR-214-3p was also generally elevated in BMSCs isolated from OP mice [31]. Furthermore, bone recombinant adeno-associated viral vector-mediated inhibition of miR-214-3p levels reversed the bone loss in OP by increasing osteoblast-mediated bone formation [32]. Furthermore, miR-214-3p inhibited bone formation in obligate spondylitis fibroblasts [33]. High expression of miR-214-3p diminished osteogenic differentiation of BMSCs in diabetic mice [34]. In the current research, it was noted that miR-214-3p levels were significantly elevated in patients with OP. Additionally, miR-214-3p levels were typically suppressed during the osteogenic differentiation of BMSCs. The suppressive impact of silencing LINC01133 on the osteogenic differentiation of hBMSCs and the enhancement of apoptosis were substantially counteracted by the lower expression of miR-214-3p. We hypothesized that LINC01133 could potentially alleviate the progression of OP by enhancing the osteogenic differentiation of hBMSCs through the regulation of miR-214-3p.
CTNNB1 is a key component of adherent junctions and a crucial regulator of the canonical Wnt signaling pathway, playing a significant role in the osteogenic differentiation of BMSCs [35, 36]. Previous studies have shown that activation of the Wnt signaling pathway accelerates bone regeneration by inducing osteoblast differentiation. Aberrant expression of CTNNB1 commonly causes altered Wnt signaling. Thus, osteoanabolic therapy targeting the Wnt pathway, particularly CTNNB1, represents a new strategy for anti-OP therapy [37]. Furthermore, in terms of bone differentiation, CTNNB1 can induce osteogenic differentiation during alveolar bone defect repair [38]. In rat periodontitis, CTNNB1 enhanced osteogenic differentiation to reduce alveolar bone loss during orthodontic tooth movement [38]. Regarding osteogenesis, deflection of CTNNB1 leads to an inflammatory decrease in bone mass, while conditional activation of CTNNB1 results in a dramatic increase in bone mass [39]. Inhibition of CTNNB1 impairs osteogenic differences in periodontal ligament stem cells [40]. More importantly, CTNNB1 polymorphisms encoding β-catenin were linked to OP [41]. Prior research has demonstrated that CTNNB1 is significantly reduced in mouse models of OP [37] and patients with OP [37]. Consistent with these findings, we also observed a significant decrease in CTNNB1 expression in patients with OP. Furthermore, we identified miR-214 as targeting CTNNB1, and the reduction in CTNNB1 expression due to LINC01133 knockdown was reversed by inhibiting miR-214-3p. Rescue experiments revealed that blocking miR-214-3p reversed the inhibitory impact of LINC01133 suppression on the osteogenic differentiation of hBMSCs by upregulating CTNNB1.
This study had some limitations that could affect the application of its findings. Firstly, although some studies have confirmed the role of CTNNB1, this study did not go further to directly observe the specific effects of CTNNB1 levels on osteogenic differentiation and bone formation through knockdown and overexpression. Secondly, relying only on a single siRNA may affect the robustness and reproducibility of the results. In future experiments, we will supplement the CTNNB1 results and use at least two siRNAs for interference and subsequent studies.
Conclusions
All things considered, there is new evidence that LINC01133 regulates OP and could serve as a reliable diagnostic biomarker. Moreover, reducing LINC01133 led to lower CTNNB1 levels by enhancing miR-214-3p, ultimately hindering the osteogenic differentiation of hBMSCs. This research offers fresh perspectives on treating bone disorders.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ALP:
-
Alkaline phosphates
- ALP:
-
Alkaline phosphatase
- DLR:
-
Dual-luciferase reporter
- DElncRNAs:
-
Differentially expressed LncRNAs
- GEO:
-
Gene Expression Omnibus
- hBMSCs:
-
Human bone marrow mesenchymal stem cells
- LncRNAs:
-
Long non-coding RNAs
- OM:
-
Osteogenic differentiation medium
- OP:
-
Osteoporosis
- OD:
-
Optical density
- PM:
-
Proliferation medium
- RT-qPCR:
-
Real-time quantitative reverse transcription PCR
- RIP:
-
RNA Immunoprecipitation
- siRNA:
-
Small interfering RNA
References
Zhong L, Sun Y, Wang C, Liu R, Ru W, Dai W, et al. SP1 regulates BMSC osteogenic differentiation through the miR-133a-3p/MAPK3 axis: SP1 regulates osteogenic differentiation of BMSCs. J Orthop Surg Res. 2024;19(1):396.
Ouyang X, Li S, Ding Y, Xin F, Liu M. Foxf1 gene increases the risk of osteoporosis in rats by inhibiting osteoblast formation and promoting osteoclast differentiation through the upregulation of NF-kappaB pathway. J Musculoskelet Neuronal Interact. 2022;22(2):242–50.
Mai Z, Liu J, Jiang X, Gu W, Wang W, Li S, et al. Long noncoding RNA KCNMA1-AS1 promotes osteogenic differentiation of human bone marrow mesenchymal stem cells by activating the SMAD9 signaling pathway. Biol Direct. 2023;18(1):81.
Jin C, Jia L, Tang Z, Zheng Y. Long non-coding RNA MIR22HG promotes osteogenic differentiation of bone marrow mesenchymal stem cells via PTEN/ AKT pathway. Cell Death Dis. 2020;11(7):601.
Li Z. LncRNA PCBP1-AS1 induces osteoporosis by sponging miR-126-5p/PAK2 axis. Bone Joint Res. 2023;12(6):375–86.
Luo W, Zhang N, Wang Z, Chen H, Sun J, Yao C, et al. LncRNA USP2-AS1 facilitates the osteogenic differentiation of bone marrow mesenchymal stem cells by targeting KDM3A/ETS1/USP2 to activate the Wnt/beta-catenin signaling pathway. RNA Biol. 2024;21(1):1–13.
Mai YX, Li ZP, Pang FX, Zhou ST, Li N, Wang YY, et al. Aucubin promotes osteogenic differentiation and facilitates bone formation through the lncRNA-H19 driven Wnt/beta-Catenin Signaling Regulatory Axis. Stem Cells Int. 2024;2024:5388064.
Li Y, Sun W, Li J, Du R, Xing W, Yuan X, et al. HuR-mediated nucleocytoplasmic translocation of HOTAIR relieves its inhibition of osteogenic differentiation and promotes bone formation. Bone Res. 2023;11(1):53.
Zhang X, Yan Q, Liu X, Gao J, Xu Y, Jin Z, et al. LncRNA00638 promotes the osteogenic differentiation of periodontal mesenchymal stem cells from periodontitis patients under static mechanical strain. Stem Cell Res Ther. 2023;14(1):177.
Tu Z, Schmollerl J, Cuiffo BG, Karnoub AE. Microenvironmental Regulation of Long Noncoding RNA LINC01133 promotes Cancer Stem Cell-Like phenotypic traits in Triple-negative breast cancers. Stem Cells. 2019;37(10):1281–92.
Li Q, Zhou H, Wang C, Zhu Z. Long non-coding RNA Linc01133 promotes osteogenic differentiation of human periodontal ligament stem cells via microRNA-30c / bone gamma-carboxyglutamate protein axis. Bioengineered. 2022;13(4):9602–12.
Ru J, Guo L, Ji Y, Niu Y. Hydrostatic pressure induces osteogenic differentiation of adipose-derived mesenchymal stem cells through increasing lncRNA-PAGBC. Aging. 2020;12(13):13477–87.
Liu LL, Liu ZR, Cao LJ, Wang J, Huang SM, Hu SG, et al. Iron accumulation induced by hepcidin1 knockout accelerates the progression of aging osteoporosis. J Orthop Surg Res. 2024;19(1):59.
Jiang C, Zhu S, Zhan W, Lou L, Li A, Cai J. Comparative analysis of bone turnover markers in bone marrow and peripheral blood: implications for osteoporosis. J Orthop Surg Res. 2024;19(1):163.
Chen F, Han J, Guo Z, Mu C, Yu C, Ji Z et al. Antibacterial 3D-Printed silver Nanoparticle/Poly lactic-co-glycolic acid (PLGA) scaffolds for bone tissue Engineering. Mater (Basel). 2023;16(11).
Yang JJ, Peng WX, Zhang MB. LncRNA KCNQ1OT1 promotes osteogenic differentiation via miR-205-5p/RICTOR axis. Exp Cell Res. 2022;415(1):113119.
Li X. LncRNA metastasis-associated lung adenocarcinoma transcript-1 promotes osteogenic differentiation of bone marrow stem cells and inhibits osteoclastic differentiation of Mo in osteoporosis via the miR-124-3p/IGF2BP1/Wnt/beta-catenin axis. J Tissue Eng Regen Med. 2022;16(3):311–29.
Li J, Lin J, Ji Y, Wang X, Fu D, Wang W, et al. A novel pyroptosis-associated lncRNA LINC01133 promotes pancreatic adenocarcinoma development via miR-30b-5p/SIRT1 axis. Cell Oncol (Dordr). 2023;46(5):1381–98.
Zhang Y, Shi W, Chen R, Gu Y, Zhao M, Song J, et al. LINC01133 regulates MARCKS expression via sponging miR-30d-5p to promote the development of lung squamous cell carcinoma. Transl Oncol. 2024;44:101931.
Yao Y, Zhang F, Liu F, Xia D. Propofol-induced LINC01133 inhibits the progression of colorectal cancer via miR-186-5p/NR3C2 axis. Environ Toxicol. 2024;39(4):2265–84.
Li Z, Xu D, Chen X, Li S, Chan MTV, Wu WKK. LINC01133: an emerging tumor-associated long non-coding RNA in tumor and osteosarcoma. Environ Sci Pollut Res Int. 2020;27(26):32467–73.
Deng DK, Li X, He XT, Sun HH, Tian BM, Chen FM. [Long non-coding RNA LINC01133 regulates cementogenic differentiation of human periodontal ligament stem cells by modulating mitochondrial functions]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2022;57(12):1209–16.
Chen Y, Wei Z, Shi H, Wen X, Wang Y, Wei R. BushenHuoxue formula promotes osteogenic differentiation via affecting hedgehog signaling pathway in bone marrow stem cells to improve osteoporosis symptoms. PLoS ONE. 2023;18(11):e0289912.
Gargano G, Oliva F, Oliviero A, Maffulli N. Small interfering RNAs in the management of human rheumatoid arthritis. Br Med Bull. 2022;142(1):34–43.
Gargano G, Asparago G, Spiezia F, Oliva F, Maffulli N. Small interfering RNAs in the management of human osteoporosis. Br Med Bull. 2023;148(1):58–69.
Giordano L, Porta GD, Peretti GM, Maffulli N. Therapeutic potential of microRNA in tendon injuries. Br Med Bull. 2020;133(1):79–94.
Oliviero A, Della Porta G, Peretti GM, Maffulli N. Corrigendum to: MicroRNA in osteoarthritis: physiopathology, diagnosis and therapeutic challenge. Br Med Bull. 2022;144(1):90.
Gargano G, Oliviero A, Oliva F, Maffulli N. Small interfering RNAs in tendon homeostasis. Br Med Bull. 2021;138(1):58–67.
Shen Y, Zhang Y, Wang Q, Jiang B, Jiang X, Luo B. MicroRNA-877-5p promotes osteoblast differentiation by targeting EIF4G2 expression. J Orthop Surg Res. 2024;19(1):134.
Wang X, Li X, Li J, Zhai L, Liu D, Abdurahman A, et al. Mechanical loading stimulates bone angiogenesis through enhancing type H vessel formation and downregulating exosomal mir-214-3p from bone marrow-derived mesenchymal stem cells. FASEB J. 2021;35(1):e21150.
Wang H, Zhou K, Xiao F, Huang Z, Xu J, Chen G, et al. Identification of circRNA-associated ceRNA network in BMSCs of OVX models for postmenopausal osteoporosis. Sci Rep. 2020;10(1):10896.
John AA, Xie J, Yang YS, Kim JM, Lin C, Ma H, et al. AAV-mediated delivery of osteoblast/osteoclast-regulating miRNAs for osteoporosis therapy. Mol Ther Nucleic Acids. 2022;29:296–311.
Ding L, Yin Y, Hou Y, Jiang H, Zhang J, Dai Z, et al. microRNA-214-3p suppresses Ankylosing Spondylitis Fibroblast Osteogenesis via BMP-TGFbeta Axis and BMP2. Front Endocrinol (Lausanne). 2020;11:609753.
Wang R, Zhang Y, Jin F, Li G, Sun Y, Wang X. High-glucose-induced mir-214-3p inhibits BMSCs osteogenic differentiation in type 1 diabetes mellitus. Cell Death Discov. 2019;5:143.
Han H, Tian T, Huang G, Li D, Yang S. The lncRNA H19/miR-541-3p/Wnt/beta-catenin axis plays a vital role in melatonin-mediated osteogenic differentiation of bone marrow mesenchymal stem cells. Aging. 2021;13(14):18257–73.
Gao Y, Chen N, Fu Z, Zhang Q. Progress of wnt signaling pathway in osteoporosis. Biomolecules. 2023;13(3).
Wang CG, Hu YH, Su SL, Zhong D. LncRNA DANCR and miR-320a suppressed osteogenic differentiation in osteoporosis by directly inhibiting the Wnt/beta-catenin signaling pathway. Exp Mol Med. 2020;52(8):1310–25.
He L, Zhou Q, Zhang H, Zhao N, Liao L. PF127 hydrogel-based delivery of Exosomal CTNNB1 from mesenchymal stem cells induces osteogenic differentiation during the repair of alveolar bone defects. Nanomaterials (Basel). 2023;13(6).
Zhang M, Hu S, Sun X. Alzheimer’s Disease and Impaired Bone Microarchitecture, Regeneration and Potential Genetic Links. Life (Basel). 2023;13(2).
Cao F, Zhan J, Chen X, Zhang K, Lai R, Feng Z. miR-214 promotes periodontal ligament stem cell osteoblastic differentiation by modulating Wnt/beta–catenin signaling. Mol Med Rep. 2017;16(6):9301–8.
Wang Y, Lu L, Niu Y, Zhang Q, Cheng C, Huang H, et al. The osteoporosis risk variant rs9820407 at 3p22.1 acts as an allele-specific enhancer to regulate CTNNB1 expression by long-range chromatin loop formation. Bone. 2021;153:116165.
Acknowledgements
None.
Funding
No funds, grants, or other support was received.
Author information
Authors and Affiliations
Contributions
Chao Tang, Lina Huang, Xiu-Quan Guo made substantial contributions to conception and design, acquisition of data, analysis and interpretation of data, and draft of the manuscript. Gang-Gang Wang and Zhigang Chen revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Prior to this study, the informed consent of the subjects was obtained and the guidelines of the Declaration of Helsinki were followed with the approval of the Zhucheng People’s Hospital Ethics Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13018_2024_5053_MOESM2_ESM.tif
Supplemental Fig. 2: RT-qPCR was employed to detect miR-214-3p expression in bone tissue of patients with OP and controls. *** P < 0.001
13018_2024_5053_MOESM3_ESM.tif
Supplemental Fig. 3: RT-qPCR was employed to detect CTNNB1 expression in the bone tissue of patients with OP and controls. *** P < 0.001
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, C., Huang, L., Guo, XQ. et al. LINC01133 promotes the osteogenic differentiation of bone marrow mesenchymal stem cells by upregulating CTNNB1 by acting as a sponge for miR-214-3p. J Orthop Surg Res 19, 572 (2024). https://doi.org/10.1186/s13018-024-05053-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-024-05053-8