Abstract
Background
Intervertebral disc degeneration (IDD) is widely known as the main contributor to low back pain which has a negative socioeconomic impact worldwide. However, the underlying mechanism remains unclear. This study aims to analyze the dataset GSE23130 using bioinformatics methods to identify the pivotal genes and pathways associated with IDD.
Material/methods
The gene expression data of GSE23130 was downloaded, and differentially expressed genes (DEGs) were extracted from 8 samples and 15 controls. GO and KEGG pathway enrichment analyses were performed. Also, protein–protein interaction (PPI) network was constructed and visualized, followed by identification of hub genes and key module.
Results
A total of 30 downregulated and 79 upregulated genes were identified. The DEGs were mainly enriched in the regulation of protein catabolic process, extracellular matrix organization, collagen fibril organization, and extracellular structure organization. Meanwhile, we found that most DEGs were primarily enriched in the PI3K-Akt signaling pathway. The top 10 hub genes were FN1, COL1A2, SPARC, COL3A1, CTGF, LUM, TIMP1, THBS2, COL5A2, and TGFB1.
Conclusions
In summary, key candidate genes and pathways were identified by using integrated bioinformatics analysis, which may provide insights into the underlying mechanisms and offer potential target genes for the treatment of IDD.
Similar content being viewed by others
Introduction
Low back pain (LBP) is increasingly recognized as a global public health problem associated with decreased quality of life and increased healthcare expenditure [1,2,3]. It is estimated that 70–80% of the adult population suffer from at least one episode of LBP during their lifetime and 10% become chronically disabled [4]. With the changing of work and lifestyle, the incidence and prevalence of low back pain increase dramatically in the past few decades.
Intervertebral disc degeneration (IDD) is one of the most common sources of low back pain [5]. Being the largest avascular structure of the human body, the intervertebral disc is a complex structure consisting of the annulus fibrosus, nucleus pulposus, and cartilage endplate. The extracellular matrix (ECM) is the main component of the intervertebral disc which is responsible for maintaining both structure and function of the intervertebral disc. Intervertebral disc degeneration is characterized by the excessive degradation of ECM, leading to reduced hydration, loss of disc height, and decreased ability to absorb mechanical force [6].
Management of IDD includes both conservative treatment and surgery. Conservative management includes physiotherapy, medication, and epidural steroids injection. On the other hand, both fusion and non-fusion surgeries have demonstrated efficacy and safety in the management of IDD. The success rates of the Main and SPORT for surgically treated LDH patients and conservative-treated patients were 80% and 60%, respectively, at 1 year follow-up [7,8,9]. Although both methods have achieved satisfactory clinical outcomes, neither conservative nor surgical treatment can completely resolve lumbar disc degeneration. Hence, it is necessary to elucidate the underlying mechanisms of IDD to find out a curative method. Previous studies showed that IDD is most likely to be multifactorial, including apoptosis [10, 11], inflammation [12, 13], aging [14, 15], and biomechanical loading [16, 17]. However, the genetic factors are regarded as the most significant contributor [18].
Nowadays, the gene chip technology and bioinformatics methods have been widely used to obtain the gene expression profile of the disease. This study uses bioinformatics methods to analyze the microarray of the nucleus pulposus (GSE23130) with the aim to identify differentially expressed genes (DEGs) and pathways related to the progression of IDD. We also investigated some hub genes involved in the progression of IDD based on the protein–protein interaction (PPI) network. For example, MMP3 has been reported as a key gene in maintaining homeostasis of the extracellular matrix, and in vivo study showed that gene therapy targeting MMP3 was an efficient way to delay intervertebral disc degeneration [19]. Hence, to screen out DEGs as potential target candidates is of significance for the prevention and treatment of IDD. This study may provide new insights into the pathogenesis of IDD and potential target candidates for new therapy.
Methods
Microarray data collection
The gene expression data of GSE23130 was downloaded from the National Center of Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database, which was based on the GPL1352 platform of Affymetrix Human X3P Array. Disc tissue samples were obtained either by the National Cancer Institute Cooperative Tissue Network (CHTN) or surgical disc procedures performed on patients with herniated discs and degenerative disc disease. Thompson grades IV and V are considered to be IDD samples while Thompson grades I, II, and III are control samples according to the Thompson grading criteria [20]. A total of 23 samples, including 15 control samples and 8 IDD samples, were contained in this dataset.
Differential expression analysis
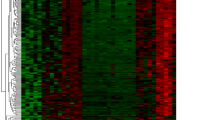
The Limma package version 3.28.21 (20) of Bioconductor 3.5 (http://www.bioconductor.org/packages/3.5/bioc/html/limma.html) was used to identify DEGs between the IDD samples and normal controls. Genes with fold change (logFC) > 1 (upregulated) or < − 1 (downregulated) and P value< 0.05 were considered differentially expressed. Both heatmap and volcano plots were constructed to present the expression profiles of differentially expressed genes using hierarchical clustering, which was performed using the R software.
KEGG and GO enrichment analyses of DEGs
GO is a major bioinformatics tool to annotate genes and analyze gene products and sequences to underlying biological phenomena, including biological process (BP), molecular function (MF), and cellular component (CC) [21]. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a knowledge base for systematic analysis, annotation, or visualization of gene functions and critical biological pathways closely related to intervertebral disc degeneration [22]. Both GO and KEGG enrichment analyses were conducted by Bioconductor. A P < 0.05 was considered to have statistical significance and to achieve significant enrichment.
PPI network construction and analysis of modules
Protein–protein interaction (PPI) enrichment analysis is useful to analyze the functional interactions between proteins which may provide insights into the mechanisms of generation or development of diseases. Online String [23] and Cytoscape [24] software were used to build the PPI network, and a confidence score > 0.4 was set as the cutoff criterion. The most significant module was identified using Molecular Complex Detection (MCODE) using the following parameters: degree cutoff = 2, node score cutoff = 0.2, k-core = 2, and maximum depth = 100. GO and KEGG pathway enrichment analyses of genes in the most significant node were subsequently performed using String.
Hub gene selection and analysis
CytoHubba was used to select hub genes of the PPI network with the Maximal Clique Centrality (MCC) method. The biological process of hub genes was then analyzed and visualized by the Biological Networks Gene Oncology tool (BiNGO; http://apps.cytoscape.org/apps/bingo) plugin.
Results
Differential expression analysis
A total of 19,703 DEGs were identified from intervertebral disc samples in GSE23130. Among them, 30 were downregulated and 79 were upregulated within the P value < 0.05 and |log2FC| > 1 criterion. A heatmap plot and a volcano plot are shown in Figs. 1 and 2, respectively, in which the red represents the upregulated genes and the green represents the downregulated genes. The top 10 upregulated and downregulated DEGs are shown in Table 1.
KEGG and GO enrichment analyses
We performed GO categories enrichment analysis to gain insights into the biological roles of the DEGs from degenerated versus non-degenerated disc samples. The DEGs were mainly enriched in skeletal system development, regulation of protein catabolic process, extracellular matrix organization, collagen fibril organization, and extracellular structure organization in terms of biological process (BP). The DEGs were mostly enriched in the collagen-containing extracellular matrix, extracellular matrix, fibrillar collagen trimer, banded collagen fibril, and complex of collagen trimers regarding cellular component (CC). The DEGs primarily participate in extracellular matrix structural constituent, collagen binding, disordered domain-specific binding, extracellular matrix structural constituent conferring tensile strength, and structural constituent of post-synapse concerning molecular function (MF). The top 5 BP, CC, and MF enrichment analyses of DEGs are summarized in Table 2. The GO enrichment-related bubble chart and circle plot are presented in Figs. 3 and 4. KEGG enrichment analysis showed that DEGs were enriched in “proteoglycans in cancer,” “platelet activation,” “PI3K-Akt signaling pathway,” “regulation of actin cytoskeleton,” and “thermogenesis.” The bubble chart and circle plot of KEGG enrichment were illustrated in Figs. 5 and 6.
PPI network construction and analysis of modules
PPI network of DEGs was downloaded from String and further analyzed by Cytoscape. The PPI network included 110 nodes and 410 edges (Fig. 7). The most significant module was identified from the PPI network using MCODE and consisted of 17 nodes and 64 edges (Fig. 8). GO and KEGG enrichment analyses of this module using String showed that these genes were mainly involved in extracellular matrix organization, skeletal system development, protein digestion and absorption, and ribosome.
Hub gene selection and analysis
The top 10 hub genes were identified by CytoHubba plugin using the Maximal Clique Centrality (MCC) method, including FN1, COL1A2, SPARC, COL3A1, CTGF, LUM, TIMP1, THBS2, COL5A2, and TGFB1 (Fig. 9). All hub genes were upregulated in the degenerated disc compared with the control group. Analysis of hub genes was summarized in Table 3.
Discussion
Despite years of numerous clinical and experimental investigations, the underlying mechanisms of intervertebral disc degeneration remain unclear, which hinders the development of curative therapy. Genetic factors, mechanical factors, aging, inflammation, and other potential factors may cause IDD whereas genetic factors play a critical role based on published literatures. Several studies indicate that genetic factors are critical contributors to the onset and progression of IDD [25, 26]. For example, COL1A1 is a key gene encoding collagen I, and polymorphisms of the COL1A1 gene have been reported to increase the risk of IDD in different population studies [27, 28]. In the present study, we identify a total of 109 DEGs between degenerative samples and controls, including 79 upregulated and 30 downregulated DEGs.
In terms of GO enrichment analysis, we found that most of DEGs were mainly involved in skeletal system development, regulation of protein catabolic process, extracellular matrix (ECM) organization, collagen fibril organization, and extracellular structure organization. The extracellular matrix (ECM) is a non-cellular three-dimensional macromolecular network predominantly composed of collagens, proteoglycans, and many other glycoproteins. ECM is crucial for maintaining the structural and functional integrity of the intervertebral disc. Previous studies showed that even though many potential mechanisms induced intervertebral disc (IVD), they led to a final common result of excessive degradation of the extracellular matrix [29]. The imbalance between anabolism and catabolism of ECM is regulated by ECM-modifying enzymes such as matrix metalloproteinases (MMPs) and their endogenous tissue inhibitors of metalloproteinases (TIMPs) [30,31,32]. Lumican (LUM) is the most significantly upregulated gene in our analysis, which is one kind of keratan sulfate proteoglycan constituents of the ECM. Several studies showed that the abundance of lumican changed with the degeneration of the intervertebral disc [33, 34]. The study conducted by Vo et al. showed that ECM degradation increased by regulation of matrix metalloproteinases (MMPs) and ADAMTSs, leading to the development of IDD [35]. On the contrary, TIMP-1 and TIMP-2 mRNA and protein expression increase in degenerated IVD tissue, antagonizing the effect of MMPs [36]. Our bioinformatics analysis also showed that TIMP-1 increased in the IVD samples than controls.
Regarding the KEGG pathway of our analysis, we found that most DEGs were primarily enriched in the PI3K-Akt signaling pathway. The PI3K-Akt signal pathway showed protective effects on the human nucleus pulposus under different pathological conditions. Activation of the PI3K-Akt pathway protects against IDD by the increase of ECM content, prevention of cell apoptosis, and induction or prevention of cell autophagy. Studies conformed that the activation of the PI3K-Akt pathway increased the SOX9 expression and activity and consequently led to the increase of the aggrecan expression in NP cells [37]. A study revealed that 17β-estradiol (E2) prevented the degradation of ECM by the activation of PI3K-Akt-FOXO3, which reduced the expression of MMP-3 and increased the expression of collagen II and aggrecan expression [38]. Many recent studies also showed that resveratrol suppressed IL-1β-mediated NP cell apoptosis through activating the PI3K-Akt pathway [39,40,41]. On the contrary, as the only known lipid phosphatase, tumor suppressor phosphatase and tensin homolog deleted from chromosome 10 (PTEN) can counteract the protective effect of the PI3K-Akt pathway. Xi et al. showed that PTEN promoted intervertebral disc degeneration by negatively influence PI3K-Akt [42]. Hence, gene therapy targeting PTEN may play an important role in treating IDD.
We further constructed a PPI network for better understanding of the interaction between DEGs. The most significant module was extracted from the PPI network using the MCODE plugin. Furthermore, the top 10 hub genes—FN1, COL1A2, SPARC, COL3A1, CTGF, LUM, TIMP1, THBS2, COL5A2, and TGFBI—were identified from this network. To be mentioned, all hub genes were also enriched in the most significant module. TGFBI is the seed DEG of the module. TGFBI is a protein secreted by many types of cells. It binds to collagen, forms part of the extracellular matrix (ECM), and interacts with integrins on cell surfaces. The study showed that TGF-β increased the expression of COL1A1, ACAN, and SOX9 genes by mediating communication between nucleus pulposus cells and mesenchymal stem cells [43]. Activation of TGF-β signaling has a protective effect on the intervertebral disc via inhibition of ECM degradation and increase of ECM synthesis, promotion of cell proliferation and inhibition of cell death, and alleviation of inflammatory response. However, excessive activation of TGF-β signaling may contribute to IVD degeneration [44]. SPARC is a matricellular glycoprotein involved in interactions between cells and matrices. Gruber showed that the deletion of the SPARC gene accelerated disc degeneration in the aging mouse [45]. Millecamps et al. demonstrated that inactivation of the SPARC gene led to the early onset of both disc degeneration and behavioral indices of LBP in mice [46]. Tajerian et al. showed that the underlying mechanism of the silence of the SPARC gene during aging may be attributed to DNA methylation [47]. A recent in vivo experimental study showed that stable expression of CTGF and TIMP1 genes by co-transfection adeno-associated virus 2 increased synthesis of aggrecan and type II collagen in the degenerated intervertebral disc, which served as a potential target gene for disc regeneration [48]. Thrombospondin proteins (THBSs) are a class of glycoproteins that function in maintaining homeostasis of ECM by regulating the level of matrix metalloproteinase-2 (MMP-2) and MMP-9 [49, 50]. A Japanese population-based genetic and functional data indicated that THBS2 played an important role in the pathogenesis of LDH by acting as a modulator of MMP-2 and MMP-9 endocytosis [51].
Conclusion
In summary, the present study provides a comprehensive analysis about the pathogenesis of IDD and offers potential target genes for the early diagnosis and treatment of intervertebral disc disease. The PI3K-Akt signal pathway and related hub genes may play important roles in the progression of IDD that needs deeper investigation. Nevertheless, further experiments are required to validate their effects and mechanisms in IDD.
Availability of data and materials
The following information was supplied regarding data availability. The raw data can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23130.
Abbreviations
- LBP:
-
Low back pain
- IVD:
-
Intervertebral disc
- IDD:
-
Intervertebral disc degeneration
- AF:
-
Annulus fibrosus
- NP:
-
Nucleus pulposus
- CEP:
-
Cartilage endplate
- miRNA:
-
MicroRNA
- ECM:
-
Extracellular matrix
- MMP:
-
Matrix metalloproteinase
- DEGs:
-
Differentially expressed genes
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Luoma K, Riihimki H, Luukkonen R, Raininko R, Viikari-Juntura E, Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine. 25(4):487–92.
Jensen CE, Riis A, Petersen KD, Jensen MB, Pedersen KM. Economic evaluation of an implementation strategy for the management of low back pain in general practice. Pain. 2017;158(5):891–9.
Lin CC, Li Q, Williams CM, et al. The economic burden of guideline-recommended first line care for acute low back pain. Eur Spine J. 2018;27(1):109–16.
Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 10(1):44–56.
Deyo RA, Weinstein JN. Low Back Pain. 1991;18(2):48.
Emilio, González, Martínez, et al. Biology and mechanobiology of the intervertebral disc.
Weinstein JN, Tosteson TD, Lurie JD, et al. Surgical vs nonoperative treatment for lumbar disk herniation: the Spine Patient Outcomes Research Trial (SPORT): a randomized trial. Jama. 2006;296(20):2441–50.
Weinstein JN, Lurie JD, Tosteson TD, et al. Surgical vs nonoperative treatment for lumbar disk herniation: the Spine Patient Outcomes Research Trial (SPORT) observational cohort. JAMA. 2006;296(20):2451–9.
Atlas SJ, Deyo RA, Keller RB, et al. The Maine Lumbar Spine Study, Part II. 1-year outcomes of surgical and nonsurgical management of sciatica. Spine (Phila Pa 1976). 1996;21(15):1777–86.
Yang S, Zhang F, Ma J, Ding W. Intervertebral disc ageing and degeneration: the antiapoptotic effect of oestrogen. Ageing Res Rev. 2020;57:100978.
Li Z, Shao Z, Chen S, et al. TIGAR impedes compression-induced intervertebral disc degeneration by suppressing nucleus pulposus cell apoptosis and autophagy. J Cell Physiol. 2020;235(2):1780–94.
Molinos M, Almeida CR, Caldeira J, Cunha C, Goncalves RM, Barbosa MA. Inflammation in intervertebral disc degeneration and regeneration. J R Soc Interface. 12(104):20141191.
Wang H, Tian Y, Wang J, et al. Inflammatory cytokines induce NOTCH signaling in nucleus pulposus cells: implications in intervertebral disc degeneration. J Biol Chem. 288(23):16761–74.
Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine. 20(11):1307–14.
Gruber HE, Ingram JA, Norton HJ, Hanley EN. Senescence in cells of the aging and degenerating intervertebral disc. Spine. 32(3):321–7.
McGill SM. The biomechanics of low back injury: implications on current practice in industry and the clinic. J Biomech. 1997;30(5):465–75.
Dougill G. Mechanical behaviours of intervertebral discs: clinical implications of loading, injury and treatment; 2016.
Ji SC, Han N, Liu Y, Li G, Sun Z, Li Z. Identification of genes associated with disc degeneration using bioinformatics. Biotech Histochem. 90(5):353–60.
Zhao Z, Li S, Huang H, Fang J, Wei H, Xi Y. In vivo delivery of MMP3-shRNA and Sox9 lentivirus cocktail enhances matrix synthesis to prevent lumbar disc degeneration. Adv Clin Exp Med. 2020;29(6):639–47.
Thompson JP, Pearce RH, Schechter MT, Adams ME, Tsang IK, Bishop PB. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine (Phila Pa 1976). 1990;15(5):411–5.
Ashburner M, Ball CA, Blake JA, Botstein D, Cherry JM. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9.
Minoru K, Susumu G. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;1:1.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Lars JJ. STRING V9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012;41(D1).
Smoot Me OKRJWPLIT. Cytoscape 2.8: new features for data integration and network visualization.
Kalb S, Martirosyan NL, Kalani MY, Broc GG, Theodore N. Genetics of the degenerated intervertebral disc. World Neurosurg. 2012;77(3-4):491–501.
Kalichman L, Hunter DJ. The genetics of intervertebral disc degeneration. Associated genes. Joint Bone Spine. 2008;75(4):388–96.
Pluijm SM, van Essen HW, Bravenboer N, et al. Collagen type I alpha1 Sp1 polymorphism, osteoporosis, and intervertebral disc degeneration in older men and women. Ann Rheum Dis. 2004;63(1):71–7.
Tilkeridis C, Bei T, Garantziotis S, Stratakis CA. Association of a COL1A1 polymorphism with lumbar disc disease in young military recruits. J Med Genet. 2005;42(7):e44.
Colombini A, Lombardi G, Corsi MM, Banfi G. Pathophysiology of the human intervertebral disc. 40(5):842.
Le Maitre CL, Freemont AJ, Hoyland JA. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J Pathol. 2004;204(1):47–54.
Li Y, Li K, Han X, et al. The imbalance between TIMP3 and matrix-degrading enzymes plays an important role in intervertebral disc degeneration. Biochem Biophys Res Commun. 2016;469(3):507–14.
Zhang JF, Wang GL, Zhou ZJ, Fang XQ, Chen S, Fan SW. Expression of matrix metalloproteinases, tissue inhibitors of metalloproteinases, and interleukins in vertebral cartilage endplate. Orthop Surg. 2018;10(4):306–11.
Sztrolovics R, Alini M, Mort JS, Roughley PJ. Age-related changes in fibromodulin and lumican in human intervertebral discs. Spine (Phila Pa 1976). 1999;24(17):1765–71.
Singh K, Masuda K, Thonar EJ, An HS, Cs-Szabo G. Age-related changes in the extracellular matrix of nucleus pulposus and anulus fibrosus of human intervertebral disc. Spine (Phila Pa 1976). 2009;34(1):10–6.
Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, Kang JD. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J Off J North Am Spine Soc. 13(3):331–41.
Bachmeier BE, Nerlich A, Mittermaier N, et al. Matrix metalloproteinase expression levels suggest distinct enzyme roles during lumbar disc herniation and degeneration. Eur Spine J. 2009;18(11):1573–86.
Cheng CC, Uchiyama Y, Hiyama A, Gajghate S, Shapiro IM, Risbud MV. PI3K/AKT regulates aggrecan gene expression by modulating Sox9 expression and activity in nucleus pulposus cells of the intervertebral disc. J Cell Physiol. 2009;221(3):668–76.
Gao XW, Su XT, Lu ZH, Ou J. 17β-Estradiol prevents extracellular matrix degradation by downregulating MMP3 expression via PI3K/Akt/FOXO3 pathway. Spine (Phila Pa 1976). 2020;45(5):292–9.
Yanhai, Jiang, Zhijie, et al. Resveratrol inhibits IL-1β-mediated nucleus pulposus cell apoptosis through regulating the PI3K/Akt pathway.
Gao J, Zhang Q, Song L. Resveratrol enhances matrix biosynthesis of nucleus pulposus cells through activating autophagy via the PI3K/Akt pathway under oxidative damage. Biosci Rep. 2018;38(4).
Wang W, Li P, Xu J, et al. Resveratrol attenuates high glucose-induced nucleus pulposus cell apoptosis and senescence through activating the ROS-mediated PI3K/Akt pathway. Biosci Rep. 2018;38(2).
Xi Y, Ma J, Chen Y. PTEN promotes intervertebral disc degeneration by regulating nucleus pulposus cell behaviors. Cell Biol Int. 2020;44(2):583–92.
Lehmann TP, Jakub G, Harasymczuk J, Jagodziński PP. Transforming growth factor β mediates communication of co-cultured human nucleus pulposus cells and mesenchymal stem cells. J Orthop Res. 2018;36(11):3023–32.
Chen S, Liu S, Ma K, Zhao L, Lin H, Shao Z. TGF-β signaling in intervertebral disc health and disease. Osteoarthr Cartil. 2019;27(8):1109–17.
Gruber EH. Targeted deletion of the SPARC gene accelerates disc degeneration in the aging mouse. J Histochem Cytochem Off J Histochem Soc. 53(9):1131–8.
Millecamps M, Czerminski JT, Mathieu AP, Stone LS. Behavioral signs of axial low back pain and motor impairment correlate with the severity of intervertebral disc degeneration in a mouse model. Spine J. 2015;15(12):2524–37.
Tajerian M, Alvarado S, Millecamps M, Dashwood T, Stone LS. DNA methylation of SPARC and chronic low back pain. Mol Pain. 2011;7(1):65.
Liu Y, Kong J, Xi Y-M, Yu T, Hu Y-G. Effects of AAV2-mediated co-transfection of CTGF and TIMP1 genes on degenerative lumbar intervertebral discs in rhesus monkeys in vivo. Am J Transl Res. 2018;10(4):1085–96.
Yuan B, Ji W, Fan B, Zhang B, Zhao Y, Li J. Association analysis between thrombospondin-2 gene polymorphisms and intervertebral disc degeneration in a Chinese Han population. Medicine. 97(2):e9586.
Hirose Y, Chiba K, Karasugi T, et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar-disc herniation. 82(5):1129.
Deguchi T, Hashizume H, Nakajima M, et al. A population-based study identifies an association of THBS2 with intervertebral disc degeneration. Osteoarthr Cartil. 2019;27(10):1501–7.
Acknowledgements
Special acknowledgment to May for the assistance and support of life and work.
Funding
There are no funders to report for this submission.
Author information
Authors and Affiliations
Contributions
Conceived and designed the study: Tao Lan and Zhe Shen. Analyzed the data: Yucheng Fu. Wrote and revised the manuscript: Tao Lan and Shiyu Hu. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study is based on microarray analysis.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to disclose in relation to this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hu, S., Fu, Y., Yan, B. et al. Analysis of key genes and pathways associated with the pathogenesis of intervertebral disc degeneration. J Orthop Surg Res 15, 371 (2020). https://doi.org/10.1186/s13018-020-01902-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-020-01902-4