Abstract
Background
Glucocorticoid-induced osteonecrosis of the femoral head (GIONFH) is a common disease in osteoarticular surgery, with a high disability rate, which brings great physical and mental pain and economic burden to patients. Its specific pathogenesis has not been fully demonstrated, and there is a lack of recognized effective biomarkers for earlier detection and prompt treatment. This has become an urgent clinical problem for orthopedic scholars.
Materials and methods
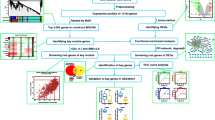
We downloaded the gene expression profile dataset GSE123568 from the Gene Expression Omnibus database, used STRING and Cytoscape to carry out module analysis and built a gene interaction network. The four core genes most related to GIONFH in this network were ultimately found out by precise analysis and animal experiment were then conducted for verification. In this verification process, thirty-six New Zealand white rabbits were randomly divided into blank control group, model group and drug group. Except for the blank control group, the animal model of GIONFH was established by lipopolysaccharide and methylprednisolone, while the drug group was given the lipid-lowering drugs for intervention as planned. The rabbits were taken for magnetic resonance imaging at different stages, and their femoral head specimens were taken for pathological examination, then the expression of target genes in the femoral head specimens of corresponding groups was detected. Validation methods included RT-PCR and pathological examination.
Results
A total of 679 differential genes were selected at first, including 276 up-regulated genes and 403 down-regulated genes. Finally, four genes with the highest degree of correlation were screened. Animal experiment results showed that ASXL1 and BNIP3L were in low expression, while FCGR2A and TYROBP were highly expressed.
Conclusion
Through animal experiments, it was confirmed that ASXL1, BNIP3L, FCGR2A and TYROBP screened from the comparative analysis of multiple genes in the database were closely related to GIONFH, which is important for early diagnosis of Glucocorticoid-induced osteonecrosis of the femoral head.
Similar content being viewed by others
Introduction
Glucocorticoids, a form of steroid hormones, are mainly used in the treatment of autoimmune diseases, such as rheumatoid arthritis, systemic lupus erythematosus and scleroderma, as well as organ transplantation and severe infection. Long-term or extensive use of glucocorticoids will bring more complications to patients, among which GIONFH is an example [1,2,3,4]. As a common type of nontraumatic osteonecrosis, GIONFH may result in the collapse of the subchondral bone structure without immediate treatment. Previous studies have shown that steroid hormones therapy can increase the risk of osteonecrosis by 4–50% [5].
In recent years, more attention has been gradually paid to GIONFH [6,7,8]. In the fight against Severe Acute Respiratory Syndrome (SARS) in 2003, glucocorticoids were used at high doses to save a large number of critically ill patients. Unfortunately, these steroid hormones also had side effects—the incidence of GIONFH was as high as 24–30% [9,10,11]. As for the novel coronavirus disease 2019 (COVID-19) epidemic, which is similar to SARS, steroid hormone is still an important medication for the treatment of infections to rescue lives, who will also be at risk of developing GIONFH in the future[12,13,14]. Therefore, early diagnosis and timely prevention and treatment of GIONFH have important clinical significance.
However, the pathogenesis of GIONFH is yet to be seen. The hypotheses proposed by researchers mainly include as follows-intravascular coagulation, vascular endothelial injury, increased intraosseous pressure, lipid metabolism disorder etc. [15]. Among them, the disorder of lipid metabolism is an important manifestation in GIONFH [16, 17]. It was confirmed in previous studies that steroid use may result in changes in the lipid metabolism, which increase the risk of fat embolism[18, 19]. Our team's studies also confirmed that GIONFH is closely related to the pathogenesis of lipid metabolism disorder induced by high-dose hormone in vivo[20,21,22]. Therefore, in order to further confirm the association between the onset of GIONFH and lipid metabolism disorder, a hypolipidemic drug group was added as comparative research to the Glucocorticoid-induced femoral head necrosis model rabbits in the animal experiment of this study.
In the current clinical research, there is a lack of methods and standards for early diagnosis of GIONFH. It is asymptomatic during the early phases. The first symptom is pain in the hip, which tends to be in proportion with lesion size, and in general precedes the beginning of femoral head collapse, which occurs on average after 8 months [23, 24]. At the moment of hip pain and dysfunction, and obvious osteonecrosis and collapse of the femoral head appear in pathological and imaging examinations, in which case surgical treatment is badly needed [25]. With the wide application of bioinformatics and the continuous development of microarray technology at the genome level, researchers have sequenced more and more disease samples with increasing fragment information, and the public functional genomics database has been greatly enriched [26, 27]. By screening and mining the massive information, we can find the mechanism of the occurrence and development of non-tumor diseases from the gene level, which helps us to screen out potential biomarkers. Then we verified the key markers screened before through animal experiments, it is expected to find new methods to facilitate the early diagnosis, detection and prevention of GIONFH.
Materials and methods
Gene chip data
GEO (http://www.ncbi.nlm.nih.gov/geo) is a public functional genomics database with high storage throughout gene expression data, chips, and microarrays [28]. The gene expression profile dataset GSE123568 was downloaded from the GEO database, and probes were converted to corresponding gene symbols based on annotation information on the platform [29]. The GSE123568 dataset contained tissue specimens from 30 GIONFH patients and 10 non-GIONFH patients. GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r), an interactive web tool, was used to screen DEGs between GIONFH and non-GIONFH samples. GEO2R allows users to compare two or more datasets in a GEO series in order to identify DEGs under certain conditions. Probe sets without corresponding gene symbols or genes with more than one probe set were removed, respectively. A log FC (fold change) > 1 or log FC < − 1 with an adjusted P value < 0.01 was considered statistically significant.
PPI network and module analysis
STRING is a database for predicting known protein interactions. Its interactions include direct (physical) associations and indirect (functional) associations. Analysis of function and interactions between proteins with STRING may provide insights into the mechanisms involved in disease development or progression [30]. In this study, the PPI network of DEGs was constructed using the STRING database, and interactions with a composite score (the support of the data) > 0.4 were considered statistically significant. Cytoscape (3.8.0 version) is an open-source software platform for visualizing complex networks and integrating them with any type of attribute data, including bioinformatics and social network analysis [31]. Cytoscape's plugin MCODE (2.0.0 version), is used to cluster a given network based on topology to find densely connected regions (highly interconnected regions) [32]. The PPI network as well as the biological process map were drawn with cytoscape, and the most important modules in the PPI network were identified with MCODE. The selection criteria were as follows: MCODE score > 5, degree cutoff = 2, node score cutoff = 0.2, maximum depth = 100, and k score = 2. Biological process analysis of central genes and protein network interaction mapping were performed by BiNGO, a plugin of cytoscape. The relationships among the nine hub genes were explored by STRING.
GO and KEGG enrichment analysis of DEGs
DAVIDv6.8 (http: //david.ncifcrf.gov/) [33], a visual integrated discovery database with annotation contains a complete update of knowledge base. KEGG is a database used for understanding advanced functions and practical experimental techniques in biological systems (e.g., cells, organisms and ecosystems) from molecular-level information (especially large-scale molecular datasets generated by genome sequencing and other high-throughput processes) [34]. The Gene Ontology (GO) knowledge base has the maximum information about gene function around the world, which can be recognized both by human and machine. It is also the basis for computational analysis of large-scale molecular biology and genetic experiments in biomedical research [35]. Functional enrichment analysis of DEGs was performed using the DAVID online database, and P < 0.05 was considered statistically significant.
Selection and analysis of key genes
UniProt (universal protein) is a database (https://www.uniprot.org/) collecting protein resources and interconnected with other databases. It has one of the most extensive protein sequences with most comprehensive annotations so far. The 18 genes we screened were consulted using the UniProt database and annotated in lists. Functional enrichment analysis of potential genes was then performed by DAVID [28] based on up-regulated genes (EPB41, BNIP3L, KLF1, SLC7A5, C10orf10, ASXL1) and down-regulated genes (CD86, FCGR3A, FCGR2B, CYBB, TLR2, TLR4, ITGAX, TLR8, TLR1, FCGR2A, TYROBP, MS4A6A). Such analysis was also performed for individual genes (BNIP3L, ASXL1, FCGR2A, TYROBP) to search for possible protein pathways.
Animal experiments
Thirty-six adult New Zealand white rabbits weighing 2.5–3.0 kg were purchased from the Animal Experimental Center of Jiangxi University of Traditional Chinese Medicine with license number of SYXK[gan]2004–0001. The 18 male and 18 female rabbits were single-housed with constant temperature at 20–23℃ under the humidity of 60%, and the light–dark cycle was 12 h each. After one week of adaptive feeding, those rabbits were randomly divided into three groups: blank control group (n = 12), model group (n = 12) and drug group (n = 12), half male and half female in each groups. Rabbits in the model group and drug group were intravenously injected with lipopolysaccharide (LPS, Sigma, USA) (10 μg/kg) via the marginal ear vein, and the second dose was administered 24 h later. After two injections of LPS, methylprednisolone sodium succinate (MSS, Huazhong Pharmaceutical, China) was injected intramuscularly at a dose of 20 mg/kg with a 24 h interval between each injection for a total of three injections. During hormone injection, 80,000 U of penicillin was intramuscularly injected into the right buttock to prevent infection (3 days) [36, 37]. After the completion of hormone injection, the drug group was intragastrical administered with simvastatin (Jingxin Pharmaceutical, China) water solution every day at a dose of 2 mg/10 ml per time for 4 weeks, the control and model groups were intragastrical administered with the same amount of saline. Magnetic resonance images of the hip joints and femoral head specimens were collected from three groups of rabbits at different time (after 2, 4, and 8 weeks). This study was conducted according to the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The protocol was approved by the Experimental Animal Ethics Committee of the Fourth Affiliated Hospital of Nanchang University.
Magnetic resonance imaging scanning
Magnetic resonance imaging (MRI) examination was performed at two, four, and eight weeks after the first hormone injection. 3% pentobarbital sodium solution (35 mg/kg) was injected intravenously into the ear margin of rabbits. After the anesthesia took effect, the bilateral femoral heads of all rabbits were scanned by a 3.0 T magnetic resonance imaging (MRI) scanner (Achieva TX, Philips). The parameters were as follows: T1WI (TR 700 ms, TE 45 ms), T2WI (TR 2600 ms, TE 200 ms), Stir (TR 1500 ms, TE 60 ms), slice thickness 1.2 mm, flip angle 90° and matrix (reconstruction 256 × matrix scan 256 mm). Then the MRI results of GIONFH in each group were compared.
Extraction and anatomy of specimens
The rabbits were sacrificed by air embolism at two, four, and eight weeks after hormone injection, and the femoral heads were harvested. Washed with ice phosphate buffer saline (PBS), the left femoral heads of all rabbits were preserved in liquid nitrogen for quantitative real-time PCR. Another femoral head was collected and immediately fixed with 4% paraformaldehyde at 4 °C for 24 h for later testing.
Pathological examination
After the femoral heads were fixed with 4% paraformaldehyde for 24 h, they were washed with 0.2 M PBS (pH 7.4) again and then decalcified in 10% ethylene diamine tetraacetic acid (EDTA) and neutralized with sodium sulphate buffer for approximately 30 days. After decalcification, the tissues were embedded in paraffin, cut into thick sections of 5 mm, and stained with hematoxylin and eosin. The osteonecrosis of the femoral head in H&E staining specimens was observed under a microscope, and the microphotos were taken. Leica Co. W550CW signal acquisition and analysis system (Weztlar, Germany) was used for observation, analysis and evaluation.
Real-time PCR
Total RNA was extracted using rnaiso plus (Takara bio, Japan) according to the manufacturer's instructions. The RNA concentration was evaluated by A260/A280 measurement, then reverse transcription was performed, and the reverse transcription product was amplified by qPCR Expression of BNIP3L, ASXL1, FCGR2A and TYROBP were analyzed using SYBR premix Ex TaqTM II (Takara bio, Japan) on ABI 7500 rapid real-time PCR system (applied biological systems, USA). Obtain the cycle threshold (CT) value and use 2-ΔΔ the relative expression of each target gene was calculated by CT method, and the data were normalized to GAPDH expression. The primer sequence is shown in Table 1.
Statistical analysis
Quantitative data are presented as mean ± standard deviation (SD). Hardy–Weinberg genetic balance method to test population representativeness, the phenotypic differences between groups were analyzed by paired t-test to estimate the relationship between each genotype and the risk of GIONFH the odds ratio (OR), 95% confidence interval (CI) and P value were calculated. Data were analyzed using SPSS 23.0 software (SPSS, USA). A level of P < 0.05 was considered to be significant.
Results
Analysis of gene arrays
Through GEO2R analysis, 679 DEGs were selected including 276 up-regulated genes and 403 down-regulated genes; the volcano plot (Fig. 1A) shows the relationship between fold change and P value in this set of microarray groups. The 10 genes with the highest weights were all down-regulated genes by Cytoscape analysis, which were CD86, FCGR3A, FCGR2B, CYBB, TLR2, TLR4, ITGAX, TLR8, FCGR2A, and TLR1 (Fig. 1B); 9 groups were found by MCODE, and there was a hub gene in each group, of which the up-regulated genes were ASXL1, BNIP3L, C10orf10, EPB41, KLF1, and SLC7A5, and the down-regulated genes were FCGR2A, MS4A6A, and TYROBP (Fig. 2A).
Functional enrichment analysis
It should be noted that the above two groups of genes have an overlapping gene (FCGR2A), so a total of 18 differential genes are synthesized from the two groups. When exploring the relationship between the nine hub genes, some genes were found to be interrelated (Fig. 2B). A protein interaction network was drawn for the 18 differential genes to provide protein expression or potential pathways and targets that may be associated with GIONFH (Fig. 3). The results of functional enrichment analysis of 18 genes mainly focus on down-regulated genes (Table 1), of which important parts have been marked (Fig. 4). The results showed that eighteen differential genes were annotated, among which BNIP3L, ASXL1, FCGR2A and TYROBP were highly correlated with osteonecrosis (Tables 2 and 3).
According to Table 3, we selected some important biological processes, cell structure, molecular functions and signal pathways from the functional enrichment analysis. A Pie chart is the relationship between items and FDR. B The relationship between bar graph entries and P-values
Magnetic resonance imaging results
Magnetic resonance imaging (MRI) is considered the most sensitive, specific modality and golden standard for the diagnosis and evaluation of GIONFH. It was used to analyze the imaging changes in the rabbits’ femoral heads. The results of T2 weighted MRI showed that in the blank control group, the shape of bilateral femoral heads was regular, the surface was smooth, no abnormal signals were found, and the bilateral hip joint space was normal (Fig. 5 A, B). As for the model group after 8 weeks, the T2-weighted images showed spotty, heterogeneous high signal intensity, slight collapse and flattening of bilateral femoral heads, edema signals in bilateral joint cavities (red arrow), and uneven joint spaces (Fig. 5 C, D). In the drug group, the joint space were normal, no obvious effusion was in the hip cavity, the epiphyseal line of the femoral heads were slightly blurred, and no obvious abnormal signals were found (Fig. 5 E, F). These results indicated that steroid hormone-induced osteonecrosis of the femoral head, however, simvastatin could exert a protective effect on GIONFH in vivo.
Analysis of bone structure of the rabbit femoral head by hip magnetic resonance coronal and traverse scanning. The representative images of rabbit bilateral femoral heads MRI scanning in the control group (A, B), model group (C, D) and drug group (E, F).The red arrow indicated slight collapse and flattening of femoral heads, edema signals in joint cavities and bone marrow
Histopathological analysis H&E staining
Based on the diagnostic criteria of GIONFH, we evaluated femoral head specimens of rabbits using H&E staining. In this study, none of the rabbits in the control group developed osteonecrosis. The bone trabeculas were regularly arrayed, with complete structure, clearly visible osteocytes and a few empty lacunaes (Fig. 6 A, B). In the model group, typical osteonecrosis occurred. The lipocytes were enlarged. Bone trabeculas turned thinner, and many empty lacunaes were observed (Fig. 6 C, D). In the drug group, there were a few lipocytes, and no apparent necrotic debris was noted. The bone trabeculas regularly arrayed, and only a few empty lacunaes were observed (Fig. 6 E, F).
Histopathological H&E staining of GIONFH in vivo. The representative images of H&E staining from the femoral heads in three groups. There were no empty lacunae in the femoral head of the control group (A, B). A large number of empty bone lacunae and necrotic bone marrow cells are visible in the model group (C, D), while there were few empty bone lacunae in the drug group (E, F). Magnification: × 200 (A, C and E), × 400 (B, D and F). The histogram shows the proportion of empty bone lacunae in the model group, which are significantly higher than those in the control group and drug group (G).The data are expressed as the means ± S.D. Significant differences between different groups are indicated as* p < 0.05 vs. the control group and # p < 0.05 vs. the model group
Expression of the key differential genes in the rabbit model of GIONFH
In order to investigate the expression of the key differential genes related to GIONFH in rabbits, real-time PCR was performed to measure the expression level of the ASXL1, BNIP3L, FCGR2A and TYROBP at different time points (after 2, 4, and 8 weeks). After 2 weeks, the results showed that the expression level of ASXL1 and BNIP3L were significantly decreased and the TYROBP robust increased in the model group compared to the blank control group (Fig. 7 A). But this effect was abolished when the rabbits were treated with simvastatin in the drug group. Similarly, after 4 and 8 weeks, the expression level of ASXL1 and BNIP3L decreased significantly in the model group compared to the blank control group, while the FCGR2A and TYROBP increased remarkable (Fig. 7 B, C). It should be noted that this effect was blocked by simvastatin in the drug group. Together, these data suggested that the four key differential genes ASXL1, BNIP3L, FCGR2A and TYROBP were significantly associated with osteonecrosis of the femoral head.
Real-time PCR assay showed the expression of ASXL1, BNIP3L, FCGR2A and TYROBP mRNA in the control group (a), model group (b) and drug group (c) at 2 weeks (A), 4 weeks (B), and 8 weeks (C). The underlined asterisk (*) represents the comparison between the model group and the control group; The asterisk (*) represents the comparison between the drug group and the control group. # means P > 0.05, * means 0.01 < P < 0.05, ** means P < 0.01, *** means P < 0.001, **** means P < 0.0001. (group a black histogram; group b light grey histogram; group c dark grey histogram)
Discussion
Although steroid hormones play an important role in the treatment of many diseases, they produce a series of side effects. For example, long-term or extensive use of steroid hormone may cause GIONFH [38]. Previous studies have shown that without early intervention, 80% of patients will develop a collapsed femoral head and significantly impaired hip function within 1–5 years of onset [39]. Such treatments for collapsed and necrotic femoral head as core decompression, Osteotomies, bone pedicled vascularized or autologous stem cell implantation are all invasive operations [40,41,42,43]. After the operations patients are very likely to have unbalanced bone resorption, which, as is well known, can be enhanced by corticosteroids [42]. Due to the negative effect in the long run, total hip arthroplasty (THA) is eventually an inevitable option. Therefore, more and more researchers are devoted to the study of early diagnosis and preventive drugs for GIONFH.
Bioinformatics analysis and microarray technology at the genome level are mostly adopted in tumor research. At first, the researchers sequenced the tumor sample cells of human, and uploaded the sequenced gene array to the gene bank. Then, a variety of sequencing results were downloaded from the gene bank by other researchers and tens of thousands of gene fragments were obtained after researchers’ classification and annotation. At last, key genes or mutated genes were identified from these gene fragments for research to discover the disease pathogenic mechanism and therapeutic targets [44, 45]. With the help of the analysis platform of lipid metabolomics technology, our research group has studied the variation trend of the metabolic profile spectrum of animal samples of GIONFH before and after the intervention, and confirmed that GIONFH is closely related to the expression of lipid metabolism gene. Moreover, animal experimental studies have confirmed that the application of lipid-lowering drugs could effectively improve the occurrence and disease progression of GIONFH [20, 21]. In this study, we obtained 679 differentially expressed genes by screening between the gene array of human GIONFH and non-GIONFH. In the screening analysis of 679 differentially expressed genes, the following two algorithm were used to identify the potential key genes. Ten weight genes were obtained by MCC (maximum clique centrality) algorithm. According to the statistics on its official website, the key networks captured by MCC algorithm will bring new insights into the basic regulatory networks and protein drug targets for experimental biologists. In addition, we also used the MCODE (molecular complex detection) plug-in to obtain nine gene groups. Despite the complexity of running the code of MCODE, this method has been used by many medical researchers for many times, which is enough to verify its reliability. Therefore, we used this method to obtain 18 potential genes to be verified.
We compared and analyzed the correlation between the related proteins expressed by the 18 genes and steroid-induced femoral head necrosis one by one. TYROBP and FCGR2A were associated with osteonecrosis at different sites in the same pathway, among which the mutation of TYROBP-DAP12 encodes membrane receptor component cells in natural killer cells and myeloid cells, which has been confirmed by Juha Paloneva et al. [46], meaning TYROBP-mediated signaling pathway played an vital role in human bone tissue. Another study by Juha Paloneva and her team [47] also demonstrated an important role for the DAP12-TREM2 signaling complex in the differentiation and function of osteoclasts. This undoubtedly highlights the importance of the high expression of TYROBP in the rabbit GIONFH model tissue. FCGR2A acts as an Fc region-binding receptor of immunoglobulin γ and has been found to be associated with many inflammatory diseases [48]. FcgammaRIIa (FcγRII) is a multivalent IgG receptor expressed primarily by myeloid cells, and its binding to lipid rafts (microdomains rich in cholesterol and sphingolipids) is critical for efficient signaling of the pathway [49]. Yang et al. [50] found in the study of myeloma that hepatocytes increase the secretion of CRP (C-reactive protein) in response to myeloma-derived cytokines, and the binding of CRP to FCGR2 on the surface of myeloma cells activates myeloma cells to promote osteoclastogenesis and bone destruction in vivo. It has also been shown that aggregation of FcgammaR on human bone marrow cells leads to tyrosine phosphorylation of inositol polyphosphate containing SHIP-2 (SRC homology 2 domain), which reveals a new regulatory role of the expression and function of inositol phosphatase SHIP-2 on FcgammaR mediated activation of human bone marrow cells [51]. These studies suggested that the FCGR2A we screened out might be highly associated with GIONFH and thus needs further study and validation. By consulting the literature, we could not find that the BNIP3L and ASXL1 were directly related to the occurrence of osteonecrosis. However, the protein encoded by BNIP3L/NIX (NIX named Bcl-2/E1B19 kDa-interacting protein 3-like, which is based on 56% homology with BNIP3) is itself a pro-apoptotic protein. BNIP3L/NIX can not be detected in normal tissues, and they were found in a variety of organelle cultures in hypoxia-induced experiments [52, 53]. The Bcl-2 gene family of hypoxia-induced NIX is expressed during erythropoiesis, and the researchers found that NIX is highly regulated during erythroid end-stage maturation [54]. This shows that NIX associated with BNIP3L can cause erythropenia under hypoxic conditions, which coincides with the ischemia and hypoxia that cause osteonecrosis of the femoral head. ASXL1 belongs to an oncogene with unstable genome and recent studies have shown that mutations in ASXL1 are found in hematopoietic cells of various myeloid tumors such as myelodysplastic syndrome and chronic myelomonocytic leukemia [55, 56]. Our bold guess here is: are patients with mutations in ASXL1 more likely to develop GIONFH? While more evidence is needed. Therefore, we selected these four core genes with the highest correlation to GIONFH as research targets.
Through the induction of hormone and endotoxin, rabbit femoral head necrosis was caused. In the experiment later, we fully proved the success of animal model establishment through the diagnostic criteria of magnetic resonance and pathological examination. In the experimental group, we took four New Zealand white rabbits at different time points of the second, fourth and eighth weeks after hormone use for MRI examination, and dissect their femoral head specimens for pathological examination and gene detection. Through the experiment, we were pleased to find that the quantitative expression of the four core genes, compared with the blank control group, in the animal samples of steroid-induced necrosis of the femoral head, the expression of ASXL1 and BNP3L is reduced, while the expression of FCGR2A and TYROBP is increased and hypolipidemic drugs can play a certain role in prevention and treatment of steroid-induced necrosis of femoral head in model animals. This result not only confirms that the core genes screened in the earlier stage do have an important relationship with the research purpose, and it is very important for early diagnosis with steroid-induced necrosis of femoral head.
The prevalence of GIONFH is increasing, and people are diagnosed with it at a much earlier age. With the loss of function of the hip joint, it has brought great harm to the physical and mental health of patients [57, 58]. We hope to find the key biomarkers of pathogenicity through bioinformatics analysis and microarray technology at genome level, so as to detect the biomarkers for early diagnosis of GIONFH or to find therapeutic targets for early prevention or effective treatment to avoid further harm.
In this study, we aimed to screen and identify DEGs that might be involved the GIONFH, 679 differential genes were screened using the public gene expression comprehensive dataset. These 18 genes might be associated with the occurrence of GIONFH. The animal model was successfully established, the expression of genes preliminarily confirmed the results of gene microarray analysis. We speculated that the expression of ASXL1, BNIP3L, FCGR2A and TYROBP were significantly associated with GIONFH, which was expected to become a new perspective for the study of the pathogenesis of GIONFH.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Buehring B, Viswanathan R, Binkley N, Busse W. Glucocorticoid-induced osteoporosis: an update on effects and management. J Allergy Clin Immunol. 2013;132(5):1019–30. https://doi.org/10.1016/j.jaci.2013.08.040.
Li X, Li YS, Li LJ, et al. Overactivated autophagy contributes to steroid-induced avascular necrosis of the femoral head. Exp Ther Med. 2017;14(1):367–72. https://doi.org/10.3892/etm.2017.4508.
Qiang H, Liu H, Ling M, Wang K, Zhang C. Early steroid-induced osteonecrosis of rabbit femoral head and panax notoginseng saponins: mechanism and protective effects. Evid Based Complement Alternat Med. 2015;2015:719370. https://doi.org/10.1155/2015/719370.
Wang MG, Huang XX, Yao D, An Q, Deng XQ. Effect of glucocorticoid combined with gamma globulin in treatment of children with myasthenia gravis and its effects on immune globulin and complement of children. Eur Rev Med Pharmacol Sci. 2016;20(11):2404–8.
Cook AM, Dzik-Jurasz AS, Padhani AR, Norman A, Huddart RA. The prevalence of avascular necrosis in patients treated with chemotherapy for testicular tumours. Br J Cancer. 2001;85(11):1624–6. https://doi.org/10.1054/bjoc.2001.2155.
Liang XZ, Luo D, Chen YR, et al. Identification of potential autophagy-related genes in steroid-induced osteonecrosis of the femoral head via bioinformatics analysis and experimental verification. J Orthop Surg Res. 2022;17(1):86. https://doi.org/10.1186/s13018-022-02977-x.
Yan Y, Wang J, Huang D, et al. Plasma lipidomics analysis reveals altered lipids signature in patients with osteonecrosis of the femoral head. Metabolomics. 2022;18(2):14. https://doi.org/10.1007/s11306-022-01872-0.
Huang C, Wen Z, Niu J, Lin S, Wang W. Steroid-induced osteonecrosis of the femoral head: novel insight into the roles of bone endothelial cells in pathogenesis and treatment. Front Cell Dev Biol. 2021;9:777697. https://doi.org/10.3389/fcell.2021.777697.
Shen J, Liang BL, Zeng QS, et al. Investigation of proximal femoral marrow with magnetic resonance imaging in recovered patients with severe acute respiratory syndrome. Zhonghua Jie He He Hu Xi Za Zhi. 2006;29(3):189–93.
Li YM, Wang SX, Gao HS, et al. Factors of avascular necrosis of femoral head and osteoporosis in SARS patients’ convalescence. Zhonghua Yi Xue Za Zhi. 2004;84(16):1348–53.
Mei R, Chen D, Zhong D, et al. Metabolic profiling analysis of the effect and mechanism of gushiling capsule in rabbits with glucocorticoid-induced osteonecrosis of the femoral head. Front Pharmacol. 2022;13:845856. https://doi.org/10.3389/fphar.2022.845856.
Ledford H. How does COVID-19 kill? Uncertainty is hampering doctors’ ability to choose treatments. Nature. 2020;580(7803):311–2. https://doi.org/10.1038/d41586-020-01056-7.
Tang C, Wang Y, Lv H, Guan Z, Gu J. Caution against corticosteroid-based COVID-19 treatment. Lancet. 2020;395(10239):1759–60. https://doi.org/10.1016/S0140-6736(20)30749-2.
Chen F, Hao L, Zhu S, et al. Potential adverse effects of dexamethasone therapy on COVID-19 patients: review and recommendations. Infect Dis Ther. 2021;10(4):1907–31. https://doi.org/10.1007/s40121-021-00500-z.
Wang A, Ren M, Wang J. The pathogenesis of steroid-induced osteonecrosis of the femoral head: a systematic review of the literature. Gene. 2018;671:103–9. https://doi.org/10.1016/j.gene.2018.05.091.
Wu Z, Ji C, Li H, Qiu G, Gao C, Weng X. Elevated level of membrane microparticles in the disease of steroid-induced vascular osteonecrosis. J Craniofac Surg. 2013;24(4):1252–6. https://doi.org/10.1097/SCS.0b013e3182902dd3.
Tong P, Wu C, Jin H, et al. Gene expression profile of steroid-induced necrosis of femoral head of rats. Calcif Tissue Int. 2011;89(4):271–84. https://doi.org/10.1007/s00223-011-9516-y.
Migliorini F, Maffulli N, Baroncini A, Eschweiler J, Tingart M, Betsch M. Prognostic factors in the management of osteonecrosis of the femoral head: a systematic review. Surgeon. 2022. https://doi.org/10.1016/j.surge.2021.12.004.
Zhang G, Qin L, Sheng H, et al. A novel semisynthesized small molecule icaritin reduces incidence of steroid-associated osteonecrosis with inhibition of both thrombosis and lipid-deposition in a dose-dependent manner. Bone. 2009;44(2):345–56. https://doi.org/10.1016/j.bone.2008.10.035.
Jiang LY, Yu X, Pang QJ. Research in the precaution of recombinant human erythropoietin to steroid-induced osteonecrosis of the rat femoral head. J Int Med Res. 2017;45(4):1324–31. https://doi.org/10.1177/0300060517707076.
Ren X, Fan W, Shao Z, Chen K, Yu X, Liang Q. A metabolomic study on early detection of steroid-induced avascular necrosis of the femoral head. Oncotarget. 2018;9(8):7984–95. https://doi.org/10.18632/oncotarget.24150.
Ren X, Shao Z, Fan W, Wang Z, Chen K, Yu X. Untargeted metabolomics reveals the effect of lovastatin on steroid-induced necrosis of the femoral head in rabbits. J Orthop Surg Res. 2020;15(1):497. https://doi.org/10.1186/s13018-020-02026-5.
Quaranta M, Miranda L, Oliva F, Aletto C, Maffulli N. Osteotomies for avascular necrosis of the femoral head. Br Med Bull. 2021;137(1):98–111. https://doi.org/10.1093/bmb/ldaa044.
Sonoda K, Yamamoto T, Motomura G, Nakashima Y, Yamaguchi R, Iwamoto Y. Outcome of transtrochanteric rotational osteotomy for posttraumatic osteonecrosis of the femoral head with a mean follow-up of 12.3 years. Arch Orthop Trauma Surg. 2015;135(9):1257–63. https://doi.org/10.1007/s00402-015-2282-y.
Migliorini F, La Padula G, Oliva F, Torsiello E, Hildebrand F, Maffulli N. Operative management of avascular necrosis of the femoral head in skeletally immature patients: a systematic review. Life. 2022. https://doi.org/10.3390/life12020179.
Bernasconi A, Canakoglu A, Masseroli M, Ceri S. The road towards data integration in human genomics: players, steps and interactions. Brief Bioinform. 2021;22(1):30–44. https://doi.org/10.1093/bib/bbaa080.
Schuster SC. Next-generation sequencing transforms today’s biology. Nat Methods. 2008;5(1):16–8. https://doi.org/10.1038/nmeth1156.
Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–5. https://doi.org/10.1093/nar/gkl887.
Li T, Zhang Y, Wang R, et al. Discovery and validation an eight-biomarker serum gene signature for the diagnosis of steroid-induced osteonecrosis of the femoral head. Bone. 2019;122:199–208. https://doi.org/10.1016/j.bone.2019.03.008.
Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15. https://doi.org/10.1093/nar/gks1094.
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. https://doi.org/10.1093/bioinformatics/btq675.
Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. https://doi.org/10.1186/1471-2105-4-2.
Huang DW, Sherman BT, Tan Q, et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. https://doi.org/10.1186/gb-2007-8-9-r183.
Kanehisa M, Symposium NF, Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–252.
Ashburner M, Ball CA, Blake JA, Botstein D, Cherry JM. Gene ontology: tool for the unification of biology. Gene Ontol Consort Nat Genet. 2000;25(1):25–9. https://doi.org/10.1038/75556.
Xu J, Gong H, Lu S, Deasey MJ, Cui Q. Animal models of steroid-induced osteonecrosis of the femoral head-a comprehensive research review up to 2018. Int Orthop. 2018;42(7):1729–37. https://doi.org/10.1007/s00264-018-3956-1.
Zhu T, Jiang M, Zhang M, et al. Construction and validation of steroid-induced rabbit osteonecrosis model. MethodsX. 2022;9:101713. https://doi.org/10.1016/j.mex.2022.101713.
Xie XH, Wang XL, Yang HL, Zhao DW, Qin L. Steroid-associated osteonecrosis: Epidemiology, pathophysiology, animal model, prevention, and potential treatments (an overview). J Orthop Translat. 2015;3(2):58–70. https://doi.org/10.1016/j.jot.2014.12.002.
Mont MA, Pivec R, Banerjee S, Issa K, Elmallah RK, Jones LC. High-dose corticosteroid use and risk of hip osteonecrosis: meta-analysis and systematic literature review. J Arthroplasty. 2015;30(9):1506-1512.e5. https://doi.org/10.1016/j.arth.2015.03.036.
Migliorini F, Maffulli N, Eschweiler J, Tingart M, Baroncini A. Core decompression isolated or combined with bone marrow-derived cell therapies for femoral head osteonecrosis. Expert Opin Biol Ther. 2021;21(3):423–30. https://doi.org/10.1080/14712598.2021.1862790.
Sadile F, Bernasconi A, Russo S, Maffulli N. Core decompression versus other joint preserving treatments for osteonecrosis of the femoral head: a meta-analysis. Br Med Bull. 2016;118(1):33–49. https://doi.org/10.1093/bmb/ldw010.
Meloni MC, Hoedemaeker WR, Fornasier V. Failed vascularized fibular graft in treatment of osteonecrosis of the femoral head. Histopathol Anal Jt. 2016. https://doi.org/10.11138/jts/2016.4.1.024.
Khan M, Abbas K, Ayling EA, Waqas Ilyas M, Dunlop DG. Autologous stem cell implantation with core decompression for avascular necrosis of the femoral head using a new device. Ann R Coll Surg Engl. 2021;103(7):508–13. https://doi.org/10.1308/rcsann.2021.0026.
Van ‘t Veer LJ, Dai H, Van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. https://doi.org/10.1038/415530a.
Liang F, Li Q, Zhou L. Bayesian neural networks for selection of drug sensitive genes. J Am Stat Assoc. 2018;113(523):955–72. https://doi.org/10.1080/01621459.2017.1409122.
Paloneva J, Manninen T, Christman G, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71(3):656–62. https://doi.org/10.1086/342259.
Paloneva J, Mandelin J, Kiialainen A, et al. DAP12/TREM2 deficiency results in impaired osteoclast differentiation and osteoporotic features. J Exp Med. 2003;198(4):669–75. https://doi.org/10.1084/jem.20030027.
Dai Y, Chen W, Huang J, et al. FCGR2A could function as a prognostic marker and correlate with immune infiltration in head and neck squamous cell carcinoma. Biomed Res Int. 2021;2021:8874578. https://doi.org/10.1155/2021/8874578.
Bournazos S, Hart SP, Chamberlain LH, Glennie MJ, Dransfield I. Association of FcgammaRIIa (CD32a) with lipid rafts regulates ligand binding activity. J Immunol. 2009;182(12):8026–36. https://doi.org/10.4049/jimmunol.0900107.
Yang J, Liu Z, Liu H, et al. C-reactive protein promotes bone destruction in human myeloma through the CD32-p38 MAPK-Twist axis. SCI Signal. 2017. https://doi.org/10.1126/scisignal.aan6282.
Pengal RA, Ganesan LP, Fang H, et al. SHIP-2 inositol phosphatase is inducibly expressed in human monocytes and serves to regulate Fcgamma receptor-mediated signaling. J Biol Chem. 2003;278(25):22657–63. https://doi.org/10.1074/jbc.M302907200.
Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW 2nd. Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117(3):396–404. https://doi.org/10.1161/CIRCULATIONAHA.107.727073.
Yussman MG, Toyokawa T, Odley A, et al. Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8(7):725–30. https://doi.org/10.1038/nm719.
Aerbajinai W, Giattina M, Lee YT, et al. The proapoptotic factor Nix is coexpressed with Bcl-xL during terminal erythroid differentiation. Blood. 2003;102(2):712–7. https://doi.org/10.1182/blood-2002-11-3324.
Sugimoto Y, Muramatsu H, Makishima H, et al. Spectrum of molecular defects in juvenile myelomonocytic leukaemia includes ASXL1 mutations. Br J Haematol. 2010;150(1):83–7. https://doi.org/10.1111/j.1365-2141.2010.08196.x.
Gelsi-Boyer V, Trouplin V, Adelaide J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. https://doi.org/10.1111/j.1365-2141.2009.07697.x.
Signorino JA, Jayaseelan DJ, Brindle K. Atypical clinical presentation of rapidly progressing femoral head avascular necrosis. J Orthop Sports Phys Ther. 2017;47(3):217. https://doi.org/10.2519/jospt.2017.6483.
Osawa Y, Seki T, Takegami Y, et al. Do femoral head collapse and the contralateral condition affect patient-reported quality of life and referral pain in patients with osteonecrosis of the femoral head? Int Orthop. 2018;42(7):1463–8. https://doi.org/10.1007/s00264-018-3867-1.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82174190, 82060017) and the Special Innovation Fund for Postgraduates of Jiangxi Province (No. YC2021-B054).
Author information
Authors and Affiliations
Contributions
XFY and KYC generated the idea, and project administration. DC and DMZ data analysis and prepared the original draft. RHM critically revised the paper. SDQ and PW experiment work; All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The animal study was reviewed and approved by the Laboratory Animal Ethics Committee of the Fourth Affiliated Hospital of Nanchang University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, D., Zhong, D., Mei, R. et al. Screening and identification of potential key biomarkers for glucocorticoid-induced osteonecrosis of the femoral head. J Orthop Surg Res 18, 28 (2023). https://doi.org/10.1186/s13018-022-03465-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-022-03465-y