Abstract
Background
The World Health Organization (WHO) classification system defines recurrent chromosomal translocations as the sole diagnostic and prognostic criteria for acute leukemia (AL). These fusion transcripts are pivotal in the pathogenesis of AL. Clinical laboratories universally employ conventional karyotype/FISH to detect these chromosomal translocations, which is complex, labour intensive and lacks multiplexing capacity. Hence, it is imperative to explore and evaluate some newer automated, cost-efficient multiplexed technologies to accommodate the expanding genetic landscape in AL.
Methods
“nCounter® Leukemia fusion gene expression assay” by NanoString was employed to detect various fusion transcripts in a large set samples (n = 94) utilizing RNA from formalin fixed paraffin embedded (FFPE) diagnostic bone marrow biopsy specimens. This series included AL patients with various recurrent translocations (n = 49), normal karyotype (n = 19), or complex karyotype (n = 21), as well as normal bone marrow samples (n = 5). Fusion gene expression data were compared with results obtained by conventional karyotype and FISH technology to determine sensitivity/specificity, as well as positive /negative predictive values.
Results
Junction probes for PML/RARA; RUNX1-RUNX1T1; BCR/ABL1 showed 100 % sensitivity/specificity. A high degree of correlation was noted for MLL/AF4 (85 sensitivity/100 specificity) and TCF3-PBX1 (75 % sensitivity/100 % specificity) probes. CBFB-MYH11 fusion probes showed moderate sensitivity (57 %) but high specificity (100 %). ETV6/RUNX1 displayed discordance between fusion transcript assay and FISH results as well as rare non-specific binding in AL samples with normal or complex cytogenetics.
Conclusions
Our study presents preliminary data with high correlation between fusion transcript detection by a throughput automated multiplexed platform, compared to conventional karyotype/FISH technique for detection of chromosomal translocations in AL patients. Our preliminary observations, mandates further vast validation studies to explore automated molecular platforms in diagnostic pathology.
Similar content being viewed by others
Background
In acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL), chromosomal abnormalities play a critical role in pathogenesis and prognosis [1]. Molecular studies of these genomic anomalies have identified specific genes implicated in the process of leukemogenesis [2]. Specific recurrent chromosomal translocations and their fusion transcripts define the current WHO classification system for acute leukemia (AL) [3]. These genetic aberrations in combination with morphology, immunophenotype and clinical features are used to diagnose distinct types of AL and thereby dictate therapeutic approaches [4]. Clinical laboratories, worldwide, utilize standard FISH technology with or without conventional karyotype to investigate these chromosomal translocations [5]. Unfortunately, FISH is a labour intensive and expensive procedure with the limited potential for “multiplexing”. These limitations impact diagnosis and clinical management. In a small number of AL sub-types, fusion transcripts can be amplified through reverse transcription, polymerase chain reaction (RT-PCR) methodology [6]. However, clinical application of RT-PCR is narrow due to variation in breakpoints resulting in several splice regions [7]. Given these limitations and the expanding new discoveries of additional recurrent chromosomal translocations, there is a need to explore newer automated technologies to “accommodate” new fusion transcript discoveries and make this significant clinical diagnostic testing cost-effective and time efficient.
Digital RNA quantification by NanoString Technologies is an automated, multiplexed platform, which can accurately measure up to 800 specific targets. “Leukemia Fusion Gene Expression Assay” by NanoString has a unique design of junction probes, which target the unique sequence spanning the fusion junction of the two exons. It has been claimed that a high degree of sensitivity and specificity has been achieved through sequence modification. However, no clinical validation data are available to determine the utility of this automated technology in routine clinical practice. The objective of this study was to evaluate the potential application and clinical utility of “Leukemia Fusion Gene Expression Assay” in the diagnostic sub-classification of acute leukemia patients. To this effect, we investigated fusion transcripts in a large cohort of AML and ALL patients utilizing genomic RNA from diagnostic bone marrow biopsies. We then correlated these results with data obtained by routine FISH and conventional karyotype. Our results provide unique preliminary data comparing a traditional platform with a novel automated technology, which provides potential promise for future adoption in the diagnosis and management of acute leukemia patients.
Methods
University of Calgary ethics committee approved this study (REB-13-0380-MODI; dated August 21, 2013). The diagnosis was confirmed according to the WHO 2008 classification [3]. We employed adequate representation of samples with recurrent chromosomal translocation (true positive) in AML (n = 23) and ALL (n = 26) while AML with normal cytogenetics (n = 19), or complex cytogenetics (n = 21) and normal bone marrow (n = 5) samples were used as negative controls (true negative).
Karyotype and fluorescence in-situ hybridization
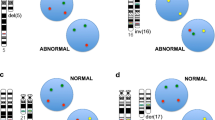
Complete karyotype analysis at diagnosis was performed on all leukemic patients and followed a standardized laboratory protocol. Banded chromosomes (20 metaphases) were examined for each patient sample utilizing the G-banding technique. All patients (100 %) with diagnosis of ALL (n = 26) were subjected to additional FISH studies utilizing a panel of four probes (TCF3-PBX1; MLL-AF4; BCR-ABL and ETV6-RUNX1). Among AML patients, FISH studies were performed only as second tier test, as required by findings on karyotype analysis (8/23; 35 %). Dual color dual fusion or dual color break part FISH probes (Abbott, Markham ON) were employed to confirm translocations as dictated by clinical requirements or the karyogram. The results of these diagnostic tests performed on aspirate material were retrieved from patient records for comparison with digital quantification of fusion transcripts by NanoString technology (see below).
Gene expression analysis
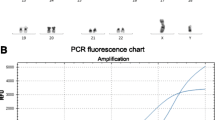
We employed nCounter® Leukemia Fusion Gene Expression Assay utilizing the NanoString platform (NanoString Technologies, Seattle, WA) to evaluate the expression of fusion transcript (Additional file 1: Table S1). RNA was isolated using the High Pure RNA Paraffin Kit (Roche, Basel, Switzerland) utilizing duplicate cores (2-mm), harvested off areas with maximum tumor cell population in the FFPE tissue block of diagnostic bone marrow specimen. All bone marrow biopsies were subjected to a standardized, short term (1 h 45 min) formic acid decalcification protocol. The RNA concentration was quantified using UV spectroscopy (Nanodrop Technologies, Wilmington, DE) and integrity assessed using a Bio-analyzer 2100 and RNA Nano Chip assay (Agilent Technologies, Wilmington, DE). The necessary probes and reagents were purchased from NanoString, and the nCounter analysis was performed according to the manufacturer’s instructions. Probes were hybridized to 800 ng of total RNA for 20 h at 65 °C. Hybridized reactions were purified using nCounter Prep Station and data collection was performed on nCounter Digital Analyzer. Raw counts were normalized using nSolver Analysis Software v3.0. Background subtraction was performed for each sample by subtracting the mean of 8 negative controls from all data points. Raw counts were further normalized to the six positive controls, included in each CodeSet, and to the two housekeeping genes TBP and GUSB.
Sensitivity, specificity, positive and negative predictive values
The sensitivity, specificity, positive and negative predictive values were determined for the seven fusion transcripts by comparing signal-to-noise ratio (SNR). The median binding value in true negative samples was considered as background or “noise”. Minimum of ten-fold or more difference between SNR was considered as the positive result and was used for further calculations.
Statistical analysis
We used nSolver software v3.0 (NanoString Technologies) for the normalization of raw counts. Heat map and principle component analyses were performed on Qlucore Omics Explorer v3.2 (Lund, Sweden). Statistical analysis was performed using SPSS software v21.0 (IBM, Armonk, NY). All computed results having two-sided P value < 0.05 were considered significant.
Results
A total of 94 (63 AML, 26 ALL and five control bone marrow) samples were analysed. AML patients included 42 men and 21 women (M: F 2:1) with a median age of 63 years (range 5–87 years). Twenty-six ALL patients comprised of 17 men and 09 women (M: F 1.8:1) with a median age of 12 years (range 2–63 years) in this cohort. Table 1 outlines the sensitivity, specificities, and positive and negative predictive values for each transcript. Figure 1 outlines, principal component analysis (PCA) plot based on digital quantification of various fusion transcripts and their correlation with chromosomal/FISH results. In AML category, digital quantification for PML/RARA and RUNX1-RUNX1T1 correlated 100 % with t(15;17) and t(8;21) respectively; while non-specific positivity was not seen in any other samples across entire cohort. CBFB-MYH11 fusion probes showed “false positivity” in AML samples with complex and normal karyotype. In 4/8 (50 %) of AML samples with inv(16) or t(16;16); false negative reaction was noted. In ALL group, BCR-ABL1 fusion probes performed seamlessly with 100 % sensitivity and specificity. TCF3-PBX1 and MLL-AF4 fusion probes showed 100 % specificity but sensitivity ranged up to 86 %. Probes for ETV6/RUNX1 fusion transcript performed poorly and showed only 50 % sensitivity but 98 % specificity.
Discussion
In WHO classification for AL, chromosomal translocations are central to diagnosis, sub classification and prognosis. Conventional cytogenetics, followed by FISH in specific cases, is the gold standard to detect these chromosomal aberrations, but poses the challenge regarding subjectivity, manual labour, cost and turnaround time. There is a need to explore automated, multiplexed, throughput and cost-efficient technologies to serve rapidly expanding discoveries of novel fusion transcripts in haematological malignancies. A large cohort of AL samples were analysed by multiplexed, automated digital quantification in parallel with conventional karyotype/FISH technology. We observed a high degree of correlation (>90 % sensitivity and specificity) for different translocations that are frequent in clinical practice. The complete correlation was noted for t(8;21) between conventional karyotype/FISH and digital fusion RNA quantification results. This high degree of correlation can be related to the generation of only two fusion transcripts by this translocation [8]. t(8;21) can also involve several chromosomes, and these variant rearrangements can be cryptic and easily overlooked by conventional G-banding technique [9]. However, it was interesting to note that none of these cryptic fusion transcripts were detected in a large cohort of AL patients with normal and complex cytogenetics. Similarly, almost complete correlation was also noted for t(15;17); t(9;22) and t(4;11) translocations. Hence, we established the potential for clinical utility of this automated throughput platform of digital quantification as a possible alternate to current and widely used FISH technique.
Multiplexed molecular techniques present opportunities in procedural efficiencies, but also pose challenges regarding cross-reactivity and false positivity [10, 11]. Our extensive series of AL sample sets with normal and complex cytogenetic provided an adequate safety net to detect any cross-reactivity of fusion probes. We noted minimal to no false positive reactions in the majority of fusion probes. These results are encouraging in our small patient sample; however, a more robust validation is required using a larger cohort. In AL patients with t(12;21) (p13;q22), we noted discordance between FISH results and ETV6-RUNX1 fusion probe. The breakpoints in t(12;21) (p13;q22) are variable within the several thousand base pairs of ETV6 as well as RUNX1 introns. In spite of micro-clustering of breakpoints in ETV6 and RUNX1 introns, it is still difficult to identify or to sequence the breakpoints by even streamlined long‐distance PCR in t(12;21) [12]. Hence, it appears that fusion transcript ETV6-RUNX1 is beyond the scope of this assay at this stage. We also noted that inv(16) showed the most variable pattern and weak correlation with FISH results. This translocation results in more than ten differently sized CBFB-MYH11 fusion transcripts, which may explain variable sensitivity [13]. The probes against CBFB-MYH11 showed cross-reactivity with few samples harbouring normal or complex karyotype. Since there is marked heterogeneity of breakpoints in MYH11 gene, false negative rates could be high [14]. False positivity in inv 16 cases has also been reported by RT-PCR [15]. These observations mandate re-designing of some probes for this assay for enhanced sensitivity.
Conclusion
Our study demonstrates initial evaluation for fusion transcript detection by a throughput automated multiplexed platform for AL patients as an alternate solution to current complex, manual and labour intensive karyotyping /FISH techniques. Our preliminary study requires validations by future studies to evaluate these emerging technologies for their potential adoption in clinical laboratories. These expression assays offer significant capacity for expansion to accommodate additional future fusion transcripts discoveries of diagnostic or prognostic value in AL patients. It also has an immense ability to include gene expression signature as a diagnostic or prognostic tool for AL patients.
Abbreviations
- WHO:
-
World Health Organization
- AL:
-
Acute leukemia
- FISH:
-
Fluorescent in-situ hybridization
- FFPE:
-
Formalin fixed paraffin embedded
- AML:
-
Acute myeloid leukemia
- ALL:
-
Acute lymphoblastic leukemia
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- SNR:
-
Signal-to-noise ratio
- SPSS:
-
Statistical package for the social sciences
- PCA:
-
Principal component analysis
References
Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18(2):115–36.
Deschler B, Lubbert M. Acute myeloid leukemia: epidemiology and etiology. Cancer. 2006;107(9):2099–107.
Arber DA BR, Le Beau M. Acute myeloid leukemia with recurrent genetic abnormalities. In: Swerdlow SH, Campo E, et al., editors. World Health Organization classification of tumours pathology and genetics of tumours of haematopoietic and lymphoid tissues. 4th ed. 2 Lyon: IARC Press; 2008. p. 110–23.
Swerdlow SH, Campo EH NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Fourthth ed. 2008.
Hou HA, Lin CC, Chou WC, Liu CY, Chen CY, Tang JL, et al. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia. 2014;28(1):50–8.
Leroy H, de Botton S, Grardel-Duflos N, Darre S, Leleu X, Roumier C, et al. Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21). Leukemia. 2005;19(3):367–72.
Salto-Tellez M, Shelat SG, Benoit B, Rennert H, Carroll M, Leonard DG, et al. Multiplex RT-PCR for the detection of leukemia-associated translocations: validation and application to routine molecular diagnostic practice. J Mol Diagn. 2003;5(4):231–6.
Reikvam H, Hatfield KJ, Kittang AO, Hovland R, Bruserud O. Acute myeloid leukemia with the t(8;21) translocation: clinical consequences and biological implications. J Biomed Biotechnol. 2011;2011:104631.
Specchia G, Albano F, Anelli L, Zagaria A, Liso A, La Starza R, et al. Insertions generating the 5′RUNX1/3′CBFA2T1 gene in acute myeloid leukemia cases show variable breakpoints. Genes Chromosomes Cancer. 2004;41(1):86–91.
McCall CM, Mosier S, Thiess M, Debeljak M, Pallavajjala A, Beierl K, et al. False positives in multiplex PCR-based next-generation sequencing have unique signatures. J Mol Diagn. 2014;16(5):541–9.
Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 2000;13(4):559–70.
Wiemels JL, Alexander FE, Cazzaniga G, Biondi A, Mayer SP, Greaves M. Microclustering of TEL-AML1 translocation breakpoints in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2000;29(3):219–28.
Claxton DF, Liu P, Hsu HB, Marlton P, Hester J, Collins F, et al. Detection of fusion transcripts generated by the inversion 16 chromosome in acute myelogenous leukemia. Blood. 1994;83(7):1750–6.
Merchant SH, Haines S, Hall B, Hozier J, Viswanatha DS. Fluorescence in situ hybridization identifies cryptic t(16;16)(p13;q22) masked by del(16)(q22) in a case of AML-M4 Eo. J Mol Diagn. 2004;6(3):271–4.
Hackwell SM, Robinson DO, Harvey JF, Ross FM. Identification of false-positive CBFbeta/MYH11 RT-PCR results. Leukemia. 1999;13(10):1617–9.
Acknowledgements
None.
Funding
This work was supported by research grants from Alberta Cancer Foundation (grant # 25999) and Calgary Laboratory Services (grant # RS-16-600).
Availability of data and material
Raw data of this study is available in supplementary section.
Authors’ contributions
AA conducted research, reviewed patient clinical data, performed statistical analyses, and wrote the manuscript. MKM, GE, GS, RZA, SS and NM organized pathology/clinical data, provided samples and assisted in data analysis/graphics. DAS and MTSR assisted in clinical data collection, manuscript writing and editing. RKF performed the cytogenetic review and assisted in manuscript writing/editing. AM designed and conducted research, performed statistical analyses, and edited the final manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study has been approved by ethics committee of University of Calgary (REB-13-0380-MODI; dated August 21, 2013).
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
Raw data. (XLS 67 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Akhter, A., Mughal, M.K., Elyamany, G. et al. Multiplexed automated digital quantification of fusion transcripts: comparative study with fluorescent in-situ hybridization (FISH) technique in acute leukemia patients. Diagn Pathol 11, 89 (2016). https://doi.org/10.1186/s13000-016-0541-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13000-016-0541-z