Abstract
Colorectal cancer (CRC) ranks among the leading causes of cancer-related mortality worldwide, posing a significant public health challenge. Despite advancements in treatment strategies, prognosis for advanced CRC remains poor. Here, we investigate the role of CLK3 and its interaction with the c-Myc signaling pathway in CRC progression. Our study reveals significant overexpression of CLK3 in CRC tumor tissues, correlating with disease advancement, and demonstrates that CLK3 promotes CRC cell proliferation, mediated by its activation of MYC signaling through upregulation of c-MYC expression. In vivo experiments confirm the oncogenic role of CLK3, with its loss resulting in decreased tumor growth and c-MYC expression. These findings highlight CLK3 as a potential therapeutic target in CRC, offering insights into novel treatment strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most common malignancy globally, ranking as the second leading cause of cancer-related deaths and the 16th leading cause of all disease and injury-related deaths worldwide [1, 2]. It has emerged as a significant public health concern posing a serious threat to human health. The main treatment modalities for CRC include surgical resection, radiotherapy, chemotherapy, targeted therapy, and immunotherapy. The selection of treatment options typically depends on the stage, location of the cancer, and the individual’s health condition [3,4,5]. Despite advancements in treatment methods, the prognosis for advanced colorectal cancer remains poor.
CLK3, also recognized as CDC-like kinase 3, is a kinase with dual specificity, targeting substrates containing serine/threonine and tyrosine residues [6]. Its involvement in RNA splicing and translation regulation is facilitated through the phosphorylation of RNA processing factors like SRSF1 and SRSF3 [6, 7]. Recent investigations have unveiled CLK3’s regulatory role in various cancer types. For instance, it has been shown to facilitate the progression of cholangiocarcinoma by altering nucleotide metabolism, impede MFAP2-induced invasion in colorectal cancer upon deletion, and contribute to the aggressive advancement of hepatocellular carcinoma as a direct target of miR-144 [8,9,10]. Despite these observations, the understanding of CLK3’s mechanisms in tumor development remains primarily superficial, with limited exploration into its underlying pathways.
The Myc signaling pathway is a critical cellular signaling network involved in regulating biological processes such as cell proliferation, growth, and metabolism [11, 12]. At the core of this pathway is the c-Myc protein, a transcription factor that regulates the expression of a large number of genes. During cancer development, c-Myc is often overactivated, leading to abnormalities in various cellular processes, including aberrant proliferation, inhibition of apoptosis, and promotion of angiogenesis, thereby facilitating tumorigenesis and progression [13, 14]. Besides, c-Myc also interacts with other signaling pathways, forming complex regulatory networks. For example, c-Myc interacts with pathways such as Wnt/β-catenin, PI3K/AKT, and MAPK, influencing cellular behaviors such as proliferation, survival, and metastasis [15,16,17]. However, the regulatory mechanisms of c-Myc have not been clearly elucidated. Further research into the c-Myc signaling pathway will contribute to a better understanding of the pathogenesis of cancer and provide new targets and strategies for cancer therapy.
In our study, we demonstrate a significant overexpression of CLK3 in CRC tumor tissues, along with its positive regulation of CRC cell proliferation. Mechanistically, we establish that CLK3 enhances MYC signaling by upregulating c-MYC expression, highlighting the essential role of c-MYC in CLK3-mediated cancer cell proliferation. In vivo experiments further validate these findings, revealing that loss of CLK3 inhibits cancer cell proliferation and reduces c-MYC expression.
Results
CLK3 demonstrates significant overexpression in colorectal cancer
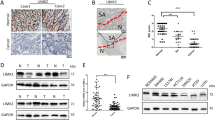
To investigate the role of CLK3 in colorectal cancer, we first collect 28 pairs of colorectal cancer tissues and adjacent normal colon tissues, and perform immunohistochemical analysis using CLK3 antibodies. The staining results are scored using the Immunoreactive Score (IRS) method. Immunohistochemical analysis reveals that CLK3 is significantly overexpressed in colorectal cancer tumor tissues (Fig. 1A, B). Additionally, we collect 71 colorectal cancer tumor tissues. Immunohistochemical analysis reveals that higher expression level of CLK3 is associated with advanced T stages in tumor tissues of patients (Fig. 1C, D). Additionally, it is observed that tumors from patients with higher clinical stages exhibit elevated levels of CLK3 expression (Fig. 1E). Analyzing the gene expression profiles of various tumors in The Cancer Genome Atlas (TCGA) database, it is found that the expression level of CLK3 is significantly upregulated in multiple types of cancers (Fig. 1F). Above all, we have demonstrated that CLK3 is significantly upregulated in colorectal cancer.
CLK3 is overexpressed in colorectal cancer. A. Representative images of IHC experiments. B. Analyzing the expression level of CLK3 in colorectal cancer and matched normal tissues from IHC experiments. C. Representative images of IHC experiments. D, E. Analyzing the expression level of CLK3 in colorectal cancer patients grouped by T (D) or clinical stage (E). F. Analysis of CLK3 expression level in multiply kinds of cancer using TCGA database. Statistical analysis was conducted using Student’s t-test
CLK3 positively regulates colorectal cancer cell proliferation
To further elucidate the role of CLK3 in colorectal cancer, we stably overexpress CLK3 in LoVo and HCT116 cell lines using lentiviral constructs. Immunoblotting confirms the successful construction of the aforementioned cell lines (Fig. 2A). Subsequently, we measure the ATP levels in LoVo and HCT116 cells expressing vector and CLK3. The results show that CLK3 overexpression significantly promoted ATP production in both cell lines (Fig. 2B). Also, CCK8 assays are performed to detect the proliferation ability of LoVo and HCT116 cells expressing vector and CLK3. Results show that elevated expression of CLK3 markedly enhances tumor cell proliferation (Fig. 2C). To further validate the proliferative role of CLK3, we employ CRISPR-cas9 technology to knockout CLK3 in LoVo and HCT116 cells. Immunoblotting experiments confirm the successful construction (Fig. 2D). Consistently, loss of CLK3 significantly inhibits ATP production in LoVo and HCT116 cells (Fig. 2E). Results from CCK8 assays further confirm that knockout of CLK3 obviously suppresses the proliferation ability of colorectal cancer cell (Fig. 2F). Overall, our data indicates a positive correlation between CLK3 expression levels and the proliferative capacity of colorectal cancer tumor cells.
CLK3 positively promotes colorectal cancer cell proliferation. A. Detecting the expression of identified proteins in Vector or CLK3 colorectal cancer cells. B. Measuring the ATP expression level in Vector or CLK3 colorectal cancer cells. C. CCK8 assays to assess the proliferation ability of Vector or CLK3 colorectal cancer cells. D. Detecting the expression of identified proteins in shnc or sgCLK3 colorectal cancer cells. E. Measuring the ATP expression level in shnc or sgCLK3 colorectal cancer cells. F. CCK8 assays to assess the proliferation ability of shnc or sgCLK3 colorectal cancer cells. Statistical analysis was conducted using Student’s t-test
CLK3 positively activates MYC signaling via upregulating c-MYC expression
We next aim to explore the potential molecular mechanisms underlying CLK3-mediated promotion of tumor proliferation. We first collect multiple colorectal cancer gene expression profiles from TCGA and GEO databases. Patients are stratified into high and low expression groups based on CLK3 expression levels, followed by GSEA analysis. Notably, Results suggest that patients with high CLK3 expression exhibit higher activation levels of the MYC signaling pathway in multiple datasets (Fig. 3A). As a key molecule in the MYC signaling pathway, c-MYC is further investigated to explore whether CLK3 can regulate its expression. Immunoblots confirm that CLK3 overexpression significantly enhances c-MYC expression, whereas loss of CLK3 markedly inhibits c-MYC expression (Fig. 3B, C). Besides, we also prove that CLK3 can bind with c-MYC (Fig. 3D). Of note, RT-PCR analysis demonstrated that CLK3 overexpression or deletion had no effect on the mRNA expression of c-MYC, while CLK3 could positively regulate the mRNA expression of c-MYC downstream gene cyclin D (Fig. 3E, F). Interesting, previous report had proved that CLK3 could regulate Wnt/ beta-catenin pathway, while we did not find this phenomenon (Fig. 3E-G and Supplementary Fig. 1A) [18]. CLK3 also did not regulate the expression level of USP13 (Supplementary Fig. 1B-C). Next, we also proved that CLK3 overexpression increased the nuclear expression level of c-MYC (Fig. 3H). Collectively, our findings reveal that CLK3 is a activator for MYC signaling.
CLK3 positively activated MYC signaling. A. GSEA analysis to reveal the relationship between CLK3 and c-MYC signaling using TCGA and GEO database. B. Detecting the expression of identified proteins in Vector or CLK3 colorectal cancer cells. C. Detecting the expression of identified proteins in shnc or sgCLK3 colorectal cancer cells. D. IP and IB assays to confirm the binding between c-MYC and CLK3. E, F. RT-PCR to detect the mRNA level of identified genes. G. Detecting the expression of Wnt/beta-catenin pathway-related proteins in shnc or sgCLK3 colorectal cancer cells. H. Isolating nuclear component in Vector or CLK3 colorectal cancer cells, and IB to detect the expression of identified proteins. Statistical analysis was conducted using Student’s t-test. All IBs were performed three times, yielding similar results
CLK3-mediated tumor proliferation depends on MYC signaling in vitro
To clarify the relationship between CLK3 signaling and MYC signaling in tumor proliferation, We established four kinds of cell lines stably expressing Vector + shnc, CLK3 + shnc, CLK3 + shc-MYC#1, and CLK3 + shc-MYC#2 respectively (Fig. 4A). Next, we detect the ATP level of four kinds of cancer cells. Results show that silencing of c-MYC can reverse the upregulation of ATP levels mediated by CLK3 (Fig. 4B). Morerover, CCK8 assays reveal that knockdown of c-MYC abrogates CLK3-mediated cancer cell proliferation (Fig. 4C). Also, knockdown of c-MYC reversed CLK3 overexpression-induced cyclin D expression increasing (Fig. 4D). Furthermore, we construct three kinds of cell lines stably expressing sgnc + Vector, sgCLK3 + Vector, and sgCLK3 + c-MYC respectively (Fig. 4E). After detecting the ATP level of these three kinds of cancer cells, we prove that gain of c-MYC can restore the decreased intracellular ATP levels caused by silencing CLK3 (Fig. 4F). Consistently, c-MYC receiving reverses the decreased proliferation ability induced by loss of CLK3 (Fig. 4G). Also, CLK3 overexpression reversed CLK3 knock-out-mediated cyclin D decreasing (Fig. 4H). It had been reported that c-MYC could transcriptionally regulate CLK3 expression in cholangiocarcinoma, which was not consistent with our data (Fig. 4D and H) [10]. Taken together, our data demonstrates that c-MYC is essential factor for CLK3-mediated cancer cell proliferation.
c-MYC is an essential factor for CLK3-mediated tumor proliferation. A. IBs to detect the expression level of related proteins in Vector + shnc, CLK3 + shnc, CLK3 + shc-MYC#1, or CLK3 + shc-MYC#2 cells. B. Measuring the ATP expression level in four kinds of colorectal cancer cells. C. CCK8 assays to assess the proliferation ability of four kinds of colorectal cancer cells. D. RT-PCR to detect the mRNA level of identified genes. E. IBs to detect the expression level of related proteins in shnc + Vector, sgCLK3#1 + Vector, or sgCLK3#1 + c-MYC cells. F. Measuring the ATP expression level in three kinds of colorectal cancer cells. G. CCK8 assays to assess the proliferation ability of three kinds of colorectal cancer cells. H. RT-PCR to detect the mRNA level of identified genes. Statistical analysis was conducted using Student’s t-test. All IBs were performed three times, yielding similar results
Loss of CLK3 inhibits tumor proliferation in vivo
To ascertain whether CLK3 promotes tumor proliferation in vivo, we subcutaneously inject LoVo cells with sgNC and sgCLK3 into the dorsal area of mice. Tumor size is measured every four days. After 20 days, mice are euthanized by cervical dislocation. Tumors are isolated. Representative images of tumors are depicted in Fig. 5A. Tumor growth curve shows that knockout of CLK3 significantly suppress tumor growth (Fig. 5B). Also, loss of CLK3 decreases the tumor weight (Fig. 5C). Additionally, we conduct another animal experiment and record the survival time of mice. The result indicates that knocking out CLK3 significantly prolongs the overall survival time of mice (Fig. 5D). Subsequently, we perform IHC assays to detect the intratumoral expression level of CLK3 and c-MYC. Results show that loss of CLK3 apparently inhibits c-MYC expression in vivo (Fig. 5E). Collectively, these results demonstrate that knockout of CLK3 obviously suppresses c-MYC expression and tumor proliferation in vivo.
loss of CLK3 inhibits tumor proliferation in vivo. A. Representative tumor images shown. B. Tumor growth curve shown. C. Statistical analysis for tumor weight. D. The survival time curve of these three group mice. E. Representative IHC results shown. Statistical analysis was conducted using Student’s t-test or Log-rank test
Discussion
Our study provides valuable insights into the role of CLK3 in colorectal cancer (CRC) progression and its interplay with the c-Myc signaling pathway. The significant overexpression of CLK3 in CRC tumor tissues underscores its potential as a key player in CRC pathogenesis. The observed positive regulation of CRC cell proliferation by CLK3 aligns with its known function in promoting cell growth and survival. Moreover, our mechanistic investigations reveal that CLK3 enhances MYC signaling by upregulating c-MYC expression, highlighting a novel regulatory mechanism in CRC. Besides, our findings revealed that CLK3 could directly bind to c-MYC, which implied that CLK3 might directly phosphorylate c-MYC to prevent the degradation of c-MYC.
The dependency of CLK3-mediated cancer cell proliferation on c-MYC further emphasizes the importance of understanding the intricate molecular networks governing CRC development. Our findings suggest that targeting CLK3 or its downstream effectors, such as c-MYC, may represent promising therapeutic strategies for CRC treatment. Inhibiting CLK3 activity or disrupting its interaction with c-MYC could potentially attenuate CRC tumor growth and improve patient outcomes. The results from our in vivo experiments provide compelling evidence supporting the role of CLK3 in CRC progression. The significant inhibition of cancer cell proliferation and reduction in c-MYC expression upon loss of CLK3 underscore the therapeutic potential of targeting CLK3 in CRC. These findings pave the way for further exploration of CLK3-targeted therapies in preclinical and clinical settings.
However, several questions remain unanswered and warrant further investigation. Firstly, the precise mechanisms underlying CLK3-mediated upregulation of c-MYC expression require elucidation. Understanding the upstream regulators and downstream effectors of CLK3-c-MYC axis could provide deeper insights into CRC pathogenesis and potential therapeutic targets. Additionally, exploring the therapeutic efficacy of CLK3 inhibition in CRC patient-derived xenograft models may better recapitulate the clinical scenario and facilitate the translation of our findings into clinical practice.
Conclusion
In conclusion, our study has unveiled the oncogenic role of CLK3 in colorectal cancer (CRC) through the activation of MYC signaling. These findings underscore the importance of both CLK3 and c-Myc signaling pathways in CRC pathogenesis, offering valuable insights into potential therapeutic avenues for CRC treatment.
Methods and material
Antibodies and cell lines
CLK3 antibodies (3256, CST), α-tubulin antibodies (2144, CST), c-MYC antibodies (18583, CST), β-catenin (A20221, ABclonal), SOX9 (A19710, ABclonal), p-GSK3β (AP1258, ABclonal) and Flag tag antibodies (14793, CST) are commercially available from their respective manufacturers. LoVo and HCT116 cell lines are obtained from the American Type Culture Collection. Cell cultures are maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum at 37 degrees Celsius in a 5% carbon dioxide atmosphere.
Cell counting Kit-8 (CCK8) assays
CCK-8 Assay Kits (RM02823) are procured from Abclonal. The CCK-8 assays are conducted following the manufacturer’s instructions.
ATP Assay
ATP Assay Kits (S0026) are procured from Beyotime. The ATP assays are conducted following the manufacturer’s instructions.
Immunoblot and immunoprecipitation
Cells are harvested, and cellular membranes or walls are disrupted using NP40 extraction buffer to liberate proteins. Protein concentration in the extracts is quantified via techniques like the BCA assay. Subsequently, protein samples are loaded onto a polyacrylamide gel, and SDS-PAGE electrophoresis is employed to fractionate proteins based on their molecular weights. The separated proteins are then transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane is probed with a specific primary antibody to bind selectively to the target protein, followed by incubation with a secondary antibody. Chemiluminescent detection (ECL) or comparable methodologies are employed to visualize and capture the protein bands’ intensity. In the case of immunoprecipitation, specific antibodies are introduced to facilitate the formation of antigen-antibody complexes with the target protein. Protein-antibody complexes are subsequently precipitated from the cell lysate using Protein A/G agarose.
Real-time PCR (RT-PCR)
Total RNAs were isolated using TRIzol (Thermo Fisher Scientific). cDNAs were synthesized using ABScript II cDNA First-Strand Synthesis Kit (RK20400, ABclonal). RT-PCR assays were conducted using ABScript III RT Master Mix for qPCR with gDNA Remover (RK20429, ABclonal). The primers were listed as followed: CLK3 F: 5- CGTACCTGAGCTACCGATGGA − 3, R: 5- TCCCTTCGGGACGGGTATC − 3; c-MYC F: 5- GGCTCCTGGCAAAAGGTCA-3, R: 5- GGCTCCTGGCAAAAGGTCA − 3; Cyclin D F: 5- TGGAGCCCGTGAAAAAGAGC − 3, R: 5- TCTCCTTCATCTTAGAGGCCAC − 3; SOX9 F: 5- AGCGAACGCACATCAAGAC − 3, R: 5- AGCGAACGCACATCAAGAC − 3.
Immunohistochemistry (IHC)
All clinical samples are obtained from The First Affiliated Hospital of Shihezi University. For immunohistochemistry experiments, tissue specimens are fixed in 4% paraformaldehyde and subsequently embedded in paraffin after dehydration. Thin sections, usually 4–5 micrometers thick, are sliced from the paraffin-embedded blocks using a microtome. These sections undergo deparaffinization, rehydration, and antigen retrieval steps. Subsequently, the samples are subjected to enzyme-based staining methods, and visualization is achieved through substrate reactions. Microscopic images of the stained sections are captured for analysis. Quantitative assessment of immunoreactivity is conducted using the Immunoreactive Score (IRS), which involves scoring both staining intensity and the percentage of positively stained cells. Staining intensity is graded on a scale from 0 (no staining) to 3 (strong staining), while the percentage of positive cells is categorized as follows: 0 (0%), 1 (1-25%), 2 (26-50%), 3 (51-75%), 4 (76-100%). The IRS is calculated as the product of the staining intensity score and the percentage of positive cells score.
Animal experiments
Six-week-old nude mice are procured from GemPharmatech LLC and subjected to a thorough health assessment before being housed in a controlled Specific Pathogen-Free (SPF) environment. Subsequently, two different cell lines are subcutaneously injected into the dorsal region of the mice. Tumor growth is monitored by measuring the long and short diameters of the tumors every 4 days. On the 20th day post-injection, the mice are humanely euthanized via cervical dislocation, and the tumors are carefully excised, weighed, and fixed for further analysis. Tumor volume is calculated using the formula: 0.5 * long diameter * short diameter * short diameter.
Statistical analysis
The Cancer Genome Atlas Program (TCGA) and the Gene Expression Omnibus (GEO) databases are accessed for data retrieval. Analysis is performed using the GSEA software to conduct Gene Set Enrichment Analysis. Statistical analysis is carried out using SPSS and GraphPad software, with the Student’s t-test utilized for comparisons. A significance level of P < 0.05 is considered statistically significant.
Data availability
No datasets were generated or analysed during the current study.
References
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–80.
Li J, Ma X, Chakravarti D, Shalapour S, DePinho RA. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 2021;35(11–12):787–820.
Zielinska A, Wlodarczyk M, Makaro A, Salaga M, Fichna J. Management of pain in colorectal cancer patients. Crit Rev Oncol Hematol. 2021;157:103122.
Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal Cancer: a review. JAMA. 2021;325(7):669–85.
Shin AE, Giancotti FG, Rustgi AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharmacol Sci. 2023;44(4):222–36.
Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241(2):300–8.
Virgirinia RP, Nakamura M, Takebayashi-Suzuki K, Fatchiyah F, Suzuki A. The dual-specificity protein kinase Clk3 is essential for Xenopus neural development. Biochem Biophys Res Commun. 2021;567:99–105.
Li H, Cui X, Hu Q, Chen X, Zhou P. CLK3 is a direct target of miR-144 and contributes to aggressive Progression in Hepatocellular Carcinoma. Onco Targets Ther. 2019;12:9201–13.
Xue M, Mi S, Zhang Z, Wang H, Chen W, Wei W, et al. MFAP2, upregulated by m1A methylation, promotes colorectal cancer invasiveness via CLK3. Cancer Med. 2023;12(7):8403–14.
Zhou Q, Lin M, Feng X, Ma F, Zhu Y, Liu X et al. Targeting CLK3 inhibits the progression of cholangiocarcinoma by reprogramming nucleotide metabolism. J Exp Med. 2020;217(8):e20191779.
Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35.
Duffy MJ, O’Grady S, Tang M, Crown J. MYC as a target for cancer treatment. Cancer Treat Rev. 2021;94:102154.
Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, et al. MYC protein interactors in gene transcription and cancer. Nat Rev Cancer. 2021;21(9):579–91.
Chen H, Liu H, Qing G. Targeting oncogenic myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5.
Das SK, Lewis BA, Levens D. MYC: a complex problem. Trends Cell Biol. 2023;33(3):235–46.
Masso-Valles D, Beaulieu ME, Soucek L. MYC, MYCL, and MYCN as therapeutic targets in lung cancer. Expert Opin Ther Targets. 2020;24(2):101–14.
Farrell AS, Sears RC. MYC degradation. Cold Spring Harb Perspect Med. 2014;4(3):a014365.
Cabel C, Thorne C, Abstract. Abstract 165: characterizing CLK3 in the colonic epithelium characterizing CLK3 in the colonic epithelium. Cancer Res. 2022;165(12Supplement):165.
Acknowledgements
Not applicable.
Funding
This study is approved by National Natural Science Foundation of China (No. 82260521), Postdoctoral Foundation of Xinjiang Production and Construction Corps (No. 308090), Xinjiang “Tianshan Talents” training plan medical and health young backbone personnel project (No. 202497), and Beijing Kechuang Medical Development Foundation project (KC2023-JX-0186-FQ037).
Author information
Authors and Affiliations
Contributions
SXL designs the experimental protocols, collects and analyzes data, drafts the manuscript. ZY conduct literature review and theoretical analysis, participate in experimental design and data analysis. ZY and MY provide equipment support, assiste with data collection and processing. MY provide theoretical guidance, participate in discussions and revise the manuscript content.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The studies involving human participants undergo review and approval by the Ethical Committee of The First Affiliated Hospital of Shihezi University. Participants provide written informed consent to participate. Approval for the animal study is obtained from the Institutional Animal Care and Use Committee at The First Affiliated Hospital of Shihezi University.
Consent for publication
The manuscript has received approval for publication from all listed authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.

Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shen, J., Zhao, Y., Man, Y. et al. CLK3 promotes tumor proliferation by activating MYC signaling. Cell Div 19, 28 (2024). https://doi.org/10.1186/s13008-024-00132-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13008-024-00132-y