Abstract
In several parts of China, there have been a large number of hydropericardium syndrome (HPS) outbreaks caused by serotype 4 fowl adenovirus (FAdV4) in broiler chickens since 2015. These outbreak-associated FAdV-4 strains were distinct from previous circulating strains which did not lead to severe HPS outbreaks. To better understand the molecular epidemiology of the currently circulating FAdV strains for effective diagnosis and treatment of HPS, we isolated 12 HPS outbreak-associated FAdV-4 strains from different regions in central China and investigated their molecular characteristics by performing phylogenetic analyses based on the hexon genes. Our results indicated the FAdV-4 strains in this study all belonged to serotype FAdV-4, species FAdV-C. And in comparison with ON1, KR5, MX-SHP95, PK-01, PJ-06 strains within the cluster where outbreak-associated FAdV-4 strains were located, the nucleotide sequence divergence were 1.31, 1.10, 1.42, 2.77 and 2.84%, respectively. Phylogenetic analyses revealed the hexon genes of the 12 outbreak-associated strains clustered to a relatively independent branch of the tree, and evolved from the same ancestor and we suggested that these outbreak-associated FAdV-4 strains originate from earlier strains in India.
Similar content being viewed by others
Introduction
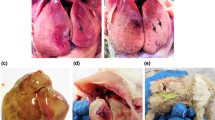
Hydropericardium syndrome (HPS) is an infectious viral disease in broiler birds at 3 to 5 weeks of age. It is caused by serotype 4 fowl adenovirus (FAdV-4) and characterized by hydropericardium and hepatic necrosis [1]. HPS was also known as “Angara Diesase”, because its first outbreak was observed in the Angara Goth, Pakistan in 1987 [2, 3]. Until now, HPS has been reported in many countries including Iraq [1], Kuwait, India [4], Mexico, Ecuador, Peru, Chile [5], USA [6], Russia [7], Japan [8, 9], and Poland [10], resulting in considerable economic losses.
Fowl adenoviruses (FAdVs) are non-enveloped double stranded DNA-viruses and belong to the genus Aviadenovirus, family Adenovirida together with other four genera: Mastadenovirus, Atadenovirus, Siadenovirus, and Ichtadenovirus [11]. Based on restriction enzyme digest pattern and serum cross-neutralization test, FAdVs have been grouped into 5 species (FAdV-A to FAdV-E) with 12 serotypes (FAdV-1 to 8a and 8b to 11) [12]. Serotype 4 fowl adenovirus (FAdV-4), the causative agent of HPS, is a member of the species Fowl Adenovirus C [4, 13]. The genome of FAdV-4 encodes a number of non-structural proteins and three structural proteins: hexon, penton and the fiber protein. The hexon gene of FAdVs is the longest and consists of hypervariable Loop L1 (HVR1-4) regions, making it a hotspot for research on taxonomy and antigenic shift of FAdVs [10, 14–16]. Hexon protein is the predominant target for induction of serotype-specific neutralizing antibodies [13].
Since 2015, clinical cases of HPS have been increasing in many regions of China, including Shandong, Hubei, Jiangsu, Anhui, Jiangxi, and Henan (Fig. 1a). These outbreaks were characterized by high mortality and no seasonal characteristics and mainly concentrated in small and medium broiler farms, rearing chickens and ducks [17]. However, to date, little is known regarding the molecular and genetic evolution characteristics of these potentially devastating FAdV strains that remain circulating in China. Hence, in this study, we aimed to investigate the molecular epidemiology of these currently circulating strains in chicken flocks in central China. Phylogenetic trees were constructed based on the hexon genes to establish the origin and genetic relationships of FAdV strains. It was found that FAdV field strains circulating after 2015 were closely related to the Indian strains PK-01 and PJ-06.
Distribution of HPS outbreaks in China (a) and in Henan province (b). Since July 2015, HPS outbreaks have been reported in Henan, Shangdong, Anhui, Jiangxi, Jiangsu, and Hubei provinces of China, which causes huge losses and continues to threaten the poultry industry (a). In this study, 12 FAdV field strains from 12 regions in central China were isolated in HPS-outbreak chicken flocks and propagated in CEF cells (b)
Materials and methods
Origin of the strains
Twelve liver samples of chickens were collected from flocks with HPS outbreaks in 12 different regions of Henan, central China, and frozen at −20 °C. All samples were confirmed to be FAdV-4 positive by polymerase chain reaction (PCR) amplifying a 632-bp fragment (Named fragment I) with primers (Table 1) based on the polymerase gene of FAdV strain MX-SHP95 (GenBank No. KP295475.1). Reactions were performed according to the following protocol: 95 °C for 5 min, followed by 31 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 50 s, and a final elongation step of 10 min at 72 °C. 24 reference FAdV strains and 12 outbreak-associated FAdV strains used for phylogenetic analyses were listed in Table 2.
Genomic DNA extraction
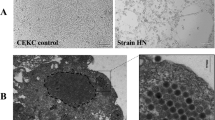
The presence of FAdV in each sample was confirmed by virus isolation followed by PCR. Initially, 5 mg of liver was cut into tiny pieces and fully ground in sterilized PBS (weight/volume = 1:3) under aseptic conditions. Following complete grinding, the suspension of liver tissue in PBS was frozen at −20 °C and thawed at 37 °C three times before centrifugation at 8000 rpm at 4 °C for 10 min. The supernatant was then collected, filtered through a 0.22 μm filter, and inoculated onto a confluent monolayer of CEF (chicken embryo fibroblast) cells prepared from 9-day SPF embryonated chicken eggs. After incubation for 1 h at 37 °C, 1.5 ml of DMEM media containing 2% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) were added to the 6-well cell culture plate. 72 h later, supernatants were collected and filtered through a 0.22 μm filter for DNA extraction. Genomic DNA from these field samples was extracted using TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver.5.0 (TaKaRa, Dalian, China), and stored at −20 °C before use. Quantification and quality of DNA extracts were determined with a Nanodrop spectrophotometer (Thermo Scientific, USA).
PCR
The hexon gene was divided into two fragments (Named fragment A and B) because of its long length in this study. Primers used for amplifying fragments A and B were designed according to the hexon genes of FAdV-4 strain MX-SHP95 (GenBank No. KP295475.1) and synthesized by Sangon Biotech (Table 1). PCR was performed in a Bio-Rad MyCycler Thermal Cycler. Fragments A and B were amplified in a 25 μl PCR reaction containing 1.5 μl genomic DNA (200–400 ng/μl), 0.25 μl Q5 High-Fidelity DNA Polymerase, 0.5 μl dNTPs (10 mM), 1.25 μl forward primer (10 pmol/μl), 1.25 μl reverse primer (10 pmol/μl), 10.25 μl sterilized water, 5 μl 5X Q5 Reaction Buffer, and 5 μl 5X Q5 High GC Enhancer. Two expected size of fragment A (1501 bp) and fragment B (1345 bp) were amplified using the following conditions: initial denaturation at 98 °C for 3 min; 31 cycles of denaturation at 98 °C for 10 s, annealing at 66 °C (for fragment A) and 57 °C (for fragment B) for 30s, extension at 72 °C for 80s; and final extension at 72 °C for 5 min. PCR products for each sample were visualized by electrophoresis in a 1.0% agarose gel containing ethidium bromide, purified according to the manufacturer’s protocol, using Universal DNA Purification Kit (TIANGEN, Beijing), and ligated into pEASY-Blunt Cloning vector using pEASY-Blunt Cloning Kit (TransGen Biotech) according to the manufacturer’s protocol. Upon transformation of each ligation reaction mixture into Trans1-T1 Phage Resistant Chemically Competent cells, a single colony was picked up and identified by PCR and restriction enzyme digestion before sequencing.
Sequencing and phylogenetic analysis
The respective bacterial clones harboring recombinant plasmids of fragment A and fragment B were sequenced by Sangon Biotech. Each sample was sequenced twice (one with the primers designed in this study and the other with the sequencing primers of the PEASY-Blunt Cloning Vector). The obtained sequences were edited using DNAStar software, and deposited into GenBank. Homology analyses to identify gene homologs were performed with the BLAST program (NCBI). Phylogenetic trees were constructed using MEGA software by the neighbor-joining analysis with 1000 bootstrap replicates, Maximum Composite Likelihood method. The phylogenetic datasets for analyses included 12 FAdV-4 isolates from this study and 24 reference strains (Table 2).
Results
Before 2014, FAdV-4 circulation also existed in China, such as ON1 (2004), JSJ13 (2013), but there were no significant HPS outbreaks. However, since July 2015, HPS outbreaks have been reported in Henan, Shandong, Anhui, Jiangxi, Jiangsu, and Hubei provinces of China (Fig. 1a), which causes huge losses and continues to threaten the poultry industry. In this study, 12 FAdV field strains from different regions in central China (Fig. 1b) were isolated in HPS-outbreak chicken flocks and propagated in CEF cells. After PCR amplification and sequencing of the hexon genes, the obtained complete hexon gene sequences were deposited into GenBank under the following accession numbers: KX640901- KX640912 (Table 2).
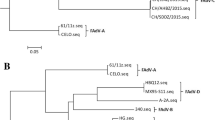
Hexon genes were used to analyze whether changes in hexon protein could explain the sudden outbreak. A phylogenetic tree based on the hexon protein included 12 outbreak-associated FAdV-4 strains from this study and 24 strains from GenBank (Table 2) produced four distinct groups: the first formed by FAdV-A (U46933.1, Z67970.1), the second formed by FAdV-C (KR5, HB1510, HN151025, ON1, JSJ13, PJ-06, PK-01, Kr-Gunwi, Kr-Yeoju, MX-SHP95, FAdV-10) and the 12 strains isolated in this study; the third formed by FAdV-D (SR48, SR49, C2B, FAdV-D), and FAdV-E (CR119, YR36, 58, TR59), and the last formed by FAdV-B (340, TR22, 09-7473-2) (Fig. 2a).
Phylogenetic analysis of nucleotide sequences of the Hexon genes (a) and Alignment of amino acid sequences (b). Phylogenetic trees were constructed using MEGA software by the neighbor-joining analysis with 1000 bootstrap replicates, Maximum Composite Likelihood method. The phylogenetic datasets for analyses included 12 FAdV-4 isolates from this study (filled triangles) and 24 reference strains (a). And alignment of amino acid sequences including those FAdV-4 strains: KR5, ON1, HNZZ (on behalf of the strains in this study), PJ-06, PK-01, MX-SHP95
The hexon genes in this study shared 100% identity both in the nucleotide and amino acid level with FAdV-4 HB1510 (China, 2015) and FAdV-4 SSDX (China, 2015), which clustered in the same branch with JSJ13 (China, 2013), PK-01and PJ-06 (India, 2008). In comparison with the hexon genes in this study, ON1, KR5, MX-SHP95, JSJ13, PK-01, PJ-06 strains within the cluster, displayed variability of 1.31, 1.10, 1.42, 25.16, 2.77 and 2.84 %, respectively at the nucleotide level. Though before 2014 FAdV-4 strain ON1 (2004, China) and JSJ13 (2011, China) had circulated in China, there were no significant HPS outbreaks. Compared with the hexon genes in this study, JSJ13 contained a deletion of 708 bases at the 5′ end albeit the rest is in full agreement, which may help explain the scare outbreaks of HPS before 2014. And alignment of amino acid sequences revealed that compared with ON1, all of the 12 outbreak-associated FAdV-4 strains contain 37 bases substitutions and as many as 13 amino acid substitutions including T to S at position 164, I to R at position 188, Q to R at position 193, E to Q to at position 195, N to D at position 238, A to T at position 240, E to N at position 243, M to I at position 263, I to V at position 264, T to A at position 410, V to I at position 574, A to P at position 797, G to A at position 842 (Fig. 2b). These mutations may help to explain the sudden outbreaks.
However, compared with Indian strains PK-01 and PJ-06, the hexon proteins only contain 2, 5 amino acid substitutions respectively, despite of 25 amino acid deletions at the N end, which do not lead to decline of the virulence. And the genetic relationship between them is very close. Thus, we postulate that ancestors of the strains is not the local strains but earlier strains in India.
Discussion
FAdVs are commonly present in chicken farms worldwide [2]. HPS is the most severe disease associated with FAdV infection, which has been attributed exclusively to serotype 4. Before 2014, FAdV-4 circulation had been reported in China, but there were no severe HPS outbreaks. However, since July 2015, outbreaks of HPS occurred with sudden high mortality rates in broilers in many small and middle chicken forms in central China, even the duck farms, leading to tremendous economic losses. Recent years, the frequency of HPS have also been increasing in many countries, such as India (2014) [18], Canada (2011) [19], Hungary (2013) [20], Korea (2012) [21], Japan (2012) [22], and Poland (2016) [10]. The ON1 and JSJ13 strains in local regions in China didn’t lead to severe clinical symptoms. The phylogenetic tree revealed that the strains circulated in China before 2014 and after July 2015 had different ancestors, which lead to this phenomenon. And the strains circulated in China now derived from earlier strains in India, with some mutations and 25 amino acid deletions; however, the exact role of the deletion in these FAdVs still needs to be elucidated in future studies.
Recently, it has been demonstrated that FAdV-4 strains circulating in China carried deletion within ORF19, which might lead to higher virulence [3]. And in comparison with JSJ13 and JH13 (isolated from China in 2013), the strains HB1510 (isolated from China in 2015) and MX-SHP95 (isolated from Mexico), KR-5 (isolated from Australia) carried a deletion of 33 nt in ORF29 [3]. This indicated that FAdV strains also carried changes in other genes and the exact of these changes in relation to antigenic variation and pathogenicity should be further investigated.
Currently, commercial vaccines against HPS have not yet been developed in China due to the lack of knowledge and understanding of FAdVs. Shengwang Liu and Huixin Li also confirmed that HPS outbreak is attributed to the emergence of FAdv strains in chickens in China, and what’s worse, FAdV species C, D, and E were co-circulating in chicken flocks in China, which made the infection more complex [23]. The molecular and phylogenetic analysis based on hexon genes in this study may further the understanding of FAdV evolution and provide relevant information for developing vaccines against the disease.
Conclusions
FAdV-4 strins is continuously affecting the poultry industry of China all over the world. In this study, we investigated the molecular epidemiology and characteristics of HPS outbreak-associated FAdV-4 strains based on the hexon genes. It showed that FAdV strains isolated in central China in this study all belonged to serotype FAdV-4, species FAdV-C. Besides, FAdV-4 strains (represented by FAdV ON1) which did not cause many HPS outbreaks before 2015 cluster together with the Austria strain KR5, Mexico strain MX-SHP95. FAdV strains now circulating in central China since 2015 are closely related to the Indian strains PK-01 and PJ-06, which revealed that the strains now originated from the earlier strains in India. This study also provides new information about the prevalence of those FAdV-4 strains currently circulating in central China, which contributes to the prevention and contral, even elimination of the strains.
Abbreviations
- BLAST:
-
Basic local alignment search tool
- CEF:
-
Chicken embryo fibroblast cells
- FAdV-4:
-
Serotype 4 fowl adenovirus
- FAdVs:
-
Fowl adenovirus
- HPS:
-
Hydropericardium syndrome
- HVR:
-
Hypervariable loop
- PCR:
-
Polymerase chain reaction
- SPF:
-
Specific pathogen free
References
abdul-Aziz TA, al-Attar MA. New syndrome in Iraqi chicks. Vet Rec. 1991;129:272.
Asthana M, Chandra R, Kumar R. Hydropericardium syndrome: current state and future developments. Arch Virol. 2013;158:921–31.
Ye J, Liang G, Zhang J, Wang W, Song N, Wang P, Zheng W, Xie Q, Shao H, Wan Z, et al. Outbreaks of serotype 4 fowl adenovirus with novel genotype, China. Emerg Microbes Infect. 2016;5:e50.
Dahiya S, Srivastava RN, Hess M, Gulati BR. Fowl adenovirus serotype 4 associated with outbreaks of infectious hydropericardium in Haryana, India. Avian Dis. 2002;46:230–3.
Toro H, Prusas C, Raue R, Cerda L, Geisse C, Gonzalez C, Hess M. Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile. Avian Dis. 1999;43:262–70.
Mendelson C, Nothelfer HB, Monreal G. Identification and characterization of an avian adenovirus isolated from a ‘spiking mortality syndrome’ field outbreak in broilers on the Delmarva Peninsula, USA. Avian Pathol. 1995;24:693–706.
Lobanov VA, Borisov VV, Borisov AV, Drygin VV, Gusev AA, Shmarov MM, Akopian TA, Naroditskii BS. Sequence analysis of hexon gene from adenovirus KR95 inducing hydropericardium syndrome in chickens. Mol Gen Mikrobiol Virusol. 2000;(1):30–6.
Nakamura K, Mase M, Yamaguchi S, Shibahara T, Yuasa N. Pathologic study of specific-pathogen-free chicks and hens inoculated with adenovirus isolated from hydropericardium syndrome. Avian Dis. 1999;43:414–23.
Abe T, Nakamura K, Tojo H, Mase M, Shibahara T, Yamaguchi S, Yuasa N. Histology, immunohistochemistry, and ultrastructure of hydropericardium syndrome in adult broiler breeders and broiler chicks. Avian Dis. 1998;42:606–12.
Niczyporuk JS. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch Virol. 2016;161:33–42.
Zhao J, Zhong Q, Zhao Y, Hu YX, Zhang GZ. Pathogenicity and complete genome characterization of fowl adenoviruses isolated from chickens associated with inclusion body hepatitis and hydropericardium syndrome in China. PLoS One. 2015;10:e0133073.
Hess M. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. 2000;29:195–206.
Schachner A, Marek A, Jaskulska B, Bilic I, Hess M. Recombinant FAdV-4 fiber-2 protein protects chickens against hepatitis-hydropericardium syndrome (HHS). Vaccine. 2014;32:1086–92.
Meulemans G, Couvreur B, Decaesstecker M, Boschmans M, van den Berg TP. Phylogenetic analysis of fowl adenoviruses. Avian Pathol. 2004;33:164–70.
Yu B, Wang C, Dong J, Zhang M, Zhang H, Wu J, Wu Y, Kong W, Yu X. Chimeric hexon HVRs protein reflects partial function of adenovirus. Biochem Biophys Res Commun. 2012;421:170–6.
Pichla-Gollon SL, Drinker M, Zhou X, Xue F, Rux JJ, Gao GP, Wilson JM, Ertl HC, Burnett RM, Bergelson JM. Structure-based identification of a major neutralizing site in an adenovirus hexon. J Virol. 2007;81:1680–9.
Chen H, Dou Y, Zheng X, Tang Y, Zhang M, Zhang Y, Wang Z, Diao Y. Hydropericardium Hepatitis Syndrome Emerged in Cherry Valley Ducks in China. Transbound Emerg Dis. 2016. [Epub ahead of print].
Mittal D, Jindal N, Tiwari AK, Khokhar RS. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virusdisease. 2014;25:114–9.
Grgic H, Yang DH, Nagy E. Pathogenicity and complete genome sequence of a fowl adenovirus serotype 8 isolate. Virus Res. 2011;156:91–7.
Kajan GL, Kecskemeti S, Harrach B, Benko M. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet Microbiol. 2013;167:357–63.
Choi KS, Kye SJ, Kim JY, Jeon WJ, Lee EK, Park KY, Sung HW. Epidemiological investigation of outbreaks of fowl adenovirus infection in commercial chickens in Korea. Poult Sci. 2012;91:2502–6.
Mase M, Nakamura K, Minami F. Fowl adenoviruses isolated from chickens with inclusion body hepatitis in Japan, 2009–2010. J Vet Med Sci. 2012;74:1087–9.
Li H, Wang J, Qiu L, Han Z, Liu S. Fowl adenovirus species C serotype 4 is attributed to the emergence of hepatitis-hydropericardium syndrome in chickens in China. Infect Genet Evol. 2016;45:230–41.
Acknowledgements
The authors highly acknowledge the support of Key Laboratory of Animal Immunology of the Ministry of Agriculture, Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences.
Funding
This work was supported by grants from National Key Research and Development Program of China (No. 2016YFD0500800).
Availability of data and materials
All relevant information is provided in this current manuscript.
Authors’ contributions
TZ and QJ conceived and designed the study. TZ, YC, PD, XL carried out the sequencing. TZ and YW analyzed the data and drafted the manuscript. QJ collected the sample and JL provided the reagents. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Informed consents were provided by all participants included in the present study. Ethical Approval was given by Key Laboratory of Animal Immunology of the Ministry of Agriculture, Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, T., Jin, Q., Ding, P. et al. Molecular epidemiology of hydropericardium syndrome outbreak-associated serotype 4 fowl adenovirus isolates in central China. Virol J 13, 188 (2016). https://doi.org/10.1186/s12985-016-0644-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-016-0644-x