Abstract
Background
The objectives of this study were to investigate the relationships between polymorphisms at the interferon lambda (IFNL) locus and CD4+:CD8+ ratio normalisation in people living with HIV (PLWH) on effective antiretroviral therapy (ART); and to examine whether these polymorphisms influence the composition of T lymphocyte compartments in long-term treated HIV-1 infection.
Methods
A cross-sectional study in PLWH enrolled into the Mater Immunology study. We performed IFNL genotyping on stored samples and evaluated the association of IFNL single-nucleotide polymorphisms (rs368234815 and rs12979860) with CD4+:CD8+ ratio normalization (> 1) and expanded CD4+ and CD8+ T-cell subsets; CD45RO+CD62L+ (central-memory), CD45RO+ CD62L−(effector-memory) and CD45RO−CD62L+ (naïve), using logistic and linear regression models, respectively.
Results
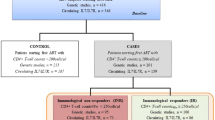
190 ambulatory PLWH recruited to the main study, 143 were included in the analysis (38 had no stored DNA and 9 no T-lymphocyte subpopulation). Of 143 included, the median age (IQR) was 45(39–48) years, 64% were male and 66% were of Caucasian ethnicity. Heterosexual-contact (36%), injecting drug-use (33%) and men who have sex with men (24%) were the most presented HIV-transmission risk groups. The majority of subjects (90.2%) were on ART with 79% of the cohort having an undetectable HIV-RNA (< 40 copies/ml) and the time since ART initiation was 7.5 (3.7–10.4) year. rs368234815 and rs12979860 displayed similar allelic frequencies, with minor alleles ΔG and T representing 39% and 42%, respectively, of circulating alleles. rs368234815 ΔG/ΔG minor homozygotes were significantly associated with increased odds for attaining a normalised CD4+:CD8+ ratio compared to rs368234815 T/T major homozygotes in PLWH virologically suppressed on effective ART (OR = 3.11; 95% CI [1.01:9.56]). rs368234815 ΔG/ΔG homozygosity was also significantly associated with lower levels of CD4+ effector memory T-cells (regression coefficient: − 7.1%, p = 0.04) and CD8+ naïve T-cell subsets were significantly higher in HIV-1 mono-infected PLWH with rs368234815 ΔG/ΔG (regression coefficient: + 7.2%, p = 0.04).
Conclusions
In virally-suppressed, long-term ART-treated PLWH, rs368234815 ΔG/ΔG homozygotes were more likely to have attained normalisation of their CD4+:CD8+ ratio, displayed lower CD4+ effector memory and higher naive CD8+ T-cells. Further studies are needed to replicate our findings in other, larger and more diverse cohorts and to determine the impact of IFNL genetic-variation on CD4+:CD8+ ratio normalisation and clinical outcomes in PLWH.
Similar content being viewed by others
Introduction
With advances in antiretroviral therapy (ART), HIV-infection has become a chronic, manageable condition. For the majority of individuals, infection with HIV-1 results in inversion of the normal CD4+:CD8+ ratio and, despite effective ART, only about 30% of subjects on ART for more than 5 years achieve normalisation of their CD4+:CD8+ T-cell ratios to > 1 [1]. The clinical implications of persistently low CD4+:CD8+ T-cell ratio are increasingly recognized, with lower ratios associated with greater risk of non-AIDS morbidity and mortality [2, 3]. As a result, attention has turned to understanding factors that influence poorer immune responses to ART, including failure to normalise the CD4+:CD8+ T-cell ratio. Initiation of ART early after HIV acquisition or at higher CD4+ T-cell counts is associated with higher subsequent CD4+:CD8+ ratios [4,5,6], and ART containing the non-nucleoside reverse transcriptase inhibitor efavirenz [7] or integrase strand transfer inhibitors (InSTI) [8] also show a stronger association with T-cell ratio normalisation compared to other ART regimens
More recently, the role of the host immune system in determining CD4+:CD8+ T-cell ratio responses to ART have also been explored. Two cohort studies have demonstrated that subjects with higher naïve CD8+ T-cell populations had higher CD4+:CD8+ T-cell ratios on ART [9, 10], while higher CD4+:CD8+ T-cell ratios prior to infection or the presence of human leukocyte antigen (HLA)-A*74:01 in an African cohort [11] have also been reported to positively influence normalisation of CD4+:CD8+ T-cell ratios. Conversely, co-infections with other viruses such as cytomegalovirus (CMV) and hepatitis C virus (HCV) [4, 5, 12], or the CD4+ T-cell apoptosis-inducing phenotype of the HIV Env glycoprotein envelope proteins and immune activation [13], all have been reported to decrease the likelihood of CD4+:CD8+ T-cell ratio normalisation.
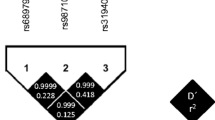
Host genetics can also influence immunological responses to viral infections. Several polymorphisms near the type III interferon lambda (IFNL3) locus, previously named interleukin 28B (IL28B), are able to predict both spontaneous clearance of hepatitis C infection by the host as well as better responses to pegylated interferon-alpha/ribavirin (PEG/RBV) therapy for HCV infection [14,15,16,17]. Similarly, the rs12979860 C>T single nucleotide polymorphism (SNP) is associated with higher HCV RNA titers and improved response to PEG/RBV therapy in those carrying the CC major homozygous genotype [14, 18, 19]. The rs12979860 polymorphism displays high linkage disequilibrium with a dinucleotide/deletion variant (TT/∆G) rs368234815 (TT>ΔG) [20]. While the TT/TT genotype represents a frameshift that abrogates IFNL4 expression, presence of ∆G allows for expression of IFNL4 [20]. IFNLs play a key role in innate immunity and antimicrobial defense of mucosal tissues [21] and recent data shows an effect on adaptive immune responses by both induction of thymic stromal lymphopoietin (TSLP) and effects on migratory dendritic cells [22]. Presence of the minor allele rs368234815 (ΔG) has been associated with poorer immune responses to CMV infection and higher risk of CMV retinitis in a cohort with advanced HIV-1 infection (nadir CD4+ T-cell counts of < 100 cells /mm3) [23], while rs368234815 (ΔG/ΔG) homozygous individuals undergoing solid organ transplant from CMV seropositive donors had a significantly higher cumulative incidence of CMV reactivation [24].
Whether expression of IFNL4 in HIV-1 infection modifies immune responses, particularly in those PLWH on effective ART, remains unclear, with contrasting observations reported. The presence of rs12979860 polymorphisms did not influence HIV acquisition in cohorts of individuals at high risk for HIV [25, 26] and was not over-represented within a cohort of African-American subjects with HIV who were classified as elite controllers [27]. In contrast, injecting drug users with HCV infection carrying the major homozygous rs368234815 (T/T) alleles were less likely to acquire HIV, [28] while presence of the rs368234815 ΔG allele was associated with higher prevalence of AIDS-defining illnesses and lower CD4+ T-cell counts in a cohort of asymptomatic, ART-naïve PLWH [29].
While IFNLs have been shown to skew T lymphocyte responses, their impact in treated HIV, in particular the potential for IFNL4 expression to modulate the CD4+:CD8+ T-cell ratio in the context of treated HIV-1 infection has not been explored to date. The objectives of the present study were to: (i) investigate relationships between polymorphisms at the IFNL locus and CD4+:CD8+ ratio normalisation in PLWH on effective ART; and (ii) examine whether these polymorphisms influence the composition of T lymphocyte compartments in long-term treated HIV-1 infection.
Methods
Study cohort and blood collection
We conducted a cross-sectional analysis exploring IFNL polymorphisms within a single-centre, prospective cohort study of PLWH designed to assess associations between CD4+ and CD8+ T lymphocyte subsets and CD4+:CD8+ T-cell ratio normalisation [9]. Briefly, the prospective cohort enrolled consecutive adult (>18 years old) PLWH attending the Mater Misericordiae University Hospital (MMUH) infectious diseases outpatients services for routine HIV clinical care, in Dublin, Ireland.
Enrolled subjects provided ethylenediaminetetraacetic acid (EDTA) blood used to determine expanded CD4+ and CD8+ T-cell subsets by flow cytometry; CD45RO-CD62L+ (naïve), CD45RO+CD62L+ (central memory) and CD45RO+CD62L− (effector memory) (Additional file 1: Figure S1).alongside routine T-cell subsets (absolute and percentage CD4+ and CD8+ counts) and samples for storage. Additional assessments included measurements of HIV-RNA and collection of demographic and treatment data. The cross-sectional analysis was conducted on data derived from study entry [9]. All enrolled subjects provided written, informed consent and the study was approved by the Mater Misericordiae University Hospital and Mater Private Hospital Institutional Review Board.
IFNL genotyping
Genomic deoxyribonucleic acid (DNA) was extracted from stored buffy coat cell pellets on the automated Magnapure 96 platform (Roche). IFNL genotyping was performed employing TaqMan SNP genotyping allelic discrimination on the ABI 7500 Fast instrument (Applied Biosystems, Warrington, United Kingdom). Assay conditions and thermocycling parameters for allelic discrimination real-time polymerase chain reaction (PCR) were as previously described for rs12979860 and validated by Sanger sequencing [18] and the rs368234815 SNP genotyping assay was supplied by Applied Biosystems. All genotyping was undertaken with blinding to clinical variables. The oligonucleotide primers and hydrolysis probes were as follows: rs12979860: rs12979860_F 5′GCCTGTCGTGTACTGAACCA; rs12979860_R 5′ GCGCGGAGTGCAATTCAAC; C allele probe (rs12979860_VIC) 5′VIC-TGGTTCGCGCCTTC-MGBNFQ; T allele probe (rs12979860_FAM) 5′FAM-CTGGTTCACGCCTTC-MGBNFQ; rs368234815: rs368234815_F 5′ CTCCAGCGAGCGGTAGTG; rs368234815_R 5′GGGTCCTGTGCACGGT; TT allele probe (ss469415590IF_VI) 5′VIC-ATCGCAGAAGGCC-MGBNFQ; ΔG allele probe (ss469415590IF_FAM) 5′FAM- ATCGCAGCGGCCC-MGBNFQ.
Statistical analysis
Subject characteristics were summarised using descriptive statistics: median and interquartile range (IQR) for continuous, non-normal variables and frequencies/percentages for categorical variables. Accordance of the genotype frequencies with Hardy–Weinberg equilibrium was confirmed using Chi square test for each SNP. Regression models for assessing associations were restricted to those PLWH virologically suppressed on effective ART (HIV-RNA < 40 copies/ml). The genotype of each SNP was scored using an additive model: 0, homozygous for a major allele, 1, heterozygous for a major allele, and 2, homozygous for a minor allele.
As both presence of HIV viraemia and hepatitis C co-infection may impact on relationships between immune markers and genetic polymorphisms of interest, we analysed three main groups; the total cohort with available DNA, a subset of the cohort who were on ART with an undetectable HIV-RNA and a smaller cohort on ART without hepatitis C co-infection. Logistic regression models were used to examine the association of CD4+:CD8+ T-cell ratio normalisation (CD4+:CD8+ T-cell ratio ≥ 1 versus CD4+:CD8+ T-cell ratio < 1) with each genetic polymorphism. Linear regression analysis was used to investigate the association of CD4+ and CD8+ T-cell subsets (naïve, central memory and effector memory T-cells) with each genetic polymorphism. All tests were two-sided and analyses were performed using Stata 13 (StataCorp, College Station, Texas).
Results
Characteristics of the study subjects
Of 190 ambulatory PLWH recruited to the main study previously described [9], 152 with stored DNA were included in the present study. Of these, 9 (5%) did not have T lymphocyte subpopulation data available and were excluded from further analysis. The demographic characteristics of the 143 remaining subjects broadly reflected the European HIV epidemic, with male and female genders (63.6% male), major HIV transmission groups (heterosexual, injecting drug-users (IDU), men who have sex with men (MSM), needle stick) as well as Caucasian (65.7%) and African (28%) ethnicities represented. The majority of subjects (90.2%) were on ART with 79% of the cohort having an undetectable HIV-RNA (< 40 copies/ml) at the time of sampling. The median (IQR) time since HIV diagnosis and ART initiation were 10 [6,7,8,9,10,11,12,13,14] years and 7.5 (3.7–10.4) years respectively.
Subjects were genotyped for polymorphisms within the IFNL locus. Of the two genotypes tested, rs368234815 and rs12979860 displayed similar allelic frequencies, with minor alleles ΔG and T representing 39% and 42%, respectively, of circulating alleles. The minor homozygotes rs368234815 ΔG/ΔG and rs12979860 T/T showed comparable frequencies of 20% and 17%, respectively. The rs12979860 genotype had a higher number of heterozygous C/T genotypes (51%), with C/C major homozygotes detected in 32% of the subjects. Conversely, rs368234815 T/T major homozygosity was present in 43% of the subjects, with the remaining 37% carrying both alleles. Allele distribution did not appreciably change between the main study population and the two sub-populations studied (Table 1). There were no gender differences in the allele distribution for both rs368234815 (p = 0.12) and rs12979860 (p = 0.21). Compared to Caucasian ethnicity, non-Caucasians had significantly higher proportion of the minor homozygotes rs368234815 ΔG/ΔG (10% vs 41%, p < 0.001) and rs12979860 T/T (9% vs 33%, p < 0.001).
Relationship between IFNL locus polymorphism and CD4+: CD8+ T-cell ratio normalisation
In the study cohort, 33/143 (23%) PLWH had CD4+:CD8+ T-cell ratio ≥ 1. Compared to those with a non-normalised CD4+:CD8+ T-cell ratio, subjects who had a normalised T-cell ratio were more likely (74.6% versus 93.6%, p = 0.02) to have an undetectable HIV-RNA (< 40 copies/ml; Table 1). Consequently, all further analyses were restricted to PLWH with virological suppression on ART (n = 111). Demographic characteristics of this subpopulation did not notably differ from the main study cohort (Table 1).
Within the virologically suppressed population, subjects homozygous for rs368234815 (ΔG/ΔG) had significantly increased odds of T-cell ratio normalisation (odds ratio (OR) = 3.11, 95% CI [1.01; 9.56]), when compared to the major homozygote (T/T; Table 2). No significant association was observed between T-cell ratio normalisation and rs12979860 polymorphisms. The association between rs368234815 (ΔG/ΔG) and CD4+:CD8+ T-cell ratio normalization was slightly abrogated after correction for age, gender and treatment duration (OR = 2.80, 95% CI [0.82; 9.54]).
Relationship between IFNL locus polymorphism and CD4+ and CD8+ T-cell subsets
In an exploratory analysis within the treated, virally suppressed population (n = 111), rs368234815 ΔG/ΔG homozygosity was also associated with significantly lower proportions of CD4+ effector memory T-cells (regression coefficient: − 7.1% (95% CI − 13.83; − 0.34) compared to the T/T genotype (Table 3). In a subpopulation of treated, virally suppressed Caucasian (n = 76), rs368234815 ΔG/ΔG homozygosity was associated with higher proportion of CD4+ and CD8 + central memory T-cells (regression coefficient: + 11.6% (95% CI 2.62; 20.50) and + 13.8 (95% CI 5.67; 21.97) respectively) compared to the T/T genotype. There was no association between the CD4 + and CD8 + T-cell subset in non-Caucasians.
Since SNPs in this IFNL3/4 region have been extensively related to HCV infection outcomes, including treatment-induced and spontaneous viral clearance, associations between CD4+ and CD8+ T-cell sub-populations with IFNL SNPs were further investigated in a subgroup of virally suppressed PLWH without HCV coinfection (n = 71). Within this sub-population, rs368234815 ΔG/ΔG was associated with significantly higher proportions of naïve CD8+ T-cells (regression coefficient: + 7.2% (95% CI 0.28; 14.06) and lower proportions of effector memory CD8+ T-cells (− 7.6% (95% CI − 16.29; 1.05), albeit of marginal statistical significance, compared to the T/T genotype (Table 4).
Discussion
This is the first study to describe associations between IFNL polymorphisms and immunological outcomes in ART-treated PLWH, individuals with undetectable viral loads on ART and homozygous for rs368234815 [ΔG/ΔG] displayed an increased likelihood of normalisation of the CD4+:CD8+ T-cell ratio, as well as higher proportions of naïve CD8+ T-cells and lower CD4+ effector memory cells in those with HIV mono-infection. Taken together with previous literature [9, 30], these data are consistent with an overall more favourable immunological profile in the setting of long-term treated HIV-1 infection in those with the rs368234815 [ΔG/ΔG] genotype; the presence of which is linked with both enhanced IFNL4 activity and Th1 responses [31,32,33].
Our findings are consistent with recent reports of more frequent rs368234815 [ΔG/ΔG] genotypes (IFNL4 expressing) among PLWH classified as long-term non-progressors [34]. While the authors of this report hypothesized that increased T-cell activation and IFN-mediated thymic dysfunction among those with the rs368234815 [TT/TT] genotype, known to be associated with higher interferon-stimulated gene expression (ISG) [20], may contribute to CD4+ T-cell decline and a loss of host immune control of HIV-1 infection, it is also possible that these observations relate to better CD8+ mediated T-cell control of viral infection in those with ΔG/ΔG who express IFNL4, a view that would be supported by our study results.
Naïve CD8+ T-cell subsets were significantly increased in those with HIV mono-infection who carried the rs368234815 ΔG/ΔG homozygous genotype. Previous data has established roles for naïve CD8+ T-cells in controlling immune responses to infection. In animal models, bacterial sepsis leads to a rapid depletion of naïve CD8+ T-cells, which persists long after resolution of the infection [35]. Previously, we, and others, have reported associations between a higher percentage of naïve CD8+ T-cells and higher CD4+:CD8+ T-cell ratio in long-term treated HIV-1 infection, consistent with better immune recovery with ART [9, 10].
A deeper understanding of how naïve CD8+ T-cells influence immune responses arising after viral infection is critical for the development of enhanced therapeutic and vaccination strategies to exploit CD8+ T-cell-mediated immunity [36], such as those proposed in HIV-cure strategies. Vaccine immunogens based on conserved HIV epitopes aim to induce CD8+ cytotoxic T lymphocytes to kill virus-infected cells and are a target for therapeutic strategies aimed at eradication of HIV reservoirs. Our data suggest that host immunogenetics at the IFNL locus may influence CD8+ T-cell responses in those with long-term treated HIV-1 infection, with those with rs368234815 [ΔG/ΔG] homozygosity having more favourable immune responses on ART. Given the limited success of HIV vaccine research [37], our data would suggest that host profiling based on IFNL genotype may identify those individuals who may respond more favorably to immune-based therapies, although this hypothesis would require further exploration in prospective studies.
Our study has limitations. The single centre, cross-sectional nature of the study design cannot determine causality, with mortality patterns and other unmeasured factors potentially introducing biases and some ethnicities underrepresented. Our findings also require validation in other more diverse cohorts, testing validity within different ethnicities and treatment environments. In addition, although we measured immune markers that have been associated with more favorable clinical outcomes, we did not include more detailed functional markers of immune function such as measurements of cytokines, ISGs and or IFN classes. Lastly, our sample size limited the ability to explore the impact of different ART regimes on observed relationships. Nevertheless, our novel findings provide further insights consistent with previous studies demonstrating an important role for IFNL4 in the control of chronic HIV-1 infection and further build on emerging data on the importance of CD8+ T-cell responses and the CD4+:CD8+ T-cell ratio in determining response to long-term treated HIV-1 infection.
Conclusion
In summary, in virally-suppressed, long-term ART-treated PLWH, rs368234815 ΔG/ΔG homozygotes were more likely to have attained normalisation of their CD4+:CD8+ T-cell ratio, displayed lower CD4+ effector memory and higher naive CD8+ T-cells. Further studies are needed to replicate our findings in other, larger and more diverse cohorts and to determine the impact of IFNL genetic variation on CD4+:CD8+ T-cell ratio normalisation and clinical outcomes in people living with HIV.
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available due to ethical approval restrictions, but are available from the corresponding author on reasonable request.
Abbreviations
- AIDS:
-
Acquired immunodeficiency syndrome
- ART:
-
Antiretroviral therapy
- CMV:
-
Cytomegalovirus
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- HCV:
-
Hepatitis C virus
- HIV:
-
Human immunodeficiency viruses
- HLA:
-
Human leukocyte antigen
- IDU:
-
Injecting drug-users
- IFNL:
-
Interferon lambda
- ISG:
-
Interferon-stimulated gene expression
- IL:
-
Interleukin
- InSTI:
-
Integrase strand transfer inhibitors
- IQR:
-
Interquartile range
- MSM:
-
Men who have sex with men
- OR:
-
Odds ratio
- PCR:
-
Polymerase chain reaction
- PEG/RBV:
-
Pegylated interferon-alpha/ribavirin
- PLWH:
-
People living with HIV
- RNA:
-
Ribonucleic acid
- SNP:
-
Single nucleotide polymorphism
- TSLP:
-
Thymic stromal lymphopoietin
References
Mussini C, Lorenzini P, Cozzi-Lepri A, Lapadula G, Marchetti G, Nicastri E, et al. CD4/CD8 ratio normalisation and non-AIDS-related events in individuals with HIV who achieve viral load suppression with antiretroviral therapy: an observational cohort study. Lancet HIV. 2015;2(3):e98–106.
Catalfamo M, Wilhelm C, Tcheung L, Proschan M, Friesen T, Park JH, et al. CD4 and CD8 T cell immune activation during chronic HIV infection: roles of homeostasis, HIV, type I IFN, and IL-7. J Immunol. 2011;186(4):2106–16.
Castilho JL, Shepherd BE, Koethe J, Turner M, Bebawy S, Logan J, et al. CD4+/CD8+ ratio, age, and risk of serious noncommunicable diseases in HIV-infected adults on antiretroviral therapy. AIDS. 2016;30(6):899–908.
Caby F, Guihot A, Lambert-Niclot S, Guiguet M, Boutolleau D, Agher R, et al. Determinants of a low CD4/CD8 ratio in HIV-1-infected individuals despite long-term viral suppression. Clin Infect Dis. 2016;62:1297–303.
Brites-Alves C, Netto EM, Brites C. Coinfection by hepatitis C is strongly associated with abnormal CD4/CD8 ratio in HIV patients under stable ART in Salvador, Brazil. J Immunol Res. 2015;2015:174215.
Cao W, Mehraj V, Trottier B, Baril JG, Leblanc R, Lebouche B, et al. Early initiation rather than prolonged duration of antiretroviral therapy in HIV infection contributes to the normalization of CD8 T-Cell counts. Clin Infect Dis. 2016;62(2):250–7.
Bellissimo F, Pinzone MR, Celesia BM, Cacopardo B, Nunnari G. Baseline CD4/CD8 T-cell ratio predicts prompt immune restoration upon cART initiation. Curr HIV Res. 2016;14:491–6.
De Salvador-Guillouet F, Sakarovitch C, Durant J, Risso K, Demonchy E, Roger PM, et al. Antiretroviral regimens and CD4/CD8 ratio normalization in HIV-infected patients during the initial year of treatment: a cohort study. PLoS ONE. 2015;10(10):e0140519.
Tinago W, Coghlan E, Macken A, McAndrews J, Doak B, Prior-Fuller C, et al. Clinical, immunological and treatment-related factors associated with normalised CD4+/CD8+ T-cell ratio: effect of naive and memory T-cell subsets. PLoS ONE. 2014;9(5):e97011.
Serrano-Villar S, Sainz T, Lee SA, Hunt PW, Sinclair E, Shacklett BL, et al. HIV-infected individuals with low CD4/CD8 ratio despite effective antiretroviral therapy exhibit altered T cell subsets, heightened CD8+ T cell activation, and increased risk of non-AIDS morbidity and mortality. PLoS Pathog. 2014;10(5):e1004078.
Tang J, Li X, Price MA, Sanders EJ, Anzala O, Karita E, et al. CD4:CD8 lymphocyte ratio as a quantitative measure of immunologic health in HIV-1 infection: findings from an African cohort with prospective data. Front Microbiol. 2015;6:670.
Zaegel-Faucher O, Bregigeon S, Cano CE, Obry-Roguet V, Nicolino-Brunet C, Tamalet C, et al. Impact of hepatitis C virus coinfection on T-cell dynamics in long-term HIV-suppressors under combined antiretroviral therapy. AIDS. 2015;29(12):1505–10.
Joshi A, Sedano M, Beauchamp B, Punke EB, Mulla ZD, Meza A, et al. HIV-1 Env glycoprotein phenotype along with immune activation determines CD4 T cell loss in HIV patients. J Immunol. 2016;196(4):1768–79.
Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401.
Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41(10):1100–4.
Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41(10):1105–9.
Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’hUigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801.
Collison M, Chin JL, Abu Shanab A, Mac Nicholas R, Segurado R, Coughlan S, et al. Homozygosity for HLA group 2 alleles predicts treatment failure with interferon-alpha and ribavirin in chronic hepatitis C virus genotype 1 infection. J Interferon Cytokine Res. 2015;35(2):126–33.
Lindh M, Lagging M, Norkrans G, Hellstrand K. A model explaining the correlations between IL28B-related genotypes, hepatitis C virus genotypes, and viral RNA levels. Gastroenterology. 2010;139(5):1794–6.
Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45(2):164–71.
Ye L, Schnepf D, Staeheli P. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat Rev Immunol. 2019;19(10):614–25.
Ye L, Schnepf D, Becker J, Ebert K, Tanriver Y, Bernasconi V, et al. Interferon-lambda enhances adaptive mucosal immunity by boosting release of thymic stromal lymphopoietin. Nat Immunol. 2019;20(5):593–601.
Bibert S, Wojtowicz A, Taffé P, Manuel O, Bernasconi E, Furrer H, et al. The IFNL3/4 ΔG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS. 2014;28(13):1885–9.
Manuel O, Wojtowicz A, Bibert S, Mueller NJ, van Delden C, Hirsch HH, et al. Influence of IFNL3/4 polymorphisms on the incidence of cytomegalovirus infection after solid-organ transplantation. J Infect Dis. 2015;211(6):906–14.
Martin MP, Qi Y, Goedert JJ, Hussain SK, Kirk GD, Hoots WK, et al. IL28B polymorphism does not determine outcomes of hepatitis B virus or HIV infection. J Infect Dis. 2010;202(11):1749–53.
Rallon NI, Restrepo C, Naggie S, Lopez M, Del Romero J, Goldstein D, et al. Interleukin-28B gene polymorphisms do not influence the susceptibility to HIV-infection or CD4 cell decline. AIDS. 2011;25(2):269–71.
Salgado M, Kirk GD, Cox A, Rutebemberwa A, Higgins Y, Astemborski J, et al. Protective interleukin-28B genotype affects hepatitis C virus clearance, but does not contribute to HIV-1 control in a cohort of African-American elite controllers/suppressors. AIDS. 2011;25(3):385–7.
Real LM, Herrero R, Rivero-Juarez A, Camacho A, Macias J, Vic S, et al. IFNL4 rs368234815 polymorphism is associated with innate resistance to HIV-1 infection. AIDS. 2015;29(14):1895–7.
Machmach K, Abad-Molina C, Romero-Sanchez MC, Dominguez-Molina B, Moyano M, Rodriguez MM, et al. IFNL4 ss469415590 polymorphism is associated with unfavourable clinical and immunological status in HIV-infected individuals. Clin Microbiol Infect. 2015;21(3):289.
Serrano-Villar S, Caruana G, Zlotnik A, Perez-Molina JA, Moreno S. The effects of maraviroc versus efavirenz in combination with zidovudine/lamivudine on the CD4/CD8 ratio in treatment-naive HIV-infected individuals. Antimicrob Agents Chemother. 2017;61:e01763–817.
Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M, et al. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8(3):254–61.
Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, et al. IL-28A (IFN-lambda2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol Med. 2011;3(6):348–61.
Srinivas S, Dai J, Eskdale J, Gallagher GE, Megjugorac NJ, Gallagher G. Interferon-lambda1 (interleukin-29) preferentially down-regulates interleukin-13 over other T helper type 2 cytokine responses in vitro. Immunology. 2008;125(4):492–502.
Dominguez-Molina B, Tarancon-Diez L, Hua S, Abad-Molina C, Rodriguez-Gallego E, Machmach K, et al. HLA-B*57 and IFNL4-related polymorphisms are associated with protection against HIV-1 disease progression in controllers. Clin Infect Dis. 2017;64(5):621–8.
Condotta SA, Rai D, James BR, Griffith TS, Badovinac VP. Sustained and incomplete recovery of naive CD8+ T cell precursors after sepsis contributes to impaired CD8+ T cell responses to infection. J Immunol. 2013;190(5):1991–2000.
La Gruta NL, Thomas PG. Interrogating the relationship between naive and immune antiviral T cell repertoires. Curr Opin Virol. 2013;3(4):447–51.
Safrit JT, Fast PE, Gieber L, Kuipers H, Dean HJ, Koff WC. Status of vaccine research and development of vaccines for HIV-1. Vaccine. 2016;34(26):2921–5.
Acknowledgements
The authors would like to thank the participants attending the Mater Misericordiae University Hospital (MMUH) infectious diseases clinic, the clinic staff and the Department of Immunology at MMUH for the immunophenotyping.
Funding
This work was supported by the Wellcome Trust (097427/Z/11/Z to I. F.) and through the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie (Grant Agreement No. 666010 to W. T.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
MJC, WWH and PWM conceived the study and contributed to the writing of the article. IF and WT performed the statistical analyses and interpretations, and contributed to the writing of the article. PWM, AGC, JM, BD, CPF, EM, JS, JSL and MJC collected clinical data contributed to the interpretation of the results and reviewed the article. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All enrolled subjects provided written, informed consent and the study was approved by the Mater Misericordiae University Hospital and Mater Private Hospital Institutional Review Board (Ref: 1/378/1391).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Gating strategy used to discriminate CD4+ and CD8+ T-cell subsets. Note: In the dot Plot of CD62L PE versus CD45RO APC, Q1 displays CD62L+CD45RO−cells (naïve cells). Q2 displays CD62L+ CD45RO+ cells (central memory cells), Q3 displays CD62L−CD45RO− cells (revertant memory cells), Q4 displays CD62L−CD45RO+ cells (effector memory cells).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Freitas, I.T., Tinago, W., Sawa, H. et al. Interferon lambda rs368234815 ΔG/ΔG is associated with higher CD4+:CD8+ T-cell ratio in treated HIV-1 infection. AIDS Res Ther 17, 13 (2020). https://doi.org/10.1186/s12981-020-00269-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12981-020-00269-0