Abstract
Foot-and-mouth disease virus (FMDV) represses host translation machinery, blocks protein secretion, and cleaves cellular proteins associated with signal transduction and the innate immune response to infection. Non-structural proteins (NSPs) and non-coding elements (NCEs) of FMDV play a critical role in these biological processes. The FMDV virion consists of capsid and nucleic acid. The virus genome is a positive single stranded RNA and encodes a single long open reading frame (ORF) flanked by a long structured 5ʹ-untranslated region (5ʹ-UTR) and a short 3ʹ-UTR. The ORF is translated into a polypeptide chain and processed into four structural proteins (VP1, VP2, VP3, and VP4), 10 NSPs (Lpro, 2A, 2B, 2C, 3A, 3B1–3, 3Cpro, and 3Dpol), and some cleavage intermediates. In the past decade, an increasing number of studies have begun to focus on the molecular pathogenesis of FMDV NSPs and NCEs. This review collected recent research progress on the biological functions of these NSPs and NCEs on the replication and host cellular regulation of FMDV to understand the molecular mechanism of host–FMDV interactions and provide perspectives for antiviral strategy and development of novel vaccines.
Similar content being viewed by others
Background
Foot-and-mouth disease (FMD), an acute highly contagious viral disease in susceptible cloven-hoofed animals, was described 100 years ago. The etiologic agent, FMD virus (FMDV), is a positive-sense, single-stranded RNA virus that belongs to the Aphthovirus genus, Picornaviridae family. FMDV is one of the most contagious viruses in cloven-hoofed animals and can cause both acute and prolonged, asymptomatic but persistent infection [1]. Upon infection of susceptible species, FMDV proliferates rapidly and causes vesicular disease in feet and mouth.
The RNA virus genome of FMDV displays a very high mutation rate because the virus-encoded RNA polymerase lacks a proofreading mechanism [2, 3]. The high mutation rate of FMDV, coupled with its rapid proliferation and extensive population, result in the rapid evolution of this virus [4], which contributes to the existence of seven main serotypes (A, O, C, Asia1, South African Territories (SAT) 1, SAT2, and SAT3). In addition, numerous variants and subtypes have been further evolved from each serotype [1]. Given that cross-reactivity varies, antigenic diversity among these serotypes have to be considered during vaccine development [5].
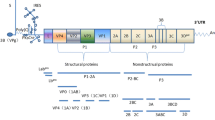
FMDV virion has a symmetric protein shell (or capsid) enclosing the genomic RNA. Genome RNA contains a positive single-strand chain approximately 8.3 kb long and encodes a single long open reading frame (ORF) of about 7 kb with two alternative initiation sites. The ORF is flanked by a long 5ʹ-untranslated region (5ʹ-UTR) and a short 3ʹ-UTR, and ends with a genetically encoded poly-(A) tail [6]. A genome-linked viral nonstructural protein (NSP), 3B (also known as VPg) containing 23–24 amino acid (aa) residues, is covalently bound to its 5ʹ end, although this protein is rapidly released into an infected cell and is deemed to play no part in translation initiation [7]. The viral ORF can be translated into a polyprotein of about 250 kDa, which is subsequently cleaved by two virus-encoded proteinases (leader (Lpro) and 3Cpro) to yield structural and NSPs [8, 9] (Fig. 1).
Schematic diagram of FMDV genome, processing of viral polypeptide and conformations of the structural proteins. FMDV genome RNA contains a single open reading frame (ORF) of about 7 kb with two alternative initiation sites. The ORF is flanked by a long 5ʹ-untranslated region (5ʹ-UTR) and a short 3ʹ-UTR. 3B (VPg) is covalently bound to its 5ʹ end. The ORF region is generally divided into four functional areas (L, P1, P2 and P3) due to the different functions of mature polypeptides. ORF-encoded polyprotein is processed into four products, Lpro, P1-2A, 2BC and P3 by Lpro, 2A and 3Cpro. The precursors P1-2A, 2BC and P3 are further processed into mature viral proteins and some cleavage intermediates with relative stability, such as VP0 or 1AB, 3ABC, 3BCD, 3AB, and 3CD by 3Cpro. Structural proteins form the biological protomer and viral capsid
The FMDV genome was completely sequenced, and all cleavage sites involved in the processing of polypeptides were also identified in the past two decades. Generally, the ORF region in FMDV genome is artificially divided into four functional areas due to the different functions of mature polypeptides [10], which are shown as follows (Fig. 1): L region, which is located at 5ʹ end to the capsid component and codes for Lpro. P1 region, encoding a precursor for capsid polypeptide, which can generate four mature capsid proteins (VP4, VP2, VP3, and VP1) upon cleavage by viral protease. P2 region encodes three viral proteins (2A, 2B, and 2C) in the middle region of the genome. And P3 region, which encodes four viral proteins: 3A, 3B, 3Cpro and 3Dpol, in which, 3C is a viral protease and 3D an RNA-dependent RNA polymerase [11]. Actually, primary polyprotein is not strictly processed into four products as the functional regions by initial protease, but Lpro, P1-2A, 2BC and P3 by Lpro, 2A and 3Cpro. The precursors P1-2A, 2BC and P3 are further processed into mature viral proteins and some cleavage intermediates with relative stability, such as VP0 or 1AB, 3ABC, 3BCD, 3AB, and 3CD by 3Cpro (Fig. 1). Usually, the intermediates may perform functions other than those of their individual constituents.
The virus capsid consists of 60 copies of each of the four structural polypeptides (VP1 to VP4), which are self-assembled into an icosahedral structure with a diameter of 30 nm [12, 13] (Fig. 1). Studies on structural information and protein interaction have shown that the structural protein or the precursor products VP0 (VP2/4 or 1AB), VP1 (1D), and VP3 (1C), which are encoded by P1 region, form immature protomers through weak chemical bond interaction. Then, pentamers are assembled by five protomers [14]. After self-assembly of pentamers to generate an empty capsid, the viral genomic RNA covalently linked to VPg at the 5ʹ end enters the capsid to produce pro-virion. Then the pro-virion is eventually processed into a mature virion following the RNA-triggered auto-cleavage of VP0 [15]. Finally, the virion particles with complete assembly are released from the infected host cells (Fig. 2).
Viral non-coding elements (NCEs) and NSPs play essential roles in FMDV-induced viral evasion. This review focus on the molecular biology of FMDV, along with the functional roles of FMDV NSPs and NCEs in viral replication and virulence, to elucidate how FMDV evades the host immune response and evolves into such an aggressive pathogen.
Functions of FMDV NCEs
5ʹ-UTR
The 5ʹ-UTR is a central element that initiates replication and translation of the picornavirus genome. Similar to other picornaviruses, the FMDV RNA genome does not contain the 5ʹ-terminal cap structure (m7GpppN…), which can be recognized by the translation initiation machinery in all eukaryotic cells [16]. Instead, a short viral protein, 3B or VPg, is covalently linked to the 5ʹ end of the viral RNA [8]. FMDV is unique among the picornaviruses as it encodes three non-identical copies of 3B in tandem (see Section 3.3).
FMDV RNA, which consists of a long 5ʹ-UTR containing over 1300 nucleotides (nt), exhibits extensive secondary structure and is commonly divided into five regions (Fig. 3). The first element of the 5ʹ end is the S-fragment with approximately 350 nt long. Its sequence is highly base-paired and capable of folding into a long stem-loop. Although this highly structured segment has not been extensively studied, this portion is presumed to prevent host exonuclease function to maintain viral genome stability and replication [8].
FMDV 5ʹ-UTR provides structural basis for initiation of viral protein translation. a Schematic representation of the structural elements within the FMDV 5ʹ-UTR. The 5ʹ-UTR is highly structured and comprises S-fragment, poly(C) tract, 3B-uridylylation site (bus) or cis-acting replication element (cre), pseudoknots (PKs), and internal ribosome entry site (IRES). The organization of the IRES element in different domains (I, II, III, IV, and V) is indicated. Domain I (the first 20 nt of the IRES) belongs to the right arm of bus. The conserved AAACA, GNRA, RAAA motifs, C-rich loop and polypyrimidine tracts are also indicated. The two different initiator codons AUG1 and AUG2 are separated by 84 nt. Note that the stems indicated are not perfectly base-paired. b Intact eIF4G is required in cap-dependent protein synthesis of cellular mRNA. The initiation factors are required for the assembly of 48S initiation complex on a capped cellular mRNA. c Involvement of translation initiation factors in IRES-dependent protein synthesis. The FMDV IRES elements are indicated. The factors shown are eIF4E (4E), eIF4G (4G), eIF4A (4A), and eIF2 (2) (as part of the ternary complex eIF2/GTP/met-tRNA), along with small ribosomal subunit (40S)
Following S-fragment is a poly(C) tract exhibiting a variable length of 150–200 nt in typical field strains. Investigations on genetically engineered viral genome revealed that a certain threshold length of poly(C) is required to rescue live virus, although no evidence has shown that poly(C) tract length is directly associated with virulence [17]. On the 3ʹ end of the poly(C) tract is a variable region folded as multiple pseudoknots (PKs) (2–4) [18, 19]. The function of this element remains unknown, but it is possibly associated with the poly(C) tract [16].
Downstream of the PK region lies a 3B-uridylylation site (bus), also known as cis-acting replication element (cre), which is a highly conserved stem-loop of 55 nt and is essential for viral genome RNA replication [20]. This element is commonly consisted in different picornavirus RNAs, and all picornavirus bus elements contain a conserved “AAACA” motif in the loop region. This motif acts as the template for uridylylation of VPg (3B) to produce VPgpU and/or VPgpUpU, collectively termed as VPgpU(pU), by the viral RNA polymerase 3D [21, 22]. VPgpU(pU) serves as a primer that initiates viral RNA synthesis. Interestingly, most known viral bus structures are embedded in protein-coding regions of the viral genome, except for that of FMDV, which has a bus structure that lies in the 5ʹ-UTR region [8]. Alteration in the consensus “AAACA” motif disrupts viral genome replication but does not significantly affect RNA translation in FMDV [20]. Moreover, FMDV is temperature sensitive (ts), and viral replication is greatly suppressed at non-permissive temperature. Mutant analysis reveals that the ts element is located within the bus sequence [23], indicating that “cre” also functions in trans.
The last portion of the 5ʹ-UTR is the internal ribosome entry site (IRES) element, a type II IRES element [24]. Consisting of approximately 450 nt, the IRES element is required for cap-independent internal translation initiation of viral RNA [20, 25]. FMDV IRES has five domains and forms multiple stem-loops (Fig. 3a). There are evidences to support that these domains are involved in translational control. Some highly conserved sequences exist in these domains, like Domain II containing a polypyrimidine tract (UCUUU) that provides a polypyrimidine tract-binding protein (PTB) binding site [26]. Domain III includes two conserved essential motifs, GNRA and RAAA (N is any nucleotide and R stands for purine) at the apical region, and a conserved C-rich loop at the middle region [27, 28]. Modification of the 5ʹ-G or 3ʹ-A residue of GNRA both greatly diminish the activity of FMDV IRES [27, 29] and mutation of RAAA shows abolished activity of FMDV IRES [28, 30], suggesting that they are important for IRES activity. This domain also plays a key role in RNA–RNA interactions and RNA–protein interactions in FMDV [31]. Domain IV is arranged into two stem-loop structures containing A-rich internal bulges in FMDV, and it is responsible for the interaction with eIF4G, an essential translation initiation factor for IRES-mediated translation initiation in FMDV [32, 33]. Domain V consists of a conserved hairpin-loop region and a polypyrimidine-rich tract sequence at 20 nt upstream of the initiation codon AUG [34], which is crucial to identify and initiate viral protein synthesis. Some mutations in this region are highly detrimental to IRES activity in FMDV [25] (Fig. 3a).
Translation of FMDV RNA
The featured IRES element is an essential structural region for initiation of protein synthesis in picornavirus genome RNA [25, 35, 36]. The IRES is a cis-acting RNA sequence that adopts diverse three-dimensional structures to recruit the translation machinery using a mechanism that is independent on the 5′end [37]. Nearly all canonical cellular translation initiation factors are required for IRES-dependent initiation of translation in FMDV except eIF4E, a cap-binding protein [16, 38], consistent with the fact that FMDV genome RNA lacks the cap structure on the 5ʹ end (Fig. 3b and c). For most cellular mRNAs, the 5′cap structure is recognized by eIF4F, a trimeric complex composed of eIF4A (an RNA helicase), eIF4E (a specific cap structure-binding protein), and eIF4G (a scaffold protein). The scaffold protein eIF4G interacts with eIF4A, PABP and the multimeric factor eIF3. The complex bound to the 40S ribosomal subunit is recruited to the mRNA along with other factors, resulting in eukaryotic protein synthesis [38] (Fig. 3b). Cleavage of eIF4G by viral Lpro removes its N-terminal portion, the binding sites for eIF4E, impairing cap-dependent protein synthesis in host cells. By contrast, the C-terminal portion of eIF4G retains the binding sites for eIF4A and eIF3, which is sufficient for FMDV IRES activity.
However, viral IRES are characterized by the presence of ignored AUGs upstream of the functional start codon, heavy RNA structure and high GC content [39, 40]. Despite being unable to direct cap-dependent translation, the C-terminal portion of eIF4G is fully efficient in FMDV IRES-driven translation [16, 41]. Reconstitution assays have demonstrated that assembly of initiation complexes into IRES element requires either the intact type or C-terminal fragment of eIF4G, in addition to eIF4A, and eIF3 [42, 43]. Moreover, eIF4G, either the intact type or the C-terminal cleavage product, directly interacts with IRES in domains III and IV [44]. eIF4A and eIF3, which bind to eIF4G, also indirectly interact with IRES to participate in IRES-directed translation initiation. eIF4B, another factor that directly interacts with IRES at domain IV, also affects the activity of IRES, although the effect is rather modest [45].
In addition to the eIFs described above, many other cellular proteins are also involved in the modulation (stimulate or repress) of IRES activity. They are all termed IRES-transacting factors (ITAFs), including PTB, PCBP2, the SR splicing factor (SRp20), the far upstream element binding protein 2 (FBP2), the lupus antigen (La), unr (upstream of N-ras), nucleolin, or Gemin5, etc [46].
The polypyrimidine PTB was the first protein identified as an ITAF [25, 47, 48]. PTB contains four RNA recognition motifs (RRM) to recognize U/C-rich sequences. PTB directly binds to polypyrimidine tracts on IRES element to stimulate IRES activity in FMDV [47]. The poly(C) binding proteins, PCBP1 and PCBP2, also recognize and interact with FMDV IRES domain II, but the depletion test showed that such an interaction site is not indispensable for FMDV IRES activity [41, 49]. Moreover, secondary protein–protein or RNA-protein bridges could facilitate IRES activity. As examples, SRp20 enhances IRES-mediated translation via its interaction with PCBP2 [50]. ITAF45 (also known as Ebp1), together with PTB, sharing the same binding region in IRES, cooperatively stimulate FMDV IRES activity [51]. Later studies found some ITAFs of IRES downregulators. For example, Gemin5, a cytoplasmic protein that binds directly to the FMDV IRES and down-regulates translation [52]. Besides, FBP2 negatively regulates EV71 IRES activity [53], and the double stranded RNA-binding protein DRBP76:NF45, is a nuclear heterodimeric protein that interacts with HRV IRES and represses its activity [54].
As a consequence of in depth RNA–protein interaction studies performed with picornavirus IRES, the list of ITAFs is still growing incessantly [55]. The Glycil tRNA synthetase (GARS) emerges as a class of novel ITAFs stimulating picornavirus IRES activity [56]. Predictably, more ITAFs will be identified in the near future, which will help to provide more details about interactions of IRES-proteins within host cells.
3ʹ-UTR
FMDV RNA 3ʹ-UTR consists of two components, a structural sequence of 90 nt folding into two separate stem-loops and a poly(A) tail with variable length [16, 57] (Fig. 1). These elements are both involved in viral replication and virulence [24]. Molecular biology studies have demonstrated that the structured 3ʹ-UTR directly binds to S-fragment and IRES elements at two distinct positions in the 5ʹ-UTR through specific long-range RNA–RNA contact pattern [57]. IRES-3ʹ-UTR interaction requires both stem–loop structures of 3ʹ-UTR, which stimulates IRES activity and is independent of the poly(A) tail. Whereas the S-fragment interacts with each of the stem–loops and is dependent on the poly(A) tail [57]. These findings indicated that the 3ʹ-UTR enhances IRES activity and determines the virulence of FMDV. Moreover, genetic evidence reveals that recombinant FMDV with a deleted structural sequence in 3ʹ-UTR cannot be recovered [58], demonstrating that the structured region in the 3ʹ-UTR is essential for FMDV infectivity and replication.
In addition to direct RNA-RNA interaction, 5′–3′ end bridges could also involve protein-protein and protein–RNA interaction. Studies found that cellular proteins PCBPs and p47 can both interact with the S region and 3ʹ-UTR by directly binding to them [57]. As mentioned in the above section, SRp20 directly interacts with PCBP2, and Ebp1 cooperates with PTB to stimulate viral genome translation, raising the possibility that secondary protein–protein bridges take important roles in RNA–RNA interaction between 5ʹ and 3ʹ ends to modulate the viral genome translation in FMDV.
Functions of FMDV NSPs
Lpro
The viral protein Lpro is a region in the polyprotein preceding the capsid precursor protein [59]. Lpro, the first protein in FMDV to be translated, is initiated at two different start codon AUGs separated by 84 nt. Lpro has two alternative forms, namely, Labpro and Lbpro. Both forms have been detected in vitro and in vivo [60, 61]. Lbpro protein (synthesized from the second AUG) is the major protein type in vivo. Viable viruses can be recovered from synthetic genomes containing mutations in the first AUG but not in the second [62, 63]. The sequence between the two AUGs are possibly involved in start codon recognition through interactions with a regulatory factor [62, 64]. FMDV Lpro is a well-characterized papain-like proteinase [65–67] that releases itself from the polyprotein via cleavage between its own C-terminus and the N-terminus of VP4 at the sequence ArgLysLeuLys ↓ GlyAlaGlySer during viral maturation [66, 68].
Lpro is an important determinant of virulence. Previous studies showed that the Lpro-deleted virus shows only a slightly slower replication rate than the wild-type (WT) virus [63] but exerts a dramatically lower ability to cause lesions during intradermal injection [69] and fails to cause clinical signs of FMD when exposed to aerosol containing high doses of leaderless virus in cattle and swine [70, 71]. Thus, Lpro is not required for viral replication but is indispensable for the pathogenesis of FMDV.
Lpro also represses host cell translation by cleaving the translation initiation factor eIF4G [8, 66]. Strong evidence confirmed that Lpro recognizes and cleaves the crucial host translation initiation factor eIF4G at the site between Gly479 and Arg480 [66, 72, 73]. eIF4G is a key scaffold protein for the attachment of other translation initiation factors to exert their functions. Cleavage of eIF4G directly shuts off host cap-dependent mRNA translation [73, 74]. By contrast, FMDV RNA, which initiates translation in a cap-independent manner via its IRES element, does not require intact eIF4G. Removal of the N-terminus of eIF4G by Lpro does not impair viral RNA translation initiation. Thus, FMDV freely uses the host protein synthesis machinery to synthesize viral protein (Fig. 3) [8].
Lpro blocks interferon (IFN) activity directly and indirectly by inhibiting the central upstream regulatory factor [75–77]. IFN induction is one of the most important host innate immune response to viral infection [78]. Secreted IFN proteins bind to neighbor cells by paracrine manner to induce the expression of a number of IFN-stimulated genes to mediate various biological responses [79], including inhibition of viral replication. Notably, infection with leaderless virus, which lacks the Lpro coding region, induces higher levels of type I IFN (IFN-α and IFN-β) mRNA level than WT virus, and type I IFN downstream signaling can only be detected in cultures with the leaderless virus infection [75, 76, 80]. Therefore, type I IFNs, including their mRNAs and proteins, are limited by viral Lpro. Lpro also inhibits dsRNA-induced IFN-λ1 expression, which is a type III IFN demonstrating significant antiviral activity against FMDV [77].
No evidence demonstrated the direct cleavage of IFNs by Lpro. Translational repression of IFNs is probably resulted from the blocking of FMDV Lpro-induced cap-dependent mRNA translation by Lpro-mediated cleavage of eIF4G [8, 81]. Further studies have attempted to investigate the molecular mechanism of Lpro virulence. Promoter activity and protein studies have shown that FMDV Lpro down-regulates interferon regulatory factor 3/7 (IRF-3/7) expression both at the transcription and translation levels [82, 83]. IRF-3/7 are important regulators in RIG-I/MDA5-induced innate immune signaling, a crucial pathway response to infections caused by picornaviruses [84]. Their inhibition suppresses the expression of type I IFNs and downstream cytokines, including IFN-α/β and CCL5, also known as RANTES [82, 83].
A recent study found that FMDV Lbpro is a novel viral deubiquitylation (DUB) enzyme [85]. Ubiquitination and deubiquitination, which are a class of important regulation patterns, are critically involved in many signaling cascades, including virus-induced type I IFN signaling [86]. Viruses are connected to ubiquitin and ubiquitin-like modifiers in a variety of ways [87, 88]. Sequence analyses revealed that catalytic residues (Cys51 and His148) are highly conserved in Lbpro, the shorter form of Lpro, in all seven serotypes. The topology of FMDV Lbpro is remarkably similar to that of some known DUBs, such as ubiquitin-specific protease 14, a cellular DUB [89], as well as severe acute respiratory syndrome coronavirus (SARS-CoV) papain-like protease (PLpro), a coronaviral DUB [90]. In addition, Biochemical and molecular evidence also revealed that FMDV Lbpro can remove ubiquitin (Ub) moieties from cellular substrates, function on both lysine-48- and lysine-63-linked polyubiquitin chains, a feature shared by other known viral DUBs, such as human cytomegalovirus UL48, herpes simplex virus type 1 UL36, and SARS-CoV PLpro [91, 92], indicating that FMDV Lbpro exhibits deubiquitinating activity. A new mechanism of Lpro blocking IFN antiviral response is revealed by the evidence that Lpro antagonizes type I IFN induction by deubiquitinating the critical signaling components RIG-I, TBK1, TRAF3, and TRAF6 [85].
To sum up, FMDV Lpro generally functions as a multifunctional protein that blocks IFN-mediated antiviral response via multiple distinct mechanisms (Fig. 4): (1) Lpro shuts off host cell translation through cleavage of the translation initiation factor eIF4G, suppressing IFN protein expression [93]; (2) Lpro represses IFN activity by inhibiting activation of central upstream regulatory factors, including NFkB and IRF-3/7 [82]; and (3) Lpro acts as a DUB and cleaves ubiquitin chains from RIG-I, TBK1, TRAF3, and TRAF6, thereby inhibiting type I IFN signaling [85].
Known functions of FMDV nonstructural proteins on cellular regulation. Purple circles: FMDV nonstructural proteins. Red arrow: activation of downstream gene expression. Orange line with vertical stub: inhibition of downstream proteins. kB, a canonical kB element response to Nuclear factor kB (NF-kB). ISRE, IFN-sensitive response element. Caspases, represent a class of genes inducing apoptosis and inflammation
2A Protein
The P2 portion in the picornavirus genome encodes three mature viral proteins, namely 2A, 2B, and 2C (Fig. 1). FMDV 2B and 2C are partially homologous to other picornavirus, whereas FMDV 2A is only an 18 aa peptide [94] and is much shorter than the other picornavirus members but highly conserved with cardiovirus at the 2A/2B junction. The FMDV 2A protein lacks any protease motifs and only contains the characteristic C-terminal motif “-Glu(x)AsnProGly(2A)/Pro(2B)-” [95]. In addition, the conserved cleavage site is located between 2A and 2B Gly(2A)/Pro(2B) [94, 96]. Mutation research confirmed that Gly (2A) is the most important amino acid for cleavage activity at the 2A/2B junction [97, 98], whereas recombinant FMDV sequence containing mutation in the 2A peptide can produce uncleaved proteins. Moreover, cleavage between 2A and 2B only occurs as a co-translational event. Thus, the 2A cleavage event occurs only during polypeptide synthesis [95], such that the 2A peptide remains connected to the P1 structural protein precursor (P1-2A) following primary cleavage of the polyprotein [99]. 2A is cleaved from the P1-2A precursor either by 3Cpro or 3CDpro [100].
The FMDV 2A peptide, along with the first aa of the 2B protein, can mediate cleavage in artificial polyprotein systems [94, 95]. However, this FMDV 2A-mediated artificial polyprotein cleavage does not work in the prokaryotic system [94]. In addition, the synthesized upstream proteins of the 2A sequence are always present in greater molar excess than the downstream proteins of the 2A sequence [95]. Thus, the 2A-2B cleavage event is not a proteolytic event but a modification of the translational machinery by the 2A peptide, allowing the release of the protein-2A from the ribosome while permitting the synthesis of the downstream proteins to proceed [95, 96].
2B Protein
Picornaviruses 2B are viroporins, a class of low-molecular-weight hydrophobic transmembrane proteins encoded by a wide range of animal viruses. The transmembrane hydrophobic domains interact with the phospholipid bilayer to induce dispersion, increasing membrane permeability and promoting the release of viral particles. Thus, viroporins are crucial for viral pathogenicity [101, 102].
Knowledge on FMDV 2B is limited. FMDV 2B encodes a 154 aa peptide, which is slightly longer than other viroporins and contains two predicted putative transmembrane domains located in 83–104 aa and 119–137 aa, respectively [103]. Topology studies have shown that FMDV 2B is located in the endoplasmic reticulum (ER) and exhibits a transmembrane topology similar to that of class IIB viroporins, which consist of two transmembrane domains and their N- and C-termini both extend to the cytoplasmic matrix [103]. Similar to other known viroporins, FMDV 2B induces damage to the integrity of the host cell’s membrane and causes Ca2+ abnormalities, activating autophagy [103]. However, this phenomenon has not yet been fully confirmed in FMDV.
Some reports have indicated that 2B functions synergistically with other viral proteins, such as 2C, in FMDV [104]. Previous findings have also confirmed that picornaviral infections block protein secretion in host cells. Rather than other picornaviruses, ER-to-Golgi apparatus protein transport in FMDV is blocked by the 2BC precursor protein but not by 2B or 2C. Microscopy and immunoprecipitation indicated that the 2BC protein blocks cellular protein transport between the ER and Golgi apparatus during FMDV infection [105], and a study showed that protein transport was abolished only in the case of both 2B and 2C co-expression [104]. This phenomenon is possibly caused by the synergistic effect of 2B and 2C.
Viroporins participate in multiple stages of the viral life cycle, such as cell entry and genome replication. Viruses with viroporin deletion cannot properly assemble and release from cells [102, 106]. Thus, viroporins are attractive targets for antiviral therapy based on recent findings on their structure and biological functions. Several viroporin inhibitors have been developed to effectively suppress viral replication through inhibition of membrane permeabilizing activities. These inhibitors include amantadine, which is used against hepatitis C virus (HCV) p7 and influenza A virus M2 protein; 4,4ʹ-diisothiocyano-2,2ʹ-stilbenedisulphonic acid [107], which is against enterovirus 71 P2B protein; 5-(N,N-hexamethylene) amiloride (HMA), against HIV-1 Vpu, HCV p7, and SARS-CoV E protein; and N-(5-(1-methyl-1H-pyrazol-4-yl)-napthalene-2-carbonyl)-guanidine (BIT225), which can also inhibit HIV-1 Vpu and HCV p7 [102, 108]. Although no reports demonstrated the effect of these drugs on FMDV replication, these findings provide potential strategies for developing antiviral drugs against FMDV viroporin 2B.
2C Protein
Protein 2C is one of the highly conserved molecules among the viral proteins encoded by FMDV, and 2C-like (2CL) proteins generally exist in many other RNA viruses across animals and plants [109]. The FMDV protein 2C is a 318 aa polypeptide that contains a predicted amphipathic helix in its N-terminus (residues 17–34) [110, 111]. It is responsible for many biological functions linked to membrane targeting. Although the reports about FMDV 2C are limited now, a number of studies have uncovered the function of other picornaviruses 2C. FMDV 2C is the largest membrane-binding component of the virus RNA replication complex and is speculated to perform a function similar to the 2C in other picornaviruses [104, 112]. 2C plays a key role in both membrane rearrangement and formation of the viral replication complex. In addition, 2C is implicated in virus-induced cytopathic effects [113–116]. Immunofluorescence studies demonstrated that protein 2C aggregates at the cellular periphery in FMDV-infected cells [117] and is inclined to bind to ER [118]. These findings are consistent with those of subcellular fractionation studies, which revealed that 2C co-localizes with the ER membrane, Golgi apparatus, and lysosomes in poliovirus [119–122]. 2C also co-localizes with the membrane-bound replication complex, suggesting that 2C acts as an important factor in viral replication. This function is also confirmed by the inhibition experiments. Guanidine hydrochloride, an antiviral compound and a molecular antagonist against protein 2C can inhibit viral RNA synthesis in picornavirus-infected cells, and virus strains containing guanidine-resistant (gr) 2C mutation cannot be inhibited by guanidine hydrochloride, providing direct evidence that FMDV 2C protein plays a key role in virus replication [123, 124]. This phenomenon is also observed in poliovirus [111, 125, 126]. Notably, FMDV 2C is only present when the replication complex forms during virus proliferation but absent from clarified virus stocks used for vaccine preparation. Thus, 2C protein can be used to differentiate potential carrier convalescent animals from vaccinated livestock [127–130].
Many other functions of 2C in other picornavirus members have been reported, including viral RNA binding activity [131, 132], NTP binding activity [109], ATPase and GTPase activities [133, 134], binding to Reticulon 3, as well as viral replication [135]. Given that 2C proteins are highly conserved among picornavirus members, FMDV 2C is speculated to demonstrate most of these activities. Indeed, some functions of FMDV 2C have been confirmed [136].
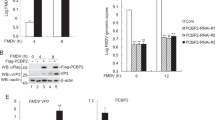
A recent report discovered that FMDV 2C is involved in apoptosis induction and type 1 IFN response. Yeast two-hybrid system and immunoprecipitation approaches revealed that FMDV 2C interacts with N-myc and STAT interacting (Nmi) protein [137], a factor involved in multiple cell signaling by interacting with many proteins, including IFN signaling and apoptosis signaling [138–140], as well as its heterodimeric complex partner, IFN-induced 35-kDa protein (IFP35) [137], a protein with potential roles in apoptosis, cytokine response, and antiviral activity [141–143]. Moreover, immunoprecipitation and immunofluorescence studies indicated that 2C can recruit Nmi and IFP35 into the intracellular membrane structure by changing their subcellular distribution, forming the 2C-Nmi-IFP35 complex. Thus, 2C likely induces apoptosis through the host protein Nmi, as well as induces a type I IFN response through the host protein IFP35 [118, 137, 144]. Nevertheless, some studies indicated that apoptosis does not occur in FMDV-infected cells [145, 146]. Further investigation is needed to clarify the underlying mechanism of FMDV and apoptosis.
Some other research reported that FMDV 2C is involved in FMDV-induced autophagy. FMDV triggers cellular autophagy and enhances viral replication [147]. Gladue et al. further investigated the interaction between FMDV 2C and cellular Beclin1 using a yeast two-hybrid model, immunoprecipitation and confocal microscopy. They found that FMDV 2C binds to Beclin1, a factor that plays dual roles in the autophagy pathway. Beclin1 is involved in initiation of autophagosome formation and fusion of autophagosome to lysosome [148, 149]. So 2C-induced Beclin1 inactivation blocks the fusion of FMDV-containing autophagosomes to lysosomes and prevents virus degradation [150]. 2C-Beclin1 interaction plays a significant role in virus replication [150].
Viral 2C protein is speculated to act as an important regulator integrating multiple cell signaling during FMDV infection, including apoptosis, immune response, and autophagy. This protein reduces the cellular killing effect against viruses and promotes virus survival and proliferation, thereby facilitating viral proliferation and release of virus particles (Fig. 4).
3A Protein
FMDV 3A protein is a 153 aa peptide, which is larger than other picornaviral 3A protein, like the 87 aa-long poliovirus 3A [8]. FMDV 3A is conserved in most FMDV strains. Half of the 3A coding region in the N-terminus (positions 1–75) encoding a hydrophilic domain and a hydrophobic domain capable of binding to membranes, is highly conserved in all FMDV strains [151]. By contrast, many mutations and deletions occur in the C-terminus of all FMDV strains.
Based on its hydrophobic motifs, FMDV 3A exerts membrane binding activity [105]. Fluorescent staining revealed that 3A in infected cells partially co-localizes with the ER marker calreticulin and with Golgi stacks protein p58 [152]. In contrast to other picornaviruses, transient expression of 3AB proteins does not induce major rearrangements of intracellular membranes as inferred from immunofluorescence and electron microscopy studies [152]. FMDV 3A preferentially localizes in small vesicles when transiently expressed [105]. Rosas et al. [153] generated BHK21 cell lines stably expressing 3A and its precursor 3AB and found that expression levels of 3A and its processors exert varying degrees of cell toxicity but do not induce cell membrane rearrangements [153]. In addition, stable expression of 3A or 3AB protein enhances FMDV replication, including increase of virus plaque formation and virus titers. But transiently expressing 3AB protein shows a decreased level of FMDV infection. These indicated a transacting role of 3A and 3AB on the FMDV multiplication cycle [153].
Reports demonstrated that 3A plays a role in virulence and host range. Some amino acids substitutions or deletions in 3A protein are associated with change in host range and tropism of FMDV [154, 155] and other picornaviruses, including poliovirus and human rhinovirus [156]. In the N-terminus of 3A, the amino acid substitution Q44 to R is sufficient to confer FMDV C-S8c1 strain adaptation to guinea pig [155]. Two kinds of natural deletion mutants were reported in the C- terminus: 10 aa deletion at positions 93–102 and 11 aa deletion at positions 133–143 [154, 157]. The 133–143 deletion in 3A was observed in both cattle and pig isolates and does not affect the host range and virulence of FMDV [151]. By contrast, the 93–102 deletion in 3A is associated with high virulence in swine and is observed in a variant of FMDV serotype O isolated in Taiwan in 1997 (O/TAW/97). This deletion severely affected swine but did not spread to cattle [154, 157, 158], and also reduced virus replication efficiency in bovine cells but not in swine cells [159]. Similar deletion mutants containing 19 aa (O1C-O/E) to 20 aa (C3R-O/E) deletions in 3A protein were also observed in egg-adapted attenuated strains. These mutants displayed reduced virulence in cattle and were used for early vaccine development [154, 158]. Another artificial mutant containing 20 aa deletion at positions 87–106 of 3A demonstrated a significantly reduced replication ability and attenuated virulence in cattle [157]. Although its underlying mechanisms remain unclear, FMDV 3A is another viral protein affecting FMDV virulence, and some positions in 3A are associated with alterations in viral virulence and host range as indicated by the aforementioned studies.
In addition to 2C protein, 3ABC, the 3A precursor, is also used to differentiate potential carrier from vaccinated animals both in cattle and swine [160]. Actually, 3ABC is the most antigenic protein among all NSPs and is the best serological indicator of infection with FMDV [160]. Various ELISAs based on 3ABC antigen or antibody have been developed for discrimination between infected and non-infected animals regardless of their vaccination status [160–163].
3B Protein
Protein 3B, which is also known as VPg, is covalently bound to the 5ʹ terminus of the genome and antigenome and primes picornavirus RNA synthesis [8]. In contrast to other picornaviruse that encode a single copy of 3B, the FMDV 3B protein is unique because it exists in three similar but non-identical copies (3B1, 3B2, and 3B3), which are 23–24 aa long [164]. No natural FMDV strains have been reported to contain fewer than three copies of 3B [165], although not all three copies of FMDV 3B are needed to maintain infectivity [164], suggesting that there is a strong selective pressure towards maintaining this redundancy.
Uridylylation of the VPg peptide primer is the first stage in the replication of the picornavirus genome (Fig. 2). The picornavirus genome has a 5ʹ terminal feature of VPgpU(pU) covalently linked, which contributes to the use of VPg as a peptide primer to synthesize viral RNA. This peptide attaches to RNA via a conserved Tyr3 residue through the action of viral RNA polymerase (3Dpol). In this process, the viral 3Dpol catalyzes the binding of two uridine monophosphate (UMP) molecules to the hydroxyl group of this Tyr3 using as template a cis-replicating element (bus/cre) in FMDV genome [166]. Three isoforms of FMDV 3B (3B1, 3B2, and 3B3) can all be uridylylated in vitro, although 3B3 is likely the most efficient substrate for 3Dpol activity [167]. VPg peptide primer uridylylation can be performed in vitro using purified components, including VPg (3B) with 3Dpol, 3CD precursor, UTP, and an RNA template containing a stem-loop structure (bus) [167]. VPgpU(pU) are produced during this reaction and synthesis of positive- and negative-sense RNAs is initiated [166, 168].
A reverse genetics study demonstrated that deletion of the 3B3 coding sequence exerts a deleterious effect on FMDV RNA replication, resulting in production of a non-infectious RNA transcript [167, 169]. Laboratory recombinant virus lacking 3B1 and 3B2 also reduces viral RNA synthesis levels and infective particle formation in vitro, attenuats disease in pigs, but not drastically [170]. These studies indicated that 3B3 is more important than 3B1 and 3B2 to maintain viral RNA replication, but co-existence of all three 3B copies exerts the best RNA replication efficiency. Whereas the underlying mechanisms of their respective roles on viral RNA replication and how the three copies are integrated during viral replication still require further investigation.
3Cpro
FMDV 3C proteinase, responsible for most cleavages of viral polyprotein, was recently identified as a chymotrypsin-like cysteine protease [8, 171], although its function and catalytic residues were reported since 1995 [172]. Crystal structure and mutagenic research in FMDV demonstrated that a featured apolar surface loop containing a β-ribbon structure that folds over peptide binding cleft and clearly contributes to substrate recognition is important for catalytic activity. In addition, Cys142 in the Cys-His-Asp/Glu catalytic triad at the tip of the β-ribbon significantly affects enzyme activity [173, 174]. Except for the autocatalytic cleavage of Lpro from P1, 2A cleavage between P1-2A and 2BC, and maturation cleavage of VP0 to VP2 and VP4, 3Cpro can efficiently process all other 10 cleavage sites in FMDV polyprotein although the rate of cleavage varies at different junctions [175]. FMDV 3Cpro cleavage sites show great heterogeneity, with cleavage occurring between multiple dipeptides, including Gln-Gly, Glu-Gly, Gln-Leu, and Glu-Ser [176]. By contrast, in other picornaviruses, such as poliovirus, the 3Cpro cleavage site located exclusively between Gln-Gly and 3CDpro is implicated as the major viral proteinase in structural protein cleavage [176, 177].
FMDV 3Cpro is also associated with inhibition of host cell transcription and translation. As mentioned above, FMDV Lpro is involved in eIF4G cleavage, which shuts off host cell gene translation. FMDV 3Cpro can also cleave eIF4A, a portion of the cap-binding complex with the function of an RNA helicase [178]. Compared with Lpro, FMDV 3Cpro cleaves eIF4G late in the infection cycle at an alternative site, although it may not be favorable for the translation of viral proteins at this stage [8]. Moreover, 3Cpro is the agent that cleaves histone H3 following FMDV infection. Histone H3 is an important component of nucleosome and is crucial in maintaining the conformation of nucleosomes, thereby affecting cellular transcription [179]. FMDV 3Cpro also removes 20 N-terminal aa residues from histone H3, resulting in inhibition of host cell transcription [180].
Similar to other picornaviruses, FMDV 3Cpro can enter nuclei through its precursor 3CD, which contains a nuclear localization sequence (NLS) in the N-terminus of the 3Dpol protein [152]. 3Cpro also cleaves multiple factors and regulators. Recent studies found that FMDV 3Cpro directly cleaves the 68 kDa Src-associated substrate during mitosis (Sam68), one of the nuclear RNA-binding proteins that participate in viral replication in cells. Immunofluorescent and immunoblot assays revealed that 3Cpro removes the NLS-containing C-terminus of the Sam68 protein (~18 kDa). The truncated Sam68 (~50 kDa) was subsequently redistributed into the cytoplasm [181]. Moreover, cytosolic Sam68 directly interacts with the FMDV IRES and enhances the translation of the viral RNA [181].
Other reports associated FMDV 3Cpro with innate immune regulation (Fig. 4). Wang et al. provided direct evidence that FMDV 3Cpro proteolytically cleaves the nuclear transcription factor kappa B (NF-kB) essential modulator (NEMO), a bridging adaptor protein essential in activating both the NF-kB and IFN-regulatory factor signaling pathways [182], to reduce RIG-I/MDA5 signaling. They found that FMDV 3Cpro specifically targets the NEMO Gln383 residue, which lies between the C-terminal leucine zipper motif and a zinc finger (ZF) domain [182]. The ZF domain plays a crucial role in fully activating NF-kB and IRFs that orchestrate immune and inflammatory responses [183, 184].
In addition, the picornavirus 3Cpro cleaves many other factors and regulators associated with cellular DNA-dependent RNA polymerases I, II, and III, such as TATA-box binding protein, octamer-binding protein, transcription activator p53, cyclic AMP-responsive element binding protein, and DNA polymerase III [7]. Although many of them have not yet been confirmed for FMDV, these data indicated that 3Cpro may widely perturb gene transcription and translation in host cells.
3Dpol
FMDV 3D protein, the virus-encoded RNA-dependent RNA polymerase (RdRP) [185], is the catalytic component of RNA replication to synthesize positive- and negative-sense genome and plays an important role in the life cycle of RNA viruses (Fig. 2). 3Dpol sequences are highly conserved among the different sero- and subtypes of FMDV [186]. Picornaviruses use a precursor 3CD as a functional intermediate in viral replication, although 3CD contains an active 3C protease component. 3Dpol remains inactive until protein processing is complete [187].
Crystal structure analysis revealed that FMDV 3Dpol shares similar structure and catalytic mechanism to all other RNA virus-encoded RdRPs of several other families [187]. The overall structure of 3Dpol imaginatively resembles a cupped “right hand” consisting of “palm,” “fingers,” and “thumb” subdomains, which determine the correct geometrical arrangement of substrate molecules and metal ions at the active catalytic site [188]. The catalytic site of all RdRPs is contained in the palm domain. This domain is the most highly conserved feature of all known polynucleotide polymerases and is composed of five motifs, a three-stranded antiparallel core β sheet, and flanked by two α helices. This domain is involved in structural integrity, nucleotide recognition and binding, phosphoryl transfer, and priming nucleotide binding. By contrast, the thumb domain consists of the C-terminal region of the polypeptide chain and exhibits the most diverse feature among the known viral RdRPs [187].
Picornavirus replication is initiated in a primer-dependent manner. The protein primer VPg provides the hydroxyl nucleophile and forms a complex with 3Dpol or 3CD, termed VPg uridylylation complex, to initiate RNA replication in picornavirus [189, 190]. Biochemical and structural studies revealed that three distinct VPg binding sites on 3Dpol are present among different members of this family [191]. In FMDV, VPg binds to the residues in the active site cleft of the polymerase in the uridylylation reaction [190]. Whereas, in coxsackievirus B3 (CVB3), VPg is bound at the base of the thumb sub-domain [192], and in EV71, VPg is found anchored at the bottom of the palm domain of the polymerase [193]. The conformation of RdRPs changes in subtle ways to accomplish three steps cycle of replication elongation process, including nucleotide selection, phosphodiester bond formation and translocation to the next nucleotide for the subsequent round of nucleotide addition (reviewed in [191]).
The structure and biochemical activity of RdRP offer the opportunity to develop very selective anti-drugs against this viral enzyme in FMDV. Reports have indicated that some base and nucleotide analogues, particularly 5-fluorouracil and ribavirin. 5-Fluorouracil, a pyrimidine analogue, is mutagenic for a number of RNA viruses, including FMDV [194]. Ribavirin is also mutagenic for viral RNA polymerases [195] and is useful to eliminate FMDV from persistently infected cells via enhanced mutagenesis [196].
Conclusions and perspective
FMDV remains a severe pathogen that hampers modern agriculture worldwide, despite the extensive work has been conducted in the past several decades to control it. Traditional vaccine development is limited by the potential risk of virus transmission. Synthetic capsid vaccine, an important novel vaccine against virus infection is limited because of the high potential for genetic and antigenic variation [1, 5]. Specific strategies against FMDV, including antiviral drugs and novel vaccines, are still required in response to repeated epidemic events.
The NSPs and NCEs of FMDV play a critical role in viral proliferation and virus–host interaction. Studies on the involvement of these proteins and elements provided important insights into the molecular mechanism of virus-induced diseases. Nearly all NSPs and NCEs of FMDV are under investigation for their involvement in either virus detection or potential anti-viral strategy development [197–205], although progress is slow. FMDV is unique among the picornavirus family because of its long 5ʹ-UTR, three copies of 3B protein and short 2A protein lacking any protease motifs. Together with our most recent finding about the molecular mechanisms of FMDV entry into host cells [206], these properties possibly contribute to the virulence and pathogenesis of FMDV. In-depth work is still required to further understand the role of these proteins and elements in FMD pathogenesis and virus–host interactions. More extensive research will be helpful to uncover effective antiviral targets.
References
Knowles NJ, Samuel AR. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 2003;91:65–80.
Haydon DT, Samuel AR, Knowles NJ. The generation and persistence of genetic variation in foot-and-mouth disease virus. Prev Vet Med. 2001;51:111–24.
Domingo E, Ruiz-Jarabo CM, Sierra S, Arias A, Pariente N, Baranowski E, Escarmis C. Emergence and selection of RNA virus variants: memory and extinction. Virus Res. 2002;82:39–44.
Domingo E, Pariente N, Airaksinen A, Gonzalez-Lopez C, Sierra S, Herrera M, Grande-Perez A, Lowenstein PR, Manrubia SC, Lazaro E, Escarmis C. Foot-and-mouth disease virus evolution: exploring pathways towards virus extinction. Curr Top Microbiol Immunol. 2005;288:149–73.
Klein J. Understanding the molecular epidemiology of foot-and-mouth-disease virus. Infect Genet Evol. 2009;9:153–61.
Grubman MJ. The 5' end of foot-and-mouth disease virion RNA contains a protein covalently linked to the nucleotide pUp. Arch Virol. 1980;63:311–5.
Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. Viral and host proteins involved in picornavirus life cycle. J Biomed Sci. 2009;16:103.
Mason PW, Grubman MJ, Baxt B. Molecular basis of pathogenesis of FMDV. Virus Res. 2003;91:9–32.
Robertson BH, Grubman MJ, Weddell GN, Moore DM, Welsh JD, Fischer T, Dowbenko DJ, Yansura DG, Small B, Kleid DG. Nucleotide and amino acid sequence coding for polypeptides of foot-and-mouth disease virus type A12. J Virol. 1985;54:651–60.
Grubman MJ, Baxt B. Translation of foot-and-mouth disease virion RNA and processing of the primary cleavage products in a rabbit reticulocyte lysate. Virology. 1982;116:19–30.
Klump W, Marquardt O, Hofschneider PH. Biologically active protease of foot and mouth disease virus is expressed from cloned viral cDNA in Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:3351–5.
Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F. The three-dimensional structure of foot-and-mouth disease virus at 2.9 A resolution. Nature. 1989;337:709–16.
Fry EE, Newman JW, Curry S, Najjam S, Jackson T, Blakemore W, Lea SM, Miller L, Burman A, King AM, Stuart DI. Structure of Foot-and-mouth disease virus serotype A10 61 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation. J Gen Virol. 2005;86:1909–20.
Fry EE, Stuart DI, Rowlands DJ. The structure of foot-and-mouth disease virus. Curr Top Microbiol Immunol. 2005;288:71–101.
Han SC, Guo HC, Sun SQ. Three-dimensional structure of foot-and-mouth disease virus and its biological functions. Arch Virol. 2015;160:1–16.
Belsham GJ. Translation and replication of FMDV RNA. Curr Top Microbiol Immunol. 2005;288:43–70.
Zibert A, Maass G, Strebel K, Falk MM, Beck E. Infectious foot-and-mouth disease virus derived from a cloned full-length cDNA. J Virol. 1990;64:2467–73.
Carrillo C, Tulman ER, Delhon G, Lu Z, Carreno A, Vagnozzi A, Kutish GF, Rock DL. High throughput sequencing and comparative genomics of foot-and-mouth disease virus. Dev Biol (Basel). 2006;126:23–30. discussion 323.
Carrillo C, Tulman ER, Delhon G, Lu Z, Carreno A, Vagnozzi A, Kutish GF, Rock DL. Comparative genomics of foot-and-mouth disease virus. J Virol. 2005;79:6487–504.
Mason PW, Bezborodova SV, Henry TM. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J Virol. 2002;76:9686–94.
Gerber K, Wimmer E, Paul AV. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2A(pro). J Virol. 2001;75:10979–90.
Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–70.
Tiley L, King AM, Belsham GJ. The foot-and-mouth disease virus cis-acting replication element (cre) can be complemented in trans within infected cells. J Virol. 2003;77:2243–6.
Garcia-Nunez S, Gismondi MI, Konig G, Berinstein A, Taboga O, Rieder E, Martinez-Salas E, Carrillo E. Enhanced IRES activity by the 3'UTR element determines the virulence of FMDV isolates. Virology. 2014;448:303–13.
Kuhn R, Luz N, Beck E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1990;64:4625–31.
Fajardo Jr T, Rosas MF, Sobrino F, Martinez-Salas E. Exploring IRES region accessibility by interference of foot-and-mouth disease virus infectivity. PLoS One. 2012;7:e41382.
Lopez de Quinto S, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3'-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–405.
Fernandez N, Fernandez-Miragall O, Ramajo J, Garcia-Sacristan A, Bellora N, Eyras E, Briones C, Martinez-Salas E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res. 2011;39:8572–85.
Robertson ME, Seamons RA, Belsham GJ. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–79.
Fernandez N, Buddrus L, Pineiro D, Martinez-Salas E. Evolutionary conserved motifs constrain the RNA structure organization of picornavirus IRES. FEBS Lett. 2013;587:1353–8.
Fernandez-Miragall O, Martinez-Salas E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–44.
Saleh L, Rust RC, Fullkrug R, Beck E, Bassili G, Ochs K, Niepmann M. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth disease virus internal ribosome entry site. J Gen Virol. 2001;82:757–63.
de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A. 2009;106:9197–202.
Pilipenko EV, Gmyl AP, Maslova SV, Svitkin YV, Sinyakov AN, Agol VI. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–31.
Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008;16:230–7.
Belsham GJ, Brangwyn JK. A region of the 5' noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol. 1990;64:5389–95.
Lozano G, Fernandez N, Martinez-Salas E. Modeling Three-Dimensional Structural Motifs of Viral IRES. J Mol Biol. 2016;428:767–76.
Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45.
Lopez-Lastra M, Ramdohr P, Letelier A, Vallejos M, Vera-Otarola J, Valiente-Echeverria F. Translation initiation of viral mRNAs. Rev Med Virol. 2010;20:177–95.
Plank TD, Kieft JS. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA. 2012;3:195–212.
Stassinopoulos IA, Belsham GJ. A novel protein-RNA binding assay: functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA. 2001;7:114–22.
Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–65.
Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res. 2015;206:62–73.
Lopez de Quinto S, Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–92.
Lopez de Quinto S, Lafuente E, Martinez-Salas E. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–26.
Lozano G, Martinez-Salas E. Structural insights into viral IRES-dependent translation mechanisms. Curr Opin Virol. 2015;12:113–20.
Luz N, Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol. 1991;65:6486–94.
Luz N, Beck E. A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett. 1990;269:311–4.
Walter BL, Nguyen JH, Ehrenfeld E, Semler BL. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–85.
Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–67.
Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–45.
Pineiro D, Fernandez N, Ramajo J, Martinez-Salas E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 2013;41:1017–28.
Lin JY, Li ML, Shih SR. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009;37:47–59.
Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol. 2006;80:6936–42.
Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R, Martinez-Salas E. RNA-protein interaction methods to study viral IRES elements. Methods. 2015;91:3–12.
Andreev DE, Hirnet J, Terenin IM, Dmitriev SE, Niepmann M, Shatsky IN. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res. 2012;40:5602–14.
Serrano P, Pulido MR, Saiz M, Martinez-Salas E. The 3' end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5' end region. J Gen Virol. 2006;87:3013–22.
Saiz M, Gomez S, Martinez-Salas E, Sobrino F. Deletion or substitution of the aphthovirus 3' NCR abrogates infectivity and virus replication. J Gen Virol. 2001;82:93–101.
Rueckert RR, Wimmer E. Systematic nomenclature of picornavirus proteins. J Virol. 1984;50:957–9.
Clarke BE, Sangar DV, Burroughs JN, Newton SE, Carroll AR, Rowlands DJ. Two initiation sites for foot-and-mouth disease virus polyprotein in vivo. J Gen Virol. 1985;66(Pt 12):2615–26.
Medina M, Domingo E, Brangwyn JK, Belsham GJ. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–9.
Cao X, Bergmann IE, Fullkrug R, Beck E. Functional analysis of the two alternative translation initiation sites of foot-and-mouth disease virus. J Virol. 1995;69:560–3.
Piccone ME, Rieder E, Mason PW, Grubman MJ. The foot-and-mouth disease virus leader proteinase gene is not required for viral replication. J Virol. 1995;69:5376–82.
Lopez de Quinto S, Martinez-Salas E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255:324–36.
Roberts PJ, Belsham GJ. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology. 1995;213:140–6.
Guarne A, Tormo J, Kirchweger R, Pfistermueller D, Fita I, Skern T. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 1998;17:7469–79.
Piccone ME, Zellner M, Kumosinski TF, Mason PW, Grubman MJ. Identification of the active-site residues of the L proteinase of foot-and-mouth disease virus. J Virol. 1995;69:4950–6.
Strebel K, Beck E. A second protease of foot-and-mouth disease virus. J Virol. 1986;58:893–9.
Mason PW, Piccone ME, McKenna TS, Chinsangaram J, Grubman MJ. Evaluation of a live-attenuated foot-and-mouth disease virus as a vaccine candidate. Virology. 1997;227:96–102.
Chinsangaram J, Mason PW, Grubman MJ. Protection of swine by live and inactivated vaccines prepared from a leader proteinase-deficient serotype A12 foot-and-mouth disease virus. Vaccine. 1998;16:1516–22.
Brown CC, Piccone ME, Mason PW, McKenna TS, Grubman MJ. Pathogenesis of wild-type and leaderless foot-and-mouth disease virus in cattle. J Virol. 1996;70:5638–41.
Kirchweger R, Ziegler E, Lamphear BJ, Waters D, Liebig HD, Sommergruber W, Sobrino F, Hohenadl C, Blaas D, Rhoads RE, et al. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4 gamma. J Virol. 1994;68:5677–84.
Devaney MA, Vakharia VN, Lloyd RE, Ehrenfeld E, Grubman MJ. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J Virol. 1988;62:4407–9.
Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–10.
Chinsangaram J, Koster M, Grubman MJ. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J Virol. 2001;75:5498–503.
Chinsangaram J, Piccone ME, Grubman MJ. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J Virol. 1999;73:9891–8.
Wang D, Fang L, Liu L, Zhong H, Chen Q, Luo R, Liu X, Zhang Z, Chen H, Xiao S. Foot-and-mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon-lambda1 pathway. Mol Immunol. 2011;49:407–12.
Friedman RM, Grimley P, Baron S. Biological effects of the interferons and other cytokines. Biotherapy. 1996;8:189–98.
Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86.
de los Santos T, Segundo FD, Zhu J, Koster M, Dias CC, Grubman MJ. A conserved domain in the leader proteinase of foot-and-mouth disease virus is required for proper subcellular localization and function. J Virol. 2009;83:1800–10.
de Los Santos T, de Avila Botton S, Weiblen R, Grubman MJ. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J Virol. 2006;80:1906–14.
Wang D, Fang L, Luo R, Ye R, Fang Y, Xie L, Chen H, Xiao S. Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced type I interferon transcription by decreasing interferon regulatory factor 3/7 in protein levels. Biochem Biophys Res Commun. 2010;399:72–8.
Wang D, Fang L, Bi J, Chen Q, Cao L, Luo R, Chen H, Xiao S. Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced RANTES transcription in PK-15 cells. Virus Genes. 2011;42:388–93.
Barral PM, Morrison JM, Drahos J, Gupta P, Sarkar D, Fisher PB, Racaniello VR. MDA-5 is cleaved in poliovirus-infected cells. J Virol. 2007;81:3677–84.
Wang D, Fang L, Li P, Sun L, Fan J, Zhang Q, Luo R, Liu X, Li K, Chen H, et al. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol. 2011;85:3758–66.
Bibeau-Poirier A, Servant MJ. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine. 2008;43:359–67.
Isaacson MK, Ploegh HL. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–70.
Zhu Z, Yang F, Zhang K, Cao W, Jin Y, Wang G, Mao R, Li D, Guo J, Liu X, Zheng H. Comparative Proteomic Analysis of Wild-Type and SAP Domain Mutant Foot-and-Mouth Disease Virus-Infected Porcine Cells Identifies the Ubiquitin-Activating Enzyme UBE1 Required for Virus Replication. J Proteome Res. 2015;14:4194–206.
Hyrskyluoto A, Bruelle C, Lundh SH, Do HT, Kivinen J, Rappou E, Reijonen S, Waltimo T, Petersen A, Lindholm D, Korhonen L. Ubiquitin-specific protease-14 reduces cellular aggregates and protects against mutant huntingtin-induced cell degeneration: involvement of the proteasome and ER stress-activated kinase IRE1alpha. Hum Mol Genet. 2014;23:5928–39.
Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79:15199–208.
Kim ET, Oh SE, Lee YO, Gibson W, Ahn JH. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J Virol. 2009;83:12046–56.
Lindner HA, Lytvyn V, Qi H, Lachance P, Ziomek E, Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466:8–14.
Foeger N, Glaser W, Skern T. Recognition of eukaryotic initiation factor 4G isoforms by picornaviral proteinases. J Biol Chem. 2002;277:44300–9.
Donnelly ML, Gani D, Flint M, Monaghan S, Ryan MD. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J Gen Virol. 1997;78(Pt 1):13–21.
Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–25.
Donnelly ML, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D, Ryan MD. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol. 2001;82:1027–41.
Sharma P, Yan F, Doronina VA, Escuin-Ordinas H, Ryan MD, Brown JD. 2A peptides provide distinct solutions to driving stop-carry on translational recoding. Nucleic Acids Res. 2012;40:3143–51.
Gao ZL, Zhou JH, Zhang J, Ding YZ, Liu YS. The silent point mutations at the cleavage site of 2A/2B have no effect on the self-cleavage activity of 2A of foot-and-mouth disease virus. Infect Genet Evol. 2014;28:101–6.
Ryan MD, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–33.
Ryan MD, Belsham GJ, King AM. Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology. 1989;173:35–45.
Ao D, Sun SQ, Guo HC. Topology and biological function of enterovirus non-structural protein 2B as a member of the viroporin family. Vet Res. 2014;45:87.
Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat Rev Microbiol. 2012;10:563–74.
Ao D, Guo HC, Sun SQ, Sun DH, Fung TS, Wei YQ, Han SC, Yao XP, Cao SZ, Liu DX, Liu XT. Viroporin Activity of the Foot-and-Mouth Disease Virus Non-Structural 2B Protein. PLoS One. 2015;10:e0125828.
Moffat K, Knox C, Howell G, Clark SJ, Yang H, Belsham GJ, Ryan M, Wileman T. Inhibition of the secretory pathway by foot-and-mouth disease virus 2BC protein is reproduced by coexpression of 2B with 2C, and the site of inhibition is determined by the subcellular location of 2C. J Virol. 2007;81:1129–39.
Moffat K, Howell G, Knox C, Belsham GJ, Monaghan P, Ryan MD, Wileman T. Effects of foot-and-mouth disease virus nonstructural proteins on the structure and function of the early secretory pathway: 2BC but not 3A blocks endoplasmic reticulum-to-Golgi transport. J Virol. 2005;79:4382–95.
Giorda KM, Hebert DN. Viroporins customize host cells for efficient viral propagation. DNA Cell Biol. 2013;32:557–64.
Sheppard HM, Verdon D, Brooks AE, Feisst V, Ho YY, Lorenz N, Fan V, Birch NP, Didsbury A, Dunbar PR. MicroRNA regulation in human CD8+ T cell subsets--cytokine exposure alone drives miR-146a expression. J Transl Med. 2014;12:292.
Scott C, Griffin S. Viroporins: structure, function and potential as antiviral targets. J Gen Virol. 2015;96:2000–27.
Gorbalenya AE, Koonin EV. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989;17:8413–40.
Teterina NL, Gorbalenya AE, Egger D, Bienz K, Rinaudo MS, Ehrenfeld E. Testing the modularity of the N-terminal amphipathic helix conserved in picornavirus 2C proteins and hepatitis C NS5A protein. Virology. 2006;344:453–67.
Klein M, Hadaschik D, Zimmermann H, Eggers HJ, Nelsen-Salz B. The picornavirus replication inhibitors HBB and guanidine in the echovirus-9 system: the significance of viral protein 2C. J Gen Virol. 2000;81:895–901.
Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64:1156–63.
Gosert R, Egger D, Bienz K. A cytopathic and a cell culture adapted hepatitis A virus strain differ in cell killing but not in intracellular membrane rearrangements. Virology. 2000;266:157–69.
Jecht M, Probst C, Gauss-Muller V. Membrane permeability induced by hepatitis A virus proteins 2B and 2BC and proteolytic processing of HAV 2BC. Virology. 1998;252:218–27.
Bolten R, Egger D, Gosert R, Schaub G, Landmann L, Bienz K. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent In situ hybridization. J Virol. 1998;72:8578–85.
Troxler M, Egger D, Pfister T, Bienz K. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology. 1992;191:687–97.
Tesar M, Berger HG, Marquardt O. Serological probes for some foot-and-mouth disease virus nonstructural proteins. Virus Genes. 1989;3:29–44.
Wang J, Wang Y, Liu J, Ding L, Zhang Q, Li X, Cao H, Tang J, Zheng SJ. A critical role of N-myc and STAT interactor (Nmi) in foot-and-mouth disease virus (FMDV) 2C-induced apoptosis. Virus Res. 2012;170:59–65.
Echeverri A, Banerjee R, Dasgupta A. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 1998;54:217–23.
Lippincott-Schwartz J. Bidirectional membrane traffic between the endoplasmic reticulum and Golgi apparatus. Trends Cell Biol. 1993;3:81–8.
Schlegel A, Giddings Jr TH, Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–88.
Teterina NL, Gorbalenya AE, Egger D, Bienz K, Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–72.
Saunders K, King AM. Guanidine-resistant mutants of aphthovirus induce the synthesis of an altered nonstructural polypeptide, P34. J Virol. 1982;42:389–94.
Saunders K, King AM, McCahon D, Newman JW, Slade WR, Forss S. Recombination and oligonucleotide analysis of guanidine-resistant foot-and-mouth disease virus mutants. J Virol. 1985;56:921–9.
Pfister T, Wimmer E. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J Biol Chem. 1999;274:6992–7001.
Barton DJ, Flanegan JB. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–9.
Lubroth J, Brown F. Identification of native foot-and-mouth disease virus non-structural protein 2C as a serological indicator to differentiate infected from vaccinated livestock. Res Vet Sci. 1995;59:70–8.
Lubroth J, Grubman MJ, Burrage TG, Newman JF, Brown F. Absence of protein 2C from clarified foot-and-mouth disease virus vaccines provides the basis for distinguishing convalescent from vaccinated animals. Vaccine. 1996;14:419–27.
Lu Z, Zhang X, Fu Y, Cao Y, Tian M, Sun P, Li D, Liu Z, Xie Q. Expression of the major epitope regions of 2C integrated with the 3AB non-structural protein of foot-and-mouth disease virus and its potential for differentiating infected from vaccinated animals. J Virol Methods. 2010;170:128–33.
Meyer RF, Babcock GD, Newman JF, Burrage TG, Toohey K, Lubroth J, Brown F. Baculovirus expressed 2C of foot-and-mouth disease virus has the potential for differentiating convalescent from vaccinated animals. J Virol Methods. 1997;65:33–43.
Banerjee R, Dasgupta A. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J Gen Virol. 2001;82:2621–7.
Rodriguez PL, Carrasco L. Poliovirus protein 2C contains two regions involved in RNA binding activity. J Biol Chem. 1995;270:10105–12.
Adams P, Kandiah E, Effantin G, Steven AC, Ehrenfeld E. Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J Biol Chem. 2009;284:22012–21.
Rodriguez PL, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–10.
Tang WF, Yang SY, Wu BW, Jheng JR, Chen YL, Shih CH, Lin KH, Lai HC, Tang P, Horng JT. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J Biol Chem. 2007;282:5888–98.
Sweeney TR, Cisnetto V, Bose D, Bailey M, Wilson JR, Zhang X, Belsham GJ, Curry S. Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism. J Biol Chem. 2010;285:24347–59.
Zheng W, Li X, Wang J, Li X, Cao H, Wang Y, Zeng Q, Zheng SJ. A critical role of interferon-induced protein IFP35 in the type I interferon response in cells induced by foot-and-mouth disease virus (FMDV) protein 2C. Arch Virol. 2014;159:2925–35.
Fillmore RA, Mitra A, Xi Y, Ju J, Scammell J, Shevde LA, Samant RS. Nmi (N-Myc interactor) inhibits Wnt/beta-catenin signaling and retards tumor growth. Int J Cancer. 2009;125:556–64.
Chen J, Naumovski L. Intracellular redistribution of interferon-inducible proteins Nmi and IFP 35 in apoptotic cells. J Interferon Cytokine Res. 2002;22:237–43.
Zhang L, Tang Y, Tie Y, Tian C, Wang J, Dong Y, Sun Z, He F. The PH domain containing protein CKIP-1 binds to IFP35 and Nmi and is involved in cytokine signaling. Cell Signal. 2007;19:932–44.
Chen J, Shpall RL, Meyerdierks A, Hagemeier M, Bottger EC, Naumovski L. Interferon-inducible Myc/STAT-interacting protein Nmi associates with IFP 35 into a high molecular mass complex and inhibits proteasome-mediated degradation of IFP 35. J Biol Chem. 2000;275:36278–84.
Lee ND, Chen J, Shpall RL, Naumovski L. Subcellular localization of interferon-inducible Myc/stat-interacting protein Nmi is regulated by a novel IFP 35 homologous domain. J Interferon Cytokine Res. 1999;19:1245–52.
Zhou X, Liao J, Meyerdierks A, Feng L, Naumovski L, Bottger EC, Omary MB. Interferon-alpha induces nmi-IFP35 heterodimeric complex formation that is affected by the phosphorylation of IFP35. J Biol Chem. 2000;275:21364–71.
Ku BK, Kim SB, Moon OK, Lee SJ, Lee JH, Lyoo YS, Kim HJ, Sur JH. Role of apoptosis in the pathogenesis of Asian and South American foot-and-mouth disease viruses in swine. J Vet Med Sci. 2005;67:1081–8.
de Los ST, Diaz-San Segundo F, Grubman MJ. Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J Virol. 2007;81:12803–15.
Diaz-San Segundo F, Salguero FJ, de Avila A, de Marco MM, Sanchez-Martin MA, Sevilla N. Selective lymphocyte depletion during the early stage of the immune response to foot-and-mouth disease virus infection in swine. J Virol. 2006;80:2369–79.
O'Donnell V, Pacheco JM, LaRocco M, Burrage T, Jackson W, Rodriguez LL, Borca MV, Baxt B. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology. 2011;410:142–50.
Gannage M, Ramer PC, Munz C. Targeting Beclin 1 for viral subversion of macroautophagy. Autophagy. 2010;6:166–7.
Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80.
Gladue DP, O'Donnell V, Baker-Branstetter R, Holinka LG, Pacheco JM, Fernandez-Sainz I, Lu Z, Brocchi E, Baxt B, Piccone ME, et al. Foot-and-mouth disease virus nonstructural protein 2C interacts with Beclin1, modulating virus replication. J Virol. 2012;86:12080–90.
Knowles NJ, Davies PR, Henry T, O'Donnell V, Pacheco JM, Mason PW. Emergence in Asia of foot-and-mouth disease viruses with altered host range: characterization of alterations in the 3A protein. J Virol. 2001;75:1551–6.
Garcia-Briones M, Rosas MF, Gonzalez-Magaldi M, Martin-Acebes MA, Sobrino F, Armas-Portela R. Differential distribution of non-structural proteins of foot-and-mouth disease virus in BHK-21 cells. Virology. 2006;349:409–21.
Rosas MF, Vieira YA, Postigo R, Martin-Acebes MA, Armas-Portela R, Martinez-Salas E, Sobrino F. Susceptibility to viral infection is enhanced by stable expression of 3A or 3AB proteins from foot-and-mouth disease virus. Virology. 2008;380:34–45.
Beard CW, Mason PW. Genetic determinants of altered virulence of Taiwanese foot-and-mouth disease virus. J Virol. 2000;74:987–91.
Nunez JI, Baranowski E, Molina N, Ruiz-Jarabo CM, Sanchez C, Domingo E, Sobrino F. A single amino acid substitution in nonstructural protein 3A can mediate adaptation of foot-and-mouth disease virus to the guinea pig. J Virol. 2001;75:3977–83.
Harris JR, Racaniello VR. Amino acid changes in proteins 2B and 3A mediate rhinovirus type 39 growth in mouse cells. J Virol. 2005;79:5363–73.
Pacheco JM, Gladue DP, Holinka LG, Arzt J, Bishop E, Smoliga G, Pauszek SJ, Bracht AJ, O'Donnell V, Fernandez-Sainz I, et al. A partial deletion in non-structural protein 3A can attenuate foot-and-mouth disease virus in cattle. Virology. 2013;446:260–7.
Giraudo AT, Beck E, Strebel K, de Mello PA, La Torre JL, Scodeller EA, Bergmann IE. Identification of a nucleotide deletion in parts of polypeptide 3A in two independent attenuated aphthovirus strains. Virology. 1990;177:780–3.
Ma X, Li P, Bai X, Sun P, Bao H, Lu Z, Cao Y, Li D, Chen Y, Qiao Z, Liu Z. Sequences outside that of residues 93-102 of 3A protein can contribute to the ability of foot-and-mouth disease virus (FMDV) to replicate in bovine-derived cells. Virus Res. 2014;191:161–71.
De Diego M, Brocchi E, Mackay D, De Simone F. The non-structural polyprotein 3ABC of foot-and-mouth disease virus as a diagnostic antigen in ELISA to differentiate infected from vaccinated cattle. Arch Virol. 1997;142:2021–33.
Brocchi E, Bergmann IE, Dekker A, Paton DJ, Sammin DJ, Greiner M, Grazioli S, De Simone F, Yadin H, Haas B, et al. Comparative evaluation of six ELISAs for the detection of antibodies to the non-structural proteins of foot-and-mouth disease virus. Vaccine. 2006;24:6966–79.
Clavijo A, Hole K, Li M, Collignon B. Simultaneous detection of antibodies to foot-and-mouth disease non-structural proteins 3ABC, 3D, 3A and 3B by a multiplexed Luminex assay to differentiate infected from vaccinated cattle. Vaccine. 2006;24:1693–704.
Sorensen KJ, de Stricker K, Dyrting KC, Grazioli S, Haas B. Differentiation of foot-and-mouth disease virus infected animals from vaccinated animals using a blocking ELISA based on baculovirus expressed FMDV 3ABC antigen and a 3ABC monoclonal antibody. Arch Virol. 2005;150:805–14.
Pacheco JM, Henry TM, O'Donnell VK, Gregory JB, Mason PW. Role of nonstructural proteins 3A and 3B in host range and pathogenicity of foot-and-mouth disease virus. J Virol. 2003;77:13017–27.
Carrillo C, Lu Z, Borca MV, Vagnozzi A, Kutish GF, Rock DL. Genetic and phenotypic variation of foot-and-mouth disease virus during serial passages in a natural host. J Virol. 2007;81:11341–51.
Paul AV, Wimmer E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015;206:12–26.
Nayak A, Goodfellow IG, Belsham GJ. Factors required for the Uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro. J Virol. 2005;79:7698–706.
Nayak A, Goodfellow IG, Woolaway KE, Birtley J, Curry S, Belsham GJ. Role of RNA structure and RNA binding activity of foot-and-mouth disease virus 3C protein in VPg uridylylation and virus replication. J Virol. 2006;80:9865–75.
Falk MM, Sobrino F, Beck E. VPg gene amplification correlates with infective particle formation in foot-and-mouth disease virus. J Virol. 1992;66:2251–60.
Pacheco JM, Piccone ME, Rieder E, Pauszek SJ, Borca MV, Rodriguez LL. Domain disruptions of individual 3B proteins of foot-and-mouth disease virus do not alter growth in cell culture or virulence in cattle. Virology. 2010;405:149–56.
Birtley JR, Knox SR, Jaulent AM, Brick P, Leatherbarrow RJ, Curry S. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J Biol Chem. 2005;280:11520–7.
Grubman MJ, Zellner M, Bablanian G, Mason PW, Piccone ME. Identification of the active-site residues of the 3C proteinase of foot-and-mouth disease virus. Virology. 1995;213:581–9.
Zunszain PA, Knox SR, Sweeney TR, Yang J, Roque-Rosell N, Belsham GJ, Leatherbarrow RJ, Curry S. Insights into cleavage specificity from the crystal structure of foot-and-mouth disease virus 3C protease complexed with a peptide substrate. J Mol Biol. 2010;395:375–89.
Sweeney TR, Roque-Rosell N, Birtley JR, Leatherbarrow RJ, Curry S. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J Virol. 2007;81:115–24.
Bablanian GM, Grubman MJ. Characterization of the foot-and-mouth disease virus 3C protease expressed in Escherichia coli. Virology. 1993;197:320–7.
Palmenberg AC. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–23.
Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–70.
Belsham GJ, McInerney GM, Ross-Smith N. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol. 2000;74:272–80.
Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26.
Tesar M, Marquardt O. Foot-and-mouth disease virus protease 3C inhibits cellular transcription and mediates cleavage of histone H3. Virology. 1990;174:364–74.
Lawrence P, Schafer EA, Rieder E. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology. 2012;425:40–52.
Wang D, Fang L, Li K, Zhong H, Fan J, Ouyang C, Zhang H, Duan E, Luo R, Zhang Z, et al. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J Virol. 2012;86:9311–22.
Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600.
Yang F, Yamashita J, Tang E, Wang HL, Guan K, Wang CY. The zinc finger mutation C417R of I-kappa B kinase gamma impairs lipopolysaccharide- and TNF-mediated NF-kappa B activation through inhibiting phosphorylation of the I-kappa B kinase beta activation loop. J Immunol. 2004;172:2446–52.
Robertson BH, Morgan DO, Moore DM, Grubman MJ, Card J, Fischer T, Weddell G, Dowbenko D, Yansura D. Identification of amino acid and nucleotide sequence of the foot-and-mouth disease virus RNA polymerase. Virology. 1983;126:614–23.
George M, Venkataramanan R, Pattnaik B, Sanyal A, Gurumurthy CB, Hemadri D, Tosh C. Sequence analysis of the RNA polymerase gene of foot-and-mouth disease virus serotype Asia1. Virus Genes. 2001;22:21–6.
Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34.
Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem. 2004;279:47212–21.
van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol. 2004;85:1077–93.
Ferrer-Orta C, Arias A, Agudo R, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. 2006;25:880–8.
Ferrer-Orta C, Ferrero D, Verdaguer N. RNA-Dependent RNA Polymerases of Picornaviruses: From the Structure to Regulatory Mechanisms. Viruses. 2015;7:4438–60.
Gruez A, Selisko B, Roberts M, Bricogne G, Bussetta C, Jabafi I, Coutard B, De Palma AM, Neyts J, Canard B. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J Virol. 2008;82:9577–90.
Chen C, Wang Y, Shan C, Sun Y, Xu P, Zhou H, Yang C, Shi PY, Rao Z, Zhang B, Lou Z. Crystal structure of enterovirus 71 RNA-dependent RNA polymerase complexed with its protein primer VPg: implication for a trans mechanism of VPg uridylylation. J Virol. 2013;87:5755–68.
Sierra S, Davila M, Lowenstein PR, Domingo E. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J Virol. 2000;74:8316–23.
Graci JD, Cameron CE. Quasispecies, error catastrophe, and the antiviral activity of ribavirin. Virology. 2002;298:175–80.
Airaksinen A, Pariente N, Menendez-Arias L, Domingo E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology. 2003;311:339–49.
Kumar R, Hosamani M, Sreenivasa BP, Kotyal A, Venkataramanan R. Expression of Foot-and-Mouth Disease Virus Non-Structural Protein, 3D in Insect Cells and its Application in Detection of Anti-FMDV Antibodies. Indian J Virol. 2012;23:326–32.
Almeida MR, Rieder E, Chinsangaram J, Ward G, Beard C, Grubman MJ, Mason PW. Construction and evaluation of an attenuated vaccine for foot-and-mouth disease: difficulty adapting the leader proteinase-deleted strategy to the serotype O1 virus. Virus Res. 1998;55:49–60.
Rodriguez-Calvo T, Diaz-San Segundo F, Sanz-Ramos M, Sevilla N. A replication analysis of foot-and-mouth disease virus in swine lymphoid tissue might indicate a putative carrier stage in pigs. Vet Res. 2011;42:22.
Horsington J, Zhang Z. Analysis of foot-and-mouth disease virus replication using strand-specific quantitative RT-PCR. J Virol Methods. 2007;144:149–55.
He C, Wang H, Wei H, Yan Y, Zhao T, Hu X, Luo P, Wang L, Yu Y. A recombinant truncated FMDV 3AB protein used to better distinguish between infected and vaccinated cattle. Vaccine. 2010;28:3435–9.
Nogueira Santos JA, Assis DM, Gouvea IE, Judice WA, Izidoro MA, Juliano MA, Skern T, Juliano L. Foot and mouth disease leader protease (Lbpro): Investigation of prime side specificity allows the synthesis of a potent inhibitor. Biochimie. 2012;94:711–8.
Guo C, Zhang C, Zheng H, Huang Y. Recombinant adenovirus expression of FMDV P1-2A and 3C protein and its immune response in mice. Res Vet Sci. 2013;95:736–41.
Dory D, Remond M, Beven V, Cariolet R, Zientara S, Jestin A. Foot-and-Mouth Disease Virus neutralizing antibodies production induced by pcDNA3 and Sindbis virus based plasmid encoding FMDV P1-2A3C3D in swine. Antiviral Res. 2009;83:45–52.
Chang Y, Dou Y, Bao H, Luo X, Liu X, Mu K, Liu Z, Liu X, Cai X. Multiple microRNAs targeted to internal ribosome entry site against foot-and-mouth disease virus infection in vitro and in vivo. Virol J. 2014;11:1.
Han SC, Guo HC, Sun SQ, Jin Y, Wei YQ, Feng X, Yao XP, Cao SZ, Xiang Liu D, Liu XT. Productive Entry of Foot-and-Mouth Disease Virus via Macropinocytosis Independent of Phosphatidylinositol 3-Kinase. Sci Rep. 2016;6:19294.
Acknowledgments
This work was supported by grants from the National Science and Technology Support Program (2013BAD12B00), International Science & Technology Cooperation Program of China (2014DFA31890), Fundamental Research Funds for the Chinese Academy of Agricultural Sciences (2015ZL062), and China Postdoctoral Science Foundation (2016M591314). We are grateful to Shichong Han (China Agricultural University) for his help with image processing. We also would like to thank Liping Yang (University of Maryland) for critical reading of this manuscript. We apologize to the people whose work cannot be cited due to space limitation.
Authors’ contributions
YG and SQ S provided the concept and study information. YG and SQ S also drafted the manuscript. HC G provided the final approval of the version for submission. All of the authors approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gao, Y., Sun, SQ. & Guo, HC. Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements. Virol J 13, 107 (2016). https://doi.org/10.1186/s12985-016-0561-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-016-0561-z