Abstract
Background
EWSR1::NFATC2 rearranged sarcomas are a group of rare round, undifferentiated sarcomas with clinicopathological features different from those of Ewing's sarcoma (ES) family and other non-ES sarcomas. We report 4 cases of this rare sarcoma and review their features.
Materials and methods
Four cases of EWSR1::NFATC2 rearranged round cell sarcoma of the bone from the Pathology Department of Peking University People's Hospital were retrospectively studied. Clinical and pathological data were summarized, and immunohistochemical staining, fluorescence in situ hybridization (FISH), and Next-generation sequencing (NGS) were performed. Relevant literature reports were also reviewed.
Results
Among the four cases of EWSR1::NFATC2 rearranged round cell sarcoma, three were male, and one was female, with the age ranged from 14 to 34 years old at diagnosis (mean age: 27.5 years). All tumors were located in the femur and ranged in size from 4 to 8cm (mean 6cm), involving the surrounding soft tissues. All four patients underwent surgical treatment, and three received chemotherapy and radiotherapy postoperatively. Follow-up results showed that all four patients were alive. Histologically, the tumors exhibited small round cell sarcoma phenotype, with the stroma rich in mucin or exhibiting a glassy appearance. The tumor cells diffusely expressed CD99, NKX2.2, NKX3.1 and focal expression of CK and EMA was observed. FISH analysis showed that EWSR1 gene rearrangement was detected in all 4 cases, accompanied by 5' locus amplification. EWSR1::NFATC2 fusion probe demonstrated multi yellow fusion signals. NGS identified EWSR1::NFATC2 breakpoints in exon 9 and exon 3 in all 4 cases. The average follow-up duration of the study group was 88 months (range from 26—180 months). One case experienced both local recurrence and metastasis to the lung and chest wall. One case presented with local recurrence. The remaining two cases did not have the recurrence or metastasis.
Conclusion
Although the disease can locally recur and metastasize to the lungs, its mortality rate is significantly lower than that of Ewing sarcoma and other high-grade small round cell undifferentiated sarcomas. Therefore, it supports to classify this tumor as a separate subtype of small round cell sarcoma.
Similar content being viewed by others
Background
Among bone and soft tissue small round cell sarcomas, the most common type is Ewing sarcoma, which is characterized by balanced translocation of the EWSR1 gene on 22q12 as a molecular genetic feature. The fusion partners of this gene include FLI-1 and ERG, accounting for 90% and 5%, respectively [1, 2]. In addition, EWSR1 can also undergo translocation with some less common genes, namely ETV1, ETV4, and FEV. With the development of molecular techniques such as RNA sequencing, rare EWSR1 fusion partners such as SP3, PATZ1, and NFATC2 have been detected in some small round cell sarcomas [4,5,6]. In the fifth edition of the classification of soft tissue and bone tumors, these tumors are classified as EWSR1-non-ETS small round cell sarcomas [3]. These cases are very rare, and only a few reports about EWSR1/FUS::NFATC2 fusion gene in small round cell sarcoma can be found in the literature [4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24]. This article reports four cases of EWSR1::NFATC2 sarcoma, reviews the literature, and explores the clinical and pathological characteristics as well as molecular genetic features of this tumor.
Materials and methods
Case selection
Four cases of EWSR1 gene rearrangement positive with red signal amplification (5’ of the EWSR1 locus) undifferentiated small round cell sarcoma were selected. Cases 1—3 were from the Pathology Department of Peking University People's Hospital, and case 4 was a consultation case from Jishuitan Hospital. The diagnoses of all the cases were confirmed by senior bone and soft tissue pathologists in the Pathology Department of Peking University People's Hospital. Clinical and follow-up data were obtained from the clinical medical database and follow-up telephone calls.
Histopathology and immunohistochemistry
The tissues were fixed with 4% neutral formaldehyde, embedded in paraffin, cut into 4μm thickness, and stained with routine hematoxylin–eosin (HE). EnVision two-step method was used for immunohistochemical detection. Antibodies used were CD99 (O13, 1:200, Beijing Jingqiao Yatu Biotechnology Co., Ltd, China.), SMA(1A4, 1:120, Beijing Jingqiao Yatu Biotechnology Co., Ltd, China.), Desmin (Ep15, 1:400, Beijing Jingqiao Yatu Biotechnology Co., Ltd, China.), Cytokeratins (AE1/3, 1:200, Fuzhou Maixin Biotechnology Development Co., Ltd, China.), S-100(15E2E2 + 4C4.9, 1:200, Beijing Zhongshan Jingqiao Biotechnology Co., Ltd, China.), Vimentin (EP21, 1:200, Beijing Zhongshan Jingqiao Biotechnology Co., Ltd, China.), EMA (E29, 1:200, Beijing Zhongshan Jingqiao Biotechnology Co., Ltd, China.), NKX2.2 (EP336, 1:200, Beijing Zhongshan Jingqiao Biotechnology Co., Ltd, China.), NKX3.1 (EP356, 1:100, Beijing Zhongshan Jingqiao Biotechnology Co., Ltd, China.) and MUC4 (8G7, 1:100, Beijing Zhongshan Jingqiao Biotechnology Co., Ltd, China.).
Fluorescence in situ hybridization (FISH)
(1) Experimental method: FISH detection was performed using 4% neutral formalin-fixed, paraffin-embedded, 3μm thick white slides. EWSR1 dual-color break-apart probe (Abbott Molecular, USA) and EWSR1::NFATC2 dual-color fusion probe (Wuhan Kanglu Biotechnology Co., Ltd, China) were used. The experiment was performed according to the instructions provided with the kit. (2) Interpretation criteria: EWSR1 break-apart probe: cells were observed under blue fluorescence, and isolated, intact and non-overlapping cell nuclei were selected for counting. Red and green signals were separated, and the distance between them was greater than twice the diameter of one signal. 100 cells were randomly counted, and if ≥ 10% of the cells showed separation signals, it was considered positive. EWSR1::NFATC2 fusion probe: similarly, isolated, intact and non-overlapping cell nuclei were selected for counting, and both red and green signals were amplified, and the red and green signals fused together, showing yellow signal amplification. If ≥ 10% of cells showed this fusion pattern, it was judged as fusion-positive.
Next-generation sequencing (NGS)
NGS analysis was performed on the patient's paraffin-embedded tumor samples. RNA was first extracted from FFPE samples using the RNA extraction kit from Qiagen, Germany. The total RNA concentration was measured using the Qubit RNA HS assay kit (ThermoFisher, catalog number: Q32852), and the quality control for integrity (DV200) was performed using the Agilent RNA 6000 Pico assay kit (Agilent, catalog number: 5067–1513). The detected total RNA amount was not less than 50 ng, with a DV200 value of not less than 30%. The extracted RNA was used for RNA library preparation. The starting amount of total RNA was at least 500 ng, and rRNA was removed using the NEBNext rRNA Depletion Kit (Human/Mouse/Rabbit) (NEB, catalog number: E6350). The RNA underwent fragmentation, primer and first-strand cDNA synthesis, second-strand synthesis, SPRI purification, end repair and A-tailing, and adapter ligation steps, followed by RNA sequencing using the Gene + Seq-2000 platform (GenePlus). The Gene + OncoBox (GenePlus) was used to identify fusion gene products. All steps were performed according to the instructions provided with the NEBNext Ultra II Directional RNA Library Prep Kit (NEB, catalog number: E7760).
Results
Clinical data
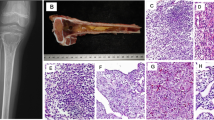
Four cases, including 3 males and 1 female, with ages ranging from 14 to 34 years (average age 27.5 years). All 4 tumors occurred in the femur and the maximum size from 4 to 8 (mean 6 cm), with one case discovered by pathological fracture and the other 3 cases discovered due to painful masses. Imaging analyses showed that all lesions were osteolytic with indistinct borders on plain X-rays (case 1). Magnetic resonance imaging showed T2WI sequences. The lesion was located in the middle and lower femur, with local bone destruction, high signal and soft tissue mass protruding from the surrounding structure (Fig. 1A, B). Patients in cases 1 to 3 received Ewing's sarcoma chemotherapy regimen after surgical resection of the tumor, with case 2 also receiving radiation therapy. Case 4 underwent complete surgical resection only and did not receive further chemotherapy or radiation therapy. The follow-up period ranged from 26 to 180 months. The follow-up results showed that case 1 developed lung and chest wall metastasis 48 months after the initial diagnosis. Case 1 and case 2 occurred local recurrence at 23 months and 72 months after initial onset respectively, while the other 2 cases (case 3 and case 4) did not experience recurrence or metastasis. All 4 patients are currently alive. In addition to the 4 cases reported in this study, a total of 61 relevant studies and reports were reviewed, and the clinical characteristics are summarized in Table 1.
Histology and immunohistochemistry
Histologically, the tumor consisted of small to medium-sized round or oval cells arranged in nests, cords, trabeculae, or pseudoglandular patterns. The stroma contained abundant mucin and hyalinization fibers, resembling myoepithelioma of soft tissue (Fig. 2A, B). The cytoplasm was either eosinophilic or pale (Fig. 2C, D). Across all 4 cases, the mitotic activity per mm2 ranged from 4 to 20 (mean 10). Immunohistochemical results are shown in Table 2. CD99 showed varying degrees of membranous staining in all tumors (Fig. 3A). Our cases showed diffuse strong nuclear expression of NKX2.2 and NKX3.1 (Fig. 3B, C). Three cases showed focal expression of CK and EMA (Fig. 3D, E).
A, B Microscopic examination reveals the tumor with abundant fibrous or mucinous stroma (10x); hematoxylin and eosin (H/E) stain. C, D Tumor cells are arranged in small beams, cords, or clusters. At high magnification, tumor cells appear as monomorphic small round cells with eosinophilic and pale-staining cytoplasm, and inconspicuous or small nucleoli (20x); hematoxylin and eosin (H/E) stain
A, B, C Immunohistochemical staining shows diffuse expression of CD99 and NKX2.2 in all tumor cells, along with diffuse nuclear positivity for NKX3.1 (20x). D, E Two cases show focal expression of CK and EMA (20x). F FISH detection using EWSR1 probe shows red and green signals simultaneously, along with amplification of the 5' of the EWSR1 locus (red signal). G FISH detection using EWSR1::NFATC2 fusion probe shows NFATC2 as the ligand gene for EWSR1, accompanied by amplification of fusion signals
Molecular genetics
In all four cases, FISH analysis using EWSR1 break apart probe showed an unusual pattern for break-apart probes with one to two fused signals, one to two green signals and several grouped and amplified red signals, indicating the rearrangement of the EWSR1 gene with the amplification of 5’ of the EWSR1 locus (Fig. 3F). The percentages of positive cells in all 4 cases were from 52 to 90% by FISH detection. The EWSR1::NFATC2 fusion probe showed multi yellow fusion signals, indicating EWSR1::NFATC2 fusion gene (Fig. 3G). Next-generation sequencing identified the breakpoints of EWSR1::NFATC2 fusion gene (Fig. 4).
Discussion
The Ewing sarcoma family of tumors is a group of malignant mesenchymal neoplasms characterized by characteristic EWSR1 gene rearrangements. They can occur in bone and soft tissue, and the pathological morphology is predominantly composed of small round cells. The fusion partners include FLI1, ERG, SP3, and others. In recent years, with the development of next-generation sequencing technology, some tumors associated with EWSR1 gene rearrangements have shown unique histological and molecular genetic characteristics, such as EWSR1/FUS::NFATC2 sarcoma. However, there is still controversy about whether to categorize it separately. EWSR1::NFATC2 sarcoma is a rare type of sarcoma, first reported by Szuhai et al. [6] in 2009, and there have been 61 reported cases to date. This tumor exhibits specific clinical and histological features. In terms of clinical characteristics, EWSR1/FUS::NFATC2 sarcoma mainly occurs in young males, with a male-to-female ratio of approximately 3:1. Long bones are the most common sites, with 26 cases occurring in the femur, 7 cases in the humerus, 6 cases in the tibia, and 3 cases in the radius. Patients often present with symptoms such as pain, swelling, and impaired mobility. The tumor has a rapid growth rate and can easily invade surrounding tissues and lymph nodes [25]. Therefore, when young individuals present with the aforementioned symptoms, the possibility of EWSR1/FUS::NFATC2 sarcoma should be considered.
In terms of histology, EWSR1/FUS::NFATC2 sarcoma typically presents as an undifferentiated small round cell sarcoma. The tumor cells are usually round or oval in shape, with finely granular chromatin, prominent nucleoli, and scant cytoplasm. The tumor cells are densely arranged, forming clusters or cords. Immunohistochemical staining shows that EWSR1/FUS::NFATC2 sarcoma often expresses markers such as CD99, NKX3.1 [9]. Markers such as S-100 and Desmin are not expressed [8,9,10]. NKX2.2, which is a relatively specific marker for diagnosing Ewing sarcoma, can also be expressed in EWSR1/FUS::NFATC2 sarcoma [11]. Therefore, histological and immunohistochemical examinations can provide important references for the diagnosis of EWSR1/FUS::NFATC2 sarcoma.
FISH and NGS play an important role in the diagnosis of EWSR1/FUS::NFATC2 sarcoma. Unlike classical Ewing sarcoma, this tumor exhibits a characteristic FISH pattern with not only the separation signal of EWSR1 but also a clear amplification at the 5' of the EWSR1 locus. This suggests the possibility of EWSR1/FUS::NFATC2 sarcoma and can be validated using EWSR1::NFATC2 or FUS::NFATC2 fusion probe and NGS. Although EWSR1/FUS::NFATC2 sarcoma has its unique FISH presentation, due to its rarity and non-exclusive molecular genetic features, the diagnosis of this tumor must be combined with histology and immunohistochemistry. For certain situations, such as EWSR1::POU5F1 sarcoma and rare myoepithelial sarcoma, validation using EWSR1/FUS::NFATC2 fusion probe is necessary. Additionally, the EWSR1/FUS::NFATC2 gene rearrangement feature can also be detected in simple bone cysts [26], but it does not harbour the amplification of the fusion gene and exhibits different clinical manifestations, histology, and prognosis compared to EWSR1/FUS::NFATC2 sarcoma. Therefore, although EWSR1/FUS::NFATC2 sarcoma has a relatively specific FISH presentation, when diagnosing tumors with EWSR1/FUS::NFATC2 gene rearrangement, it is necessary to consider factors such as the clinical presentation, histological changes, treatment and prognosis. In our review of the four cases reported in this study and previously published cases, almost all reported cases of EWSR1::NFATC2 sarcoma exhibited the FISH pattern with separation of red and green signals accompanied by 5' amplification of the EWSR1 locus. However, other tumors such as EWSR1::POU5F1 sarcoma and rare myoepithelial sarcoma may also show separation with amplification signals in FISH testing, but they are accompanied by amplification at the 3' end or other locations [27]. Therefore, differentiation between these tumors can be achieved by combining clinical manifestations, histology, and various molecular testing methods for validation.
The most important differential diagnosis for EWSR1/FUS::NFATC2 sarcoma is classical Ewing's sarcoma. Morphologically, these two tumors exhibit similar pathological features and are both small round cell tumors. However, EWSR1/FUS::NFATC2 sarcoma cells form small fascicular or cord-like structures, with an interstitium that is rich in mucin or undergoes collagen degeneration. Additionally, this tumor also needs to be distinguished from the less common sclerosing epithelioid fibrosarcoma. In our case selection, we encountered a challenging case: FISH analysis detected EWSR1 gene rearrangement, showing separation of red and green signals with 5' amplification, thus being included in this study. However, when verification was performed using the EWSR1::NFATC2 fusion probe, the result was negative. Subsequent NGS confirmed the presence of EWSR1::CREB3L1 rearrangement, and on top of this result, we performed additional immunohistochemical staining for MUC4, which showed positive expression. Considering the combined results of pathological morphology, immunohistochemistry, and molecular changes, the final diagnosis for this case was sclerosing epithelioid fibrosarcoma. Therefore, when FISH analysis shows separation of red and green signals with 5' amplification in EWSR1 gene rearrangement, the possibility of sclerosing epithelioid fibrosarcoma should also be considered, and differential diagnosis can be made based on indicators such as positive expression of MUC4 and positive rearrangement of EWSR1::CREB3L1 fusion probe in FISH analysis [28]. Due to the focal expression of epithelial markers such as CK and EMA in some cases of EWSR1/FUS::NFATC2 sarcoma, combined with morphological features, it can be easily misdiagnosed as an myoepithelioma [29]. Furthermore, approximately 50% of myoepitheliomas have translocations involving the EWSR1 gene, such as EWSR1::POU5F1 sarcoma, and myoepithelioma can also exhibit positive expression of CD99 and NKX3.1, posing a significant diagnostic challenge. Immunohistochemical staining for NKX2.2 is helpful in distinguishing between the two, as myoepithelioma shows negative or focal weak positive expression of NKX2.2 [11]. In addition, extraskeletal myxoid chondrosarcoma (EMC) needs to be differentiated from EWSR1/FUS::NFATC2 sarcoma. EMC typically occurs in soft tissues outside the bone, but there have also been reports of cases occurring in bone [30, 31]. The clinical presentation, histology, and immunohistochemical marker expression of bone EMC are similar to those occurring in deep soft tissues. Morphologically, EMC exhibits a round or slightly spindle-shaped arrangement of tumor cells forming cord-like structures, distributed in myxoid stroma, with large nuclei and eosinophilic cytoplasm. Its molecular hallmark is rearrangement of the NR4A3 gene, with the most common fusion partner being EWSR1. Based on its molecular features, it can be distinguished from EWSR1/FUS::NFATC2 sarcoma. In summary, the differential diagnosis of EWSR1/FUS::NFATC2 sarcoma is complex with many pitfalls. Diagnosis cannot rely solely on FISH analysis. Even when FISH analysis shows positive separation of EWSR1 gene, amplification, further verification with specific fusion probes is still necessary. If conditions permit, additional NGS testing should be conducted, and a comprehensive diagnosis should be made based on the combination of morphological features, immunohistochemistry, and molecular analysis.
In terms of prognosis, EWSR1/FUS::NFATC2 sarcoma typically requires treatment such as surgical resection, radiation therapy, and chemotherapy. Some studies have shown that the prognosis of EWSR1/FUS::NFATC2 sarcoma is related to factors such as age, tumor size, lymph node metastasis, and surgical resection. However, due to the rarity and heterogeneity of this tumor, assessing prognosis remains challenging. In our four cases, three underwent postoperative chemotherapy while one did not receive further treatment after surgery. The follow-up period ranged from 26 to 180 months, and two cases experienced local recurrence and lung metastasis, while the other two did not show recurrence or metastasis. Among the 30 cases with follow-up results, including the four cases reported in our study, the mortality rate of EWSR1/FUS::NFATC2 sarcoma is not high, even though it can exhibit local recurrence and lung metastasis. Whether adjuvant chemotherapy can achieve good clinical results is still controversial, and more clinical cases need to be further studied. The latest research findings and novel treatment approaches for EWSR1/FUS::NFATC2 sarcoma involve molecular pathology research, immunotherapy, targeted therapy, and gene-editing techniques. These research results and novel treatment approaches provide new insights and directions for the treatment and management of EWSR1/FUS::NFATC2 sarcoma. The fusion breakpoints and fusion types in EWSR1/FUS::NFATC2 sarcoma may impact the tumor's biological behavior and prognosis. FUS::NFATC2 sarcoma has a worse prognosis than EWSR1::NFATC2 sarcoma [8]. In recent years, immunotherapy has made significant progress in the field of cancer treatment, including some studies on soft tissue sarcoma, such as round cell sarcoma. Studies by Seligson et al. have shown that EWSR1::NFATC2 sarcoma has a lower response to Ewing sarcoma-specific chemotherapy and suggest that drugs targeting the mTOR pathway may become targeted therapies for treating this disease [32].
Conclusion
In terms of clinical, radiological, histological, and FISH detection findings, EWSR1::NFATC2 fusion sarcoma exhibits distinct histopathological and molecular genetic features. Its clinical prognosis is different from classical Ewing sarcoma.
Availability of data and materials
No datasets were generated or analysed during the current study.
Abbreviations
- FISH:
-
Fluorescence in situ hybridization
- NGS:
-
Next-generation sequencing
- EMC:
-
Extraskeletal myxoid chondrosarcoma
- FFPE:
-
Formalin-fixed, paraffin-embedded
References
Machado I, Yoshida A, Morales MGN, Abrahao-Machado LF, Navarro S, Cruz J, et al. Review with novel markers facilitates precise categorization of 41 cases of diagnostically challenging, "undifferentiated small round cell tumors". A clinicopathologic, immunophenotypic and molecular analysis. Ann Diagn Pathol. 2018;34:1–12. https://doi.org/10.1016/j.anndiagpath.2017.11.011.
Cantile M, Marra L, Franco R, Ascierto P, Liguori G, Chiara AD, Botti G. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med Oncol. 2013;30:412. https://doi.org/10.1007/s12032-012-0412-8.
Classification WHO, of Tumours Editorial Board, editors. World Health Organization classification of soft tissue and bone tumours. 5th ed. Lyon: IARC; 2020.
Mastrangelo T, Modena P, Tornielli S, Bullrich F, Testi MA, Mezzelani A, et al. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene. 2000;19:3799–804. https://doi.org/10.1038/sj.onc.1203762.
Wang L, Bhargava R, Zheng T, Wexler L, Collins MH, Roulston D, et al. Undifferentiated small round cell sarcomas with rare EWS gene fusions: identification of a novel EWS-SP3 fusion and of additional cases with the EWS-ETV1 and EWS-FEV fusions. J Mol Diagn. 2007;9:498–509. https://doi.org/10.2353/jmoldx.2007.070053.
Szuhai K, Ijszenga M, de Jong D, Karseladze A, Tanke HJ, Hogendoorn PCW. The NFATc2 gene is involved in a novel cloned translocation in a Ewing sarcoma variant that couples its function in immunology to oncology. Clin Cancer Res Off J Am Assoc Cancer Res. 2009;15:2259–68. https://doi.org/10.1158/1078-0432.CCR-08-2184.
Makise N, Yoshida KI, Lijima T, Yoshida A, Ushiku T, Ishida T. Skeletal EWSR1-NFATC2 sarcoma previously diagnosed as Ewing-like adamantinoma: A case report and literature review emphasizing its unique radiological features. Pathol int. 2021;71:614–20. https://doi.org/10.1111/pin.13135.
Perret R, Escuriol J, Velasco V, Mayeur L, Soubeyran I, Delfour C, et al. NFATc2-rearranged sarcomas: clinicopathologic, molecular, and cytogenetic study of 7 cases with evidence of AGGRECAN as a novel diagnostic marker. Mod Pathol. 2020;33:1930–44. https://doi.org/10.1038/s41379-020-0542-z.
Yoshida KI, Machado I, Parafioriti A, Lacambra M, Ichikawa H, et al. NKX3–1 is a useful immunohistochemical marker of EWSR1-NFATC2 sarcoma and mesenchymal chondrosarcoma. Am J Surg Pathol. 2020;44:719–28. https://doi.org/10.1097/PAS.0000000000001441.
Diaz-Perez JA, Nielsen GP, Antonescu C, Tayler MS, Lozano-Caldenron SA, Rosenberg A. EWSR1/FUS-NFATc2 rearranged round cell sarcoma: clinicopathological series of 4 cases and literature review. Hum Pathol. 2019;90:45–53. https://doi.org/10.1016/j.humpath.2019.05.001.
Wang GY, Thomas DG, Davis JL, Ng T, Patel RM, Harms PW, et al. EWSR1::NFATC2 translocation-associated sarcoma clinicopathologic findings in a rare aggressive primary bone or soft tissue tumor. Am J Surg Pathol. 2019;43:1112–22. https://doi.org/10.1097/PAS.0000000000001260.
Koelsche C, Kriegsmann M, Kommoss FKF, Stichel D, Kriegsmann K, Vokuhl C, et al. DNA methylation profiling distinguishes Ewing like sarcoma with EWSR1-NFATC2 fusion from Ewing sarcoma. J Cancer Res Clin Oncol. 2019;145:1273–81. https://doi.org/10.1007/s00432-019-02895-2.
Mantilla JG, Ricciotti RW, Chen E, Hoch BL, Liu Yj. Detecting disease-defining gene fusions in unclassified round cell sarcomas using anchored multiplex PCR/targeted RNA next‐generation sequencing—Molecular and clinicopathological characterization of 16 cases. Genes Chromosom Cancer. 2019;58:713–22. https://doi.org/10.1002/gcc.22763.
Bode-Lesniewska B, Fritz C, Exner GU, Wagner U, Fuchs B. EWSR1-NFATC2 and FUS-NFATC2 gene fusion-associated mesenchymal tumors: clinicopathologic correlation and literature review. Sarcoma. 2019;9386390. https://doi.org/10.1155/2019/9386390.
Yau DTW, Chan JKC, Bao S, Zheng Z, Lau GTC, Chan ACL. Bone sarcoma with EWSR1-NFATC2 fusion: sarcoma with varied morphology and amplification of fusion gene distinct from Ewing sarcoma. Int J Surg Pathol. 2019;27:561–7. https://doi.org/10.1177/1066896919827093.
Watson S, Perrin V, Guillemot D, Reynaud S, Coindre JM, Karanian M, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245:29–40. https://doi.org/10.1002/path.5053.
Toki S, Wakai S, Sekimizu M, Mori T, Ichikawa H, Kawai A, et al. PAX7 immunohistochemical evaluation of Ewing sarcoma and other small round cell tumours. Histopathology. 2018;73:645–52. https://doi.org/10.1111/his.13689.
Antonescu C. Round cell sarcomas beyond Ewing: emerging entities. Histopathology. 2014;64:26–37. https://doi.org/10.1111/his.12281.
Machado I, Yoshida A, Morales MGN, Abrahao-Machado LF, Navarro S, Cruz J, et al. Review with novel markers facilitates precise categorization of 41 cases of diagnostically challenging, ‘undifferentiatedsmall round cell tumors’. A clinicopathologic, immunophenotypic and molecular analysis. Ann Diagn Pathol. 2018;34:1–12.https://doi.org/10.1016/j.anndiagpath.2017.11.011.
Cohen JN, Sabnis AJ, Krings G, Cho SJ, Horvai AE, Davis JL. EWSR1-NFATC2 gene fusion in a soft tissue tumor withepithelioid round cell morphology and abundant stroma: a case report and review of the literature. Hum Pathol. 2018;81:281–90. https://doi.org/10.1016/j.humpath.2018.03.020.
Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475. https://doi.org/10.1371/journal.pgen.1004475.
Kinkor Z, Vaneček T, Svajdler M Jr, Mukensnabl P, Vesely K, Baxa J, et al. Where does Ewing sarcoma end and begin - two cases of unusual bone tumors with t(20;22)(EWSR1-NFATC2) alteration. Cesk Patol. 2014;50:87–91.
Sadri N, Barroeta J, Pack SD, Abdullaev Z, Chatterjee B, Puthiyaveettil R, et al. Malignant round cell tumor of bone with EWSR1-NFATC2 gene fusion. Virchows Arch. 2014;465:233–9. https://doi.org/10.1007/s00428-014-1613-7.
Romeo S, Bovée JV, Kroon HM, Tirabosco R, Natali C, Anatta L, et al. Malignant fibrous histiocytoma and fibrosarcoma of bone: a re-assessment in the light of currently employed morphological, immunohistochemical and molecular approaches. Virchows Arch. 2012;461:561–70. https://doi.org.https://doi.org/10.1007/s00428-012-1306-z.
Brcic I, Scheipl S, Bergovec M, Leithner A, Szkandera J, Sotlar K, et al. Implementation of Copy Number Variations-Based Diagnostics in Morphologically challenging EWSR1/FUS::NFATC2 Neoplasms of the Bone and Soft Tissue. Int J Mol Sci. 2022;23:16196. https://doi.org/10.3390/ijms232416196.
Pižem J, Šekoranja D, Zupan A, Boštjančič E, Matjasic A, Mavcic B, et al. FUS-NFATC2 or EWSR1-NFATC2 Fusions Are Present in a Large Proportion of Simple Bone Cysts. Am J Surg Pathol. 2020;12:1623–34. https://doi.org/10.1097/PAS.0000000000001584.
Antonescu CA, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, et al. EWSR1-POU5F1 Fusion in Soft Tissue Myoepithelial Tumors. A Molecular Analysis of Sixty-Six Cases, Including Soft Tissue, Bone, and Visceral Lesions, Showing Common Involvement of the EWSR1 gene. Gene Chromosomes Cancer. 2010;49:1114–1124. https://doi.org/10.1002/gcc.20819.
Tsuda Y, Dickson BC, Dry SM, Federman N, Suurmeijer AJH, Swanson D, et al. Clinical and molecular characterization of primary sclerosing epithelioid fibrosarcoma of bone and review of the literature. Genes Chromosomes Cancer. 2019;59:217–24. https://doi.org/10.1002/gcc.22822.
Song W, Flucke U, Suurmeijer AJH. Myoepithelial tumors of bone. Surg Pathol Clin. 2017;10:657–74. https://doi.org/10.1016/j.path.2017.04.010.
Finos L, Righi A, Frisoni T, Gambarotti M, Ghinelli C, Benini S, et al. Primary extraskeletal myxoid chondrosarcoma of bone: report of three cases and review of the literature. Pathol Res Pract. 2017;213:461–6. https://doi.org/10.1016/j.prp.2017.02.008.
Oliveira AM, Sebo TJ, McGrory JE, Gaffey TA, Rock MG, Nascimento AG. Extraskeletal myxoid chondrosarcoma: a clinicopathologic, immunohistochemical, and ploidy analysis of 23 cases. Mod Pathol. 2000;13:900–8. https://doi.org/10.1038/modpathol.3880161.
Seligson ND, Maradiaga RD, Stets CM, Katzenstein HM, Millis SZ, Rogers A, et al. Multiscale-omic assessment of EWSR1-NFATC2 fusion positive sarcomas identifies the mTOR pathway as a potential therapeutic target. Precision Oncol. 2021;5:43. https://doi.org/10.1038/s41698-021-00177-0.
Author information
Authors and Affiliations
Contributions
LLL, HTY, and DHS designed the research. LLL analysed FISH data, carried out the literature search, generated figures and drafted the manuscript. FZK performed FISH and IHC analyses. WFM performed FISH analysis. LLL, LL, and YD collected and interpreted pathological and clinical data. HTY interpreted FISH data and provided critical review of the data and manuscript. DHS and HTY supervised the study. All authors reviewed the paper and had final approval of the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and conssent to participate
We herein affirm that the manuscript fully complies with ethical standards.
Consent for publication
All patients provided consent for information to be published.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, L., Li, L., Ding, Y. et al. Report and literature review of four cases of EWSR1::NFATC2 round cell sarcoma. Diagn Pathol 19, 19 (2024). https://doi.org/10.1186/s13000-024-01443-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13000-024-01443-y