Abstract
Background
In COVID-19 patients, undetected co-infections may have severe clinical implications associated with increased hospitalization, varied treatment approaches and mortality. Therefore, we investigated the implications of viral and bacterial co-infection in COVID-19 clinical outcomes.
Methods
Nasopharyngeal samples were obtained from 48 COVID-19 patients (29% ICU and 71% non-ICU) and screened for the presence of 24 respiratory pathogens using six multiplex PCR panels.
Results
We found evidence of co-infection in 34 COVID-19 patients (71%). Influenza A H1N1 (n = 17), Chlamydia pneumoniae (n = 13) and human adenovirus (n = 10) were the most commonly detected pathogens. Viral co-infection was associated with increased ICU admission (r = 0.1) and higher mortality (OR 1.78, CI = 0.38–8.28) compared to bacterial co-infections (OR 0.44, CI = 0.08–2.45). Two thirds of COVID-19 critically ill patients who died, had a co-infection; and Influenza A H1N1 was the only pathogen for which a direct relationship with mortality was seen (r = 0.2).
Conclusions
Our study highlights the importance of screening for co-infecting viruses in COVID-19 patients, that could be the leading cause of disease severity and death. Given the high prevalence of Influenza co-infection in our study, increased coverage of flu vaccination is encouraged to mitigate the transmission of influenza virus during the on-going COVID-19 pandemic and reduce the risk of severe outcome and mortality.
Similar content being viewed by others
Background
The newly emerged Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) continues to circulate outside of Wuhan, China since December 2019, and now exported to different countries all over the world [1]. At the time of writing this report, there were nearly a quarter of a million of Coronavirus Disease-19 (COVID-19) confirmed cases ranking Saudi Arabia as the 14th highest in the world [2]. Most hospitalized patients needed admission to intensive care unit (ICU) and mortality reaches up to 50% in some cases [3]. Until now, twenty-two studies have reported co-infection in COVID-19 and of these 16 have evidence of viral co-infection [4]. The prevalence of critical cases with viral co-infection has been reported up to 35% [5]. Early literature reported that 50% of the patients who died had coexisting bacterial infection [6]. This is higher than what was previously seen during the influenza pandemic in 2009 when 25% of patients with influenza infection had secondary bacterial co-infection [7].
SARS-CoV-2 is a single stranded RNA Betacoronavirus and belongs to a corona virus family called Coronaviridae [8]. Phylogenetic analysis has revealed that SARS-CoV-2 is closely related to SARS-CoV-1 and genetically distinct from Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [9]. SARS-CoV-2 utilizes Angiotensin-Converting Enzyme 2 (ACE-2) receptors in the lower airways which are also cellular receptors for other viruses in this group i.e. SARS-CoV and MERS-CoV [10]. Despite similar expression of ACE-2 receptors in different organs of the body, the most affected site is the lung tissue [11]. Influenza strains also cause lung damage by ACE-2 receptor mediated effects [12]. On the other hand, since the ACE-2 receptor used by SARS-CoV-2 is an interferon‐stimulated gene, it was hypothesized that type I and III interferons produced after bacterial infection may facilitate SARS‐CoV‐2 attachment [13].
Influenza A virus (IAV), a member of the Orthomyxoviridae, is a respiratory pathogen. The genome consists of eight segmented negative-sense RNAs. The viral ribonucleoproteins (vRNPs) consist of replicase complex (PA, PB1, and PB2) and various viral nucleoproteins (NPs) to form viral ribonucleoproteins (vRNPs). Newly assembled vRNPs may also contain matrix 1 (M1) and nuclear export protein (NEP). The lipid bilayers of influenza A virus particles contain three viral integral membrane proteins, hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2) [14]. The HA, NA, M1, and M2 proteins are important for influenza A virus assembly and budding [15]. Amino acid changes in the M1 protein of influenza viruses are known to increase virus budding to add lethal mutations in the M2 component in the cytoplasmic tail [16].
During pandemics, the detection of the novel virus may lead to underreporting of other pathogens that could be the etiological agent contributing to the disease severity. Indeed, during the influenza A (H1N1) pdm09 pandemic, 44.3% of patients had unreported respiratory viruses [17]. Earlier studies indicated that common viral co-infections reported in COVID-19 patients include Influenza viruses, RSV and adenovirus [5, 18]. Bacterial co-infections is more frequent than viral co-infections and it is homogeneously distributed in mild, moderate or severe illness [19]. The commonly known COVID-19 co-infecting bacteria are Mycoplasma pneumoniae, Pseudomonas aeruginosa, Heamophilus influenzae and Chlamydia pneumoniae [20]. These findings clearly emphasize the importance of screening for other clinically important co-circulating respiratory pathogens contributing to the etiology of the disease.
The novelty of SARS-CoV-2 and the complexity of profound etiology of co-infection urged for consideration of comorbidities. COVID-19 patients with an underlying condition such as hypertension, diabetes, chronic kidney disease, and heart failure have been associated with COVID-19 disease severity [21]. Cardiovascular disease has a strong association with COVID-19 pneumonia (14.4%) [7, 21] and other common comorbidities found in patients with SARS-CoV-2 include hypertension (18.6%) and diabetes (11.9%) [22]. Comorbidities were also linked with increased hospitalization, prolonged stay in ICU, and mortality. Hypertension was more prevalent in severe cases (47%) compared to diabetes (24%) and Respiratory diseases (10%) among other underlying conditions [21].
In conclusion, extensive evidence revealed that viral infections predispose patients to subsequent bacterial co‐infections [7]. This knowledge gap is puzzling as limited number of reports have described prevalence of bacterial and viral co-infections simultaneously. We hypothesized that undetected co-infections might have severe clinical implications associated with increased hospitalization, prolonged stay in ICU, and mortality. Therefore, our aim was, to investigate the presence of viral and bacterial co-infections in ICU and non-ICU COVID-19 patients.
Methods
Patients' demography
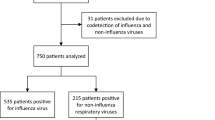
The study specimens were obtained retrospectively from a population (n = 48) admitted to King Fahad Hospital, Medina, Saudi Arabia. The number of critical cases needing admission to the ICU was 14, and 34 were mild cases. Nine patients died, (all were admitted to the ICU), and the rest survived. Thirteen patients were Saudi citizens and the rest were non Saudis (Table 1).
RNA extraction and SARS-CoV-2 PCR detection
The handling of respiratory samples, preparation of aliquots and viral RNA extraction were performed in accordance with relevant guidelines and regulations. SARS-CoV-2 RNA was extracted by MagNA Pure 96 instrument, using the MagNA Pure 96 DNA and Viral NA small volume kit, (Roche, Germany). Reverse Transcription Polymerase Chain Reaction (RT-PCR) was performed by Roche LightCycler® 480 II instrument, using the RNA Process Control Kit Trial Pack (Roche, Germany) with an internal, positive, and negative controls.
Real time PCR confirmation of SARS-CoV-2
RT-PCR assay targeting the E gene and RdRp (ORF1ab) of SARS-CoV-2 was carried out in 7500 Fast Real-Time PCR instrument (Thermo Fisher scientific, USA) with the Superscript III one-step RT-PCR kit (Invitrogen, Darmstadt, Germany) and performed as previously described [23, 24]. Thermal cycling conditions were 55 °C for 10 min, then 95 °C for 3 min, followed by 45 cycles of 95 °C for 15 s, and lastly 58 °C for 30 s.
The primers used for RdRp gene were as follows: RdRp_SARSr-F (GTGARATGGTCATGTGTGGCGG), RdRP_SARSr-P1: (FAM-CCAGGTGGWACRTCATCMGGTGATGC-BBQ) and RdRp_SARSr-R: (CARATGTTAAASACACTATTAGCATA). The primers used for E gene were as follows: E_Sarbeco_F (ACAGGTACGTTAATAGTTAATAGCGT), E_Sarbeco_P1 (FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ) and E_Sarbeco_R: (ATATTGCAGCAGTACGCACACA).
Real time PCR panel for co-infections
The quantitative Real-Time PCR assay for the co-infecting respiratory pathogens was performed on 7500 Fast Real-Time PCR instrument (Thermo Fisher scientific, USA). Extracted RNA was reinvestigated by RT/q-PCR with Fast Track Diagnostic (FTD) Respiratory pathogens 21 plus kit (Biomerieux, Luxemburg) according to the manufacturer’s instructions using 6 multiplex PCR amplifications for respiratory viruses and bacteria. The included viruses were Flu A (H1N1); Flu B; rhinovirus; seasonal Coronaviruses NL63, 229E, OC43 and HKU1; parainfluenza 1, 2, 3 and 4; metapneumoviruses A and B; bocavirus; RSV A and B; adenovirus; enterovirus and parechovirus. The screened bacteria were Chlamydia pneumoniae; Mycoplasma pneumoniae; Staphylococcus aureus; Streptococcus pneumoniae and Heamophilus influenzae B. This multiplex PCR assay also included six positive controls: five for each viral panel and one for bacterium. It also incorporated six negative controls provided in the kit. Briefly, 10 µl of the extracted RNA was added to 12.5 μl of buffer, 1 μl of enzyme, 2 pmol of each primer and probe (corresponding to each pathogen). Thermal cycling conditions include reverse transcription for 15 min at 42 °C, denaturation for 3 min at 94 °C followed by 40 cycles of primers' annealing for 8 s at 94 °C, and 34 s of template extension at 60 °C. Positivity or negativity of results were determined according to the manufacturer’s interpretation guide, and 12 samples were randomly chosen and repeated for re-confirmation purpose.
Data collection
Demographic and clinical data (Table 1) were obtained without any personally identifiable information, including the following clinical laboratory results: age, gender, history of chronic illness, Ct value, d-dimer, CK, CK-MB, Trop, HB, Platelet, RBC, WBC, Neutrophil, Lymph, CRP, Pro calciponin, Glucose, ESR, LDL, AST, ALT, Urea, Creatinine, LDH, Albumin, Total Protein and blood group. The clinical datasets used and analyzed during this study are available from the corresponding author on request.
Statistical analysis
Minitab version 19.0 software was used for statistical analysis. All data was expressed as continuous variables. Continuous data was expressed by median for normally distributed variable i.e. age; whereas absolute numbers were expressed as percentages. The paired t-test was used to compare continuous variables of normal distribution and non-normal distribution, respectively. We used Pearson Coefficient to show association between the variables. Relationship of one response and multiple predictors was examined using linear regression with best fitted model. Mortality was evaluated for bacterial and viral co-infections by using binary logistic expression and expressed as odds ratio. MANOVA (multivariate analysis of variance) was used to analyze mortality in the presence of co-infection and comorbidities and it was expressed as p-value. The patients were grouped by disease severity, comorbidities and co-infection or not. Factors were adjusted for age and gender. A 2-sided α of less than 0.05 was considered statistically significant.
Results
We investigated co-infection in 48 COVID-19 patients (including 37 males and 11 females), the male to female ratio was 3:1. Median age of our study population was 52 years (1–92). Fourteen patients (29%) needed admission to intensive care unit (ICU cases). The remaining 34 patients (71%) did not require ICU admission and were classified as non-ICU cases. We found co-infections in thirty-four (71%) patients. Although severity of disease was negatively correlated (r = -0.09) with the presence of a co-infection (p = 0.53), it had a positive correlation with the co-infecting viruses (r = 0.1, p = 0.42) by Pearson Coefficient as shown in Fig. 1. Furthermore, statistically significant inverse association was observed (r = -0.28, p = 0.04) between bacterial co-infection and ICU admission. In other words, this association indicates a lower likelihood of ICU admission with bacterial co-infection (Fig. 1). The most commonly found co-infecting virus was influenza A H1N1 in 17 patients (36%). Chlamydia pneumoniae was the most prevalent co-infecting bacteria found in 13 patients (28%). Other organisms detected were adenovirus in 10 patients and S. aureus in 4 patients. It was noticed that 4/17 (23.5%) patients with H1N1 had coexisting Chlamydia pneumoniae suggesting that 62% (21/34) may have triple co-infection.
Frequency of coexistence of pathogens in COVID-19 patients. The figure shows the frequency of viral vs bacterial co-infections in COVID-19 patients. The number of viruses detected in 14 ICU patients was 9 (6 H1N1 and 3 Adenovirus) compared to 2 bacteria (1 Chlamydia pneumoniae and 1 Staphylococcus aureus) which indicates a higher likelihood of ICU admission with viral co-infection. In 34 non-ICU patients, 36 coexisting pathogens were detected namely 15 bacteria (12 Chlamydia pneumoniae and 3 Staphylococcus aureus) and 21 viruses (11 H1N1, 7 Adenovirus, 1 metapneumovirus, 1 parainfluenza-3, and 1 influenza B) although none of them were involved in mortality
Binary logistic regression was used to analyze the mortality association with viral and bacterial co-infections. The mortality rate was 19% (9/48), all of them were critically ill COVID-19 patients admitted to the ICU, and two thirds of SARS-CoV-2 critically ill patients who died had a co-infection (6/9). The likelihood of an admission into the ICU and the probability of detecting a co-infection among different age groups revealed an increasing trend with aging (Fig. 2). We found that viral co-infections (OR = 1.78, CI = 0.38–8.28) had higher mortality compared to bacterial co-infections (OR = 0.44, CI = 0.08–2.45) in COVID-19 patients. We also observed that there was a positive correlation between co-infecting influenza H1N1 virus and mortality (r = 0.2). On the other hand, co-infection with Chlamydia pneumoniae (r = -0.17) did not have any correlation with mortality in SARS-CoV-2 infected patients (Table 2).
The binary fitted line plot shows the correlation between age and the probability of ICU admission and co-infection among COVID-19 patients. (A) The probability of an admission into the ICU among different age groups revealed an increasing trend with aging. (B) The probability of detection of a coinfection also exhibited a moderate linear correlation with age
In terms of comorbidities, the prevalence of diabetes was 54% (26/48), cardiovascular disease 4% (2/48) and chronic kidney disease (CKD) 10.4% (5/48). Other comorbidities including lobar pneumonia, cancer, acute kidney failure, and rheumatoid arthritis were found in 13 (27%) of patients despite the occurrence of multiple comorbidities in one patient. There was no significant association of co-infection with diabetes (p = 0.25), cardiovascular disease (p = 0.24) nor CKD (p = 0.7). However, when we used MANOVA test to look at associations of death and co-infection with diabetes, cardiovascular disease or CKD, it showed (Table 1) that statistically significant correlation was present only between diabetes and death (p = 0.02).
We also investigated the importance of different blood markers in COVID-19 patients (Table 1). Specifically, we examined association of d-dimer, lactate dehydrogenase (LDH) and Troponin T with the severity of disease. These markers have been interchangeably used to predict disease severity and the potential of ICU admission. Using linear regression, we found that Troponin T was strongly related (p = 0.001) with disease severity compared to LDH (p = 0.12) and d-dimer (p = 0.25). This finding may imply that Troponin T could be used as a predictor for disease severity.
Discussion
In this study, we investigated the presence of co-infections in COVID-19 cases and analyzed their clinical and epidemiological characteristics. We found evidence of co-infection in 34 COVID-19 patients (71%). Influenza A H1N1 was the most common detected among the co-infecting viruses found in 64% of patients. Viral and/or bacterial co-infections have been linked to disease severity, both directly, indirectly and through immunological response [25, 26]. Several studies have partially reported the prevalence of COVID-19 pneumonia and influenza co-infection [27,28,29]. However, data on clinical significance of influenza A H1N1 co-infections with COVID-19 is limited. The similarity of clinical manifestations between the circulating respiratory viruses such as influenza A H1N1 and SARS-CoV-2 makes the differentiation very difficult [30, 31]. Influenza A H1N1 dominance in our study population implies simultaneous spread of two viruses and clearly emphasizes on the importance of screening for other clinically important co-circulating respiratory pathogens. Besides, numerous studies have shown viral co-infections being associated with disease severity, acute respiratory distress syndrome (ARDS) and even death. These studies show higher intensive care admission rates in patients presenting with co-infections [5, 28, 32]. In this study, influenza A H1N1‐COVID‐19 co-infected cases were more severe and required ICU admission. Our results also showed a high case fatality rate among COVID-19 viral co-infections (r = 0.2) as two thirds of SARS-CoV-2 critically ill patients who died had co-infection (6/9). The severity and higher case fatality among COVID‐19 viral co-infected patients may be attributed to influenza A H1N1, which is known to induce a strong inflammatory cytokine/chemokine response (cytokine storm). Thus, the H1N1‐COVID‐19 co-infection could accelerate and play a major role in ARDS development. Our data showed that, during pandemics, focusing on the detection of the novel virus may lead to underreporting of other pathogens that could be the etiological agents contributing to disease severity.
The reporting of COVID‐19 and Influenza co-infection in the literature has been low despite high rates of single infection. Some studies have reported the difficulty in differentiating clinical characteristics between the co-infection of SARS‐CoV‐2 and Influenza [33, 34]. Regular SARS‐CoV‐2 respiratory screening is not sufficient to rule out the possibility of co-infection. The influence of the ongoing COVID‐19 pandemic should enhance utilization of a better diagnostic approach and testing of other respiratory pathogens.
Unfortunately, the topic of co-infection is usually embedded within the characteristics of patients in COVID-19 studies. However, our study investigated the coexistence of a full panel of respiratory viruses and bacteria simultaneously in order to investigate whether viral infection predisposes patients to subsequent bacterial co‐infection or not. Indeed, secondary bacterial co-infection is identified as the main cause of death in patients with viral pneumonia [7, 35]. A study of common respiratory pathogens presenting as co-infections with COVID-19 from China revealed that the Mycoplasma pneumoniae and Legionella pneumophila were the most common bacteria detected among COVID-19 patients [36]. In this study, bacterial co-infection was present in 36% of patients and the most common bacterial co-infection among COVID-19 patients was Chlamydia pneumoniae with infection rate of (27%). Our findings appeared to be inconsistent with previous findings from China [36] which could be attributed to the diversity of geographical distribution of circulating respiratory bacteria. Nevertheless, the Chlamydia pneumoniae infection is a common cause of acute respiratory infections with seroprevalence of 34.1% in patients with fatal COVID-19 [37]. However, inverse association was observed between bacterial co-infection and disease severity. This association indicates a lower likelihood of ICU admission with bacterial co-infection which may be attributed to empirical use of antibiotics during the early onset of COVID-19 disease. It could be argued that COVID-19 patients co-infected with Chlamydia pneumoniae who are treated with antibiotics may have suppressed the opportunistic growth of potentially fatal secondary bacterial infections decreasing the likelihood of ICU admission.
Many risk factors including older age, diabetes mellitus, cardiovascular disease, elevated LDH levels, high levels of d‐dimer and elevated inflammatory cytokines/ chemokines have been associated with adverse outcomes in COVID-19. In our study, the prevalence of diabetes was 54% and significant correlated was present between death and co-infection with diabetes (p = 0.02). This is expected as poor glycemic control predisposes to impaired innate and adaptive immunity which might lead to decreased viral clearance [38]. The Troponin complex is a predictor for coronary syndrome and myocardial infarction. The high levels of Troponin are significantly associated with acute myocardial infarction [39]. In this study, high levels of Troponin T were detected among COVID-19 patients. We found that Troponin T was strongly related (p = 0.001) with disease severity compared to LDH (p = 0.12) and d-dimer (p = 0.25). This is explained by presence of ACE-2 receptors on myocardial cells and presence of myocardial injury in SARS-CoV-2 infection [40]. Several studies have revealed that the higher Troponin levels were increased in COVID-19 patients' ICU admission and in-hospital death [19, 41, 42]. Our results confirm the important role of Troponin in the COVID-19 severity. We think the Troponin levels can be used as a marker of COVID-19 severity and a predictor of cardiovascular events.
There have been few reports since the start of pandemic about interaction of influenza and COVID-19 most probably due to social distancing causing low incidence of influenza viruses [31]. Other studies have indicated the severity of infection in patients found to be co-infected with influenza and SARS-CoV-2 [43] and surprisingly mild symptoms in outpatients having co-infection [44]. Similar to our study, Wuhan, China in particular has seen a tremendous increase (57.3%) in the incidence of influenza and SARS-CoV-2 co-infection [45]. Therefore, both SARS-CoV-2 and influenza virus co-infection coupled with variable clinical prognosis poses a greater challenge from public health stand point. Our study showed a high case fatality rate among COVID-19 viral co-infections (r = 0.2). The severity and higher case fatality among COVID‐19 viral co-infected patients could be attributed to influenza A co-infection, since it was recently reported that SARS-CoV-2 infection followed by influenza both in-vivo and in-vitro increased the severity of infection [46].
Our study has some limitations. First, only 48 COVID-19 patients were included. Second, our study did not include asymptomatic or pre-symptomatic cases or healthy non- COVID‐19 controls. Future studies to overcome these limitations need to be considered.
Conclusions
In conclusion, the similarity in clinical presentation for both COVID-19 and Influenza makes it difficult to assess their impact on ICU admission and mortality. Our study highlights the importance of screening for co-infecting viruses in COVID-19 patients, given the high prevalence of Influenza viruses. The detection of co-infections in COVID-19 cases shows the importance of flu vaccination and warrants its increased coverage to reduce the hospitalization and associated mortality.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- COVID-19:
-
Coronavirus Disease-19
- ICU:
-
Intensive Care Unit
- OR:
-
Odd Ratio
- SARS-CoV-2:
-
Severe Acute Respiratory Syndrome Coronavirus 2
- MERS-CoV:
-
Middle East Respiratory Syndrome Coronavirus
- ACE-2:
-
Angiotensin-Converting Enzyme 2
- Pdm09:
-
The 2009 H1N1 Pandemic
- LDH:
-
Lactate Dehydrogenase
- RT-PCR:
-
Reverse Transcription Polymerase Chain Reaction
- CK:
-
Creatine Kinase
- Trop:
-
Troponin
- HB:
-
Hemoglobin
- RBC:
-
Red Blood Cell
- WBC:
-
White Blood Cells
- CRP:
-
C-Reactive Protein
- ESR:
-
Erythrocyte Sedimentation Rate
- LDL:
-
Low-Density Lipoproteins
- AST:
-
Aspartate Aminotransferase
- ALT:
-
Alanine Aminotransferase
- CKD:
-
Chronic Kidney Disease
- ARDS:
-
Acute Respiratory Distress Syndrome
- IRB:
-
Institutional Review Board
References
WHO announces COVID-19 outbreak a pandemic. https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic . Accessed 16 July 2020.
Saudi Arabia Coronavirus: 209,509 Cases and 1,916 Deaths—Worldometer. https://www.worldometers.info/coronavirus/country/saudi-arabia/. Accessed 16 July 2020.
Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364–74.
Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020;323(20):2085–6.
Zhu X, Ge Y, Wu T, Zhao K, Chen Y, Wu B, Zhu F, Zhu B, Cui L. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005.
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–62.
MacIntyre CR, Chughtai AA, Barnes M, Ridda I, Seale H, Toms R, Heywood A. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a(H1N1)pdm09. BMC Infect Dis. 2018;18(1):637.
Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020; 5, 536–544.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–74.
Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53(3):425–35.
Guo G, Ye L, Pan K, Chen Y, Xing D, Yan K, et al. New insights of emerging SARS-CoV-2: epidemiology, etiology, clinical features, clinical treatment, and prevention. Front Cell Dev Biol. 2020;8:410.
Chen L, Hao G. The role of angiotensin-converting enzyme 2 in coronaviruses/influenza viruses and cardiovascular disease. Cardiovasc Res. 2020:cvaa093.
Bengoechea JA, Bamford CG. SARS-CoV-2, bacterial co-infections, and AMR: the deadly trio in COVID-19? EMBO Mol Med. 2020;12(7):e12560.
Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–53.
Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–36.
Liu H, Grantham ML, Pekosz A. Mutations in the influenza A virus M1 protein enhance virus budding to complement lethal mutations in the M2 cytoplasmic tail. J Virol. 2017;92(1):e00858-e917.
Ratnamohan VM, Taylor J, Zeng F, McPhie K, Blyth CC, Adamson S, et al. Pandemic clinical case definitions are non-specific: multiple respiratory viruses circulating in the early phases of the 2009 influenza pandemic in New South Wales, Australia. Virol J. 2014;11:113.
Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–75.
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506.
Lai CC, Wang CY, Hsueh PR. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J Microbiol Immunol Infect. 2020;53(4):505–12.
Gold MS, Sehayek D, Gabrielli S, Zhang X, McCusker C, Ben-Shoshan M. COVID-19 and comorbidities: a systematic review and meta-analysis. Postgrad Med. 2020; 1–7.
Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020:ciaa270.
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045.
Alosaimi B, Naeem A, Alghoribi MF, Okdah L, Hamed ME, AlYami AS, et al. Structural mapping of mutations in Spike, RdRp and Orf3a genes of SARS-CoV-2 in Influenza Like Illness (ILI) patients. Viruses. 2021;13(1):136.
DaPalma T, Doonan BP, Trager NM, Kasman LM. A systematic approach to virus-virus interactions. Virus Res. 2010;149(1):1–9.
Alosaimi B, Hamed ME, Naeem A, Alsharef AA, AlQahtani SY, AlDosari KM, et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. 2020;126:154895.
D’Abramo A, Lepore L, Palazzolo C, Barreca F, Liuzzi G, Lalle E, et al. Acute respiratory distress syndrome due to SARS-CoV-2 and Influenza A co-infection in an Italian patient: mini-review of the literature. Int J Infect Dis. 2020;97:236–9.
Ding Q, Lu P, Fan Y, Xia Y, Liu M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J Med Virol. 2020. https://doi.org/10.1002/jmv.25781.
Konala VM, Adapa S, Gayam V, Naramala S, Daggubati SR, Kammari CB, et al. Co-infection with Influenza A and COVID-19. Eur J Case Rep Intern Med. 2020;7(5):001656.
Wu X, Cai Y, Huang X, Yu X, Zhao L, Wang F, et al. Co-infection with SARS-CoV-2 and Influenza A virus in patient with pneumonia, China. Emerg Infect Dis. 2020;26(6):1324–6.
Cuadrado-Payán E, Montagud-Marrahi E, Torres-Elorza M, Bodro M, Blasco M, Poch E, et al. SARS-CoV-2 and influenza virus co-infection. Lancet. 2020;395(10236):e84.
Hashemi SA, Safamanesh S, Ghafouri M, Taghavi MR, Mohajer Zadeh Heydari MS, Namdar Ahmadabad H, et al. Co-infection with COVID-19 and influenza A virus in two died patients with acute respiratory syndrome, Bojnurd, Iran. J Med Virol. 2020. https://doi.org/10.1002/jmv.26014.
Antony SJ, Almaghlouth NK, Heydemann EL. Are coinfections with COVID-19 and influenza low or underreported? An observational study examining current published literature including three new unpublished cases. J Med Virol. 2020;92(11):2489–97.
Brehm TT, van der Meirschen M, Hennigs A, Roedl K, Jarczak D, Wichmann D, et al. Comparison of clinical characteristics and disease outcome of COVID-19 and seasonal influenza. Sci Rep. 2021;11(1):5803.
Guo L, Wei D, Zhang X, Wu Y, Li Q, Zhou M, et al. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol. 2019;10:2752. https://doi.org/10.3389/fmicb.2019.02752.Erratum.In:FrontMicrobiol.2020Jun,09(11),pp.1304.
Xing Q, Li GJ, Xing YH, Chen T, Li WJ, Ni W, et al. Precautions are needed for COVID-19 patients with co-infection of common respiratory pathogens. Lancet. 2020. https://doi.org/10.2139/ssrn.3550013.
Du Y, Tu L, Zhu P, Mu M, Wang R, Yang P, et al. Clinical features of 85 Fatal Cases of COVID-19 from Wuhan: a retrospective observational study. Am J Respir Crit Care Med. 2020;201(11):1372–9.
Gupta R, Hussain A, Misra A. Diabetes and COVID-19: evidence, current status and unanswered research questions. Eur J Clin Nutr. 2020;74:864–70.
Omland T, de Lemos JA, Sabatine MS, Christophi CA, Rice MM, Jablonski KA, et al. Prevention of Events with Angiotensin Converting Enzyme Inhibition (PEACE) Trial Investigators: a sensitive cardiac troponin T assay in stable coronary artery disease. N Engl J Med. 2009;361(26):2538–47.
Babapoor-Farrokhran S, Gill D, Walker J, Rasekhi RT, Bozorgnia B, Amanullah A. Myocardial injury and COVID-19: possible mechanisms. Life Sci. 2020;253:117723.
Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, et al. Cardiovascular implications of fatal outcomes of patients with Coronavirus Disease 2019 (COVID-19). JAMA Cardiol. 2020;5(7):811–8.
Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, et al. Association of Cardiac Injury With Mortality in Hospitalized Patients With COVID-19 in Wuhan, China. JAMA Cardiol. 2020:e200950.
Lee DY, Kim KA, Yu YG, Kim KS. Substitution of aspartic acid with glutamic acid increases the unfolding transition temperature of a protein. Biochem Biophys Res Commun. 2004;320(3):900–6.
Yamaguchi Y, Nukui Y, Kotaki A, Sawabe K, Saijo M, Watanabe H, et al. Characterization of a serine-to-asparagine substitution at position 123 in the Japanese encephalitis virus E protein. J Gen Virol. 2013;94(Pt 1):90–6.
Olsen SJ, Azziz-Baumgartner E, Budd AP, Brammer L, Sullivan S, Pineda RF, et al. decreased influenza activity during the COVID-19 pandemic—United States, Australia, Chile, and South Africa, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(37):1305–9.
Bai L, Zhao Y, Dong J, Liang S, Guo M, Liu X, et al. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021;31(4):395–403.
Acknowledgements
The authors thank the Research Center at King Fahad Medical City for funding this study. Special thanks to Dr. Omar Alhazmi, Mr. Mohammad A Alturkostani, Mr. Abdulaziz A Taleb, and Mr. Saeed Albalawi from the Regional Lab in Medina for their contribution in sample collection.
Funding
This work was supported by the Research Center at King Fahad Medical city (Grant No 20–022). The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Author information
Authors and Affiliations
Contributions
B.A. contributed to study design, data analysis, results discussion, and manuscript writing and review; A.N. performed sequencing and contributed to data analysis and manuscript writing; M.H. contributed to experimental design and work, and manuscript writing; H.A. conducted the experimental work; T.A. contributed to sample and data collection; S.A. performed results discussion and clinical interpretation; A.M. contributed to sample collection and data analysis; A.Z. contributed to Study design, clinical analysis of data, results discussion, manuscript writing and review. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The institutional review board at King Fahad Medical City reviewed and approved the study protocol (IRB Log No. 20–160). Informed consent to participate was not required and was waived by IRB since only anonymized left‐over specimens were retrospectively obtained without any personally identifiable information. All methods in the study were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Alosaimi, B., Naeem, A., Hamed, M.E. et al. Influenza co-infection associated with severity and mortality in COVID-19 patients. Virol J 18, 127 (2021). https://doi.org/10.1186/s12985-021-01594-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-021-01594-0