Abstract
Background
It has been proved that mutations in the PADI6 gene can cause early embryo arrest. This study describes a newly discovered mutation in PADI6 that expands the genetic spectrum of early embryo arrest.
Methods
Peripheral blood of a patient diagnosed with early embryo arrest was collected for whole-exome sequencing. Sanger sequencing was performed to confirm this mutation. The effects of the variant were investigated in human embryonic kidney 293T (HEK293T) cells using western blotting, real-time quantitative polymerase chain reaction, and immunofluorescence.
Results
A novel homozygous mutation in PADI6 was identified in the proband. The patient carried a frameshift insertion mutation c.558dupA (p.Thr187Asnfs*48), which was located in the protein arginine deiminase middle domain. The variant destroyed PADI6 protein expression and reduced PADI6 mRNA expression in HEK293T cells.
Conclusions
The newly identified mutation in PADI6 accounts for early embryo arrest. It expands the spectrum of genetic causes and phenotypes of infertility in humans. These findings also provide an additional possible diagnostic marker for patients with recurrent in vitro fertilization/intracytoplasmic sperm injection failure.
Plain Language Summary
Some infertile patients experience multiple in vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI) failure owing to recurrent early embryo arrest. However, the underlying mechanisms remain largely unknown. Due to the development of whole-exome sequencing, early embryo arrest has been confirmed as a type of Mendelian disease. This study aimed to identify the genetic cause of early embryo arrest in patients and to expand the genetic spectrum. Furthermore, it can help doctors offer better suggestions to such patients and prevent patients from suffering from multiple IVF/ICSI failures.
Similar content being viewed by others
Background
Infertility affects 70 million couples of reproductive age in the world [1]. The only way they can produce their own babies is through assisted reproductive technology. To date, approximately 8 million babies have been produced by in vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI) [2]. However, some couples with infertility experience multiple IVF/ICSI failures for unknown reasons. Successful establishment of human pregnancy requires that mature oocytes to fuse with sperms. Not all zygotes have the opportunity to develop viable embryos. It is estimated that only about 40–70% of human embryos develop into viable embryos [3]. This phenomenon is early embryo arrest referred to as preimplantation embryo lethality, and arrested cleaved embryos cannot form blastocysts [4]. Unexplained early embryonic arrest is the major cause.
Previously, genes related to early embryo arrest were only investigated in animal models. The cause in humans remains largely elusive due to ethical concerns. Contributing to the development of IVF/ICSI, investigators can observe human oocytes/embryos in vitro. Furthermore, several genes, including PAT1 homolog 2 (PATL2), tubulin beta 8 class VIII (TUBB8), TLE family member 6 (TLE6), cell division cycle 20 (CDC20), peptidyl arginine deiminase 6 (PADI6), NLR family pyrin domain containing 2 (NLRP2), NLR family pyrin domain containing 5 (NLRP5), KH domain containing 3 like (KHDC3L), REC114 meiotic recombination protein (REC114) and meiotic double-stranded break formation protein 1 (MEI1) [4,5,6], have been proved to cause early embryo arrest in human and then female infertility.
Maternal effect factors are essential for embryogenesis. Early human embryos (before the eight-cell stages) are transcriptionally silent and utilize maternally provided transcripts and proteins during the maturation process of growing oocytes [7]. Human subcortical maternal complex (SCMC) is a multiprotein complex uniquely located in the subcortex of the mammalian oocytes and embryos, which mainly consists of eight proteins, including TLE6, NLRP2, NLRP5, NLRP7, oocyte permitting embryonic development (OOEP), KHDC3L and PADI6, and zinc finger BED-type containing 3 (ZBED3) [8, 9]. SCMC is an important maternal-effect factor. They are highly expressed in the oocytes and early embryos, and usually begin to degrade due to lack of transcriptional compensation after the embryonic genome becomes active [10], playing an important role in orchestrating oocyte-embryo transition.
A series of studies have shown that SCMC genes are involved in reproductive failure, such as early embryonic arrest, fertilization failure, and recurrent hydatidiform moles [11,12,13]. However, this can only account for a small number of patients. Therefore, novel mutations need to be investigated to establish the role of SCMC in embryogenesis and lay the foundation for the genetic diagnosis of female infertility.
In the present study, a novel mutation in the SCMC gene PADI6 was identified using whole-exome sequencing (WES) in a patient with multiple IVF/ICSI failures. This study broadened our understanding of the genetic and phenotypic characteristics of early embryonic arrest.
Methods
Human subjects
One patient diagnosed with early embryo arrest during common IVF treatment was recruited from the Department of Reproductive Medicine, Xiangya Hospital, Central South University. Participants with normal embryo development in IVF/ICSI cycles were also recruited as the control group. The study was approved by the Institutional Review Board of the Department of Reproductive Medicine, Xiangya Hospital (No.2021005). Informed consent was obtained from all the recruited participants.
DNA extraction
Venous blood samples of patients and 100 controls were collected in EDTA blood tubes. Genomic DNA was extracted according to the standard protocol of the QIAamp DNA Blood Midi Kit (QIAGEN). The DNA concentration and purity were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
WES
The Exome was captured using xGen Exome Research Panel v2 (22307, Integrated DNA Technologies, San Diego, California, USA) and sequencing was performed with the Novaseq system (PE150, Illumina, San Diego, California, USA). Sequences were aligned to a human reference sequence (human genome 19, hg19) using Bowtie2. After filtering the unmapped variants, single nucleotide polymorphism (SNP) and insertion and deletion (InDel) polymorphisms were detected using FreeBayes. The variants were annotated using the ANNOVAR software. The frequency of corresponding mutations was determined using the 1000 Genomes Project (1000G), Exome Aggregation Consortium (ExAC) and Exome Aggregation Consortium_East Asian (ExAC_EAS) databases, and the functional effects of the mutations were predicted using the in silico algorithms MutationTaster. The filtering criteria were as followed: (1) variants with an allele frequency lower than 0.1% in the 1000G and ExAC databases; (2) exonic nonsynonymous, splicing, or coding indels; and (3) variants with a previously reported embryogenesis-related function. The American College of Medical Genetics and Genomics (ACMG) was used to assess the mutations. HomozygosityMapper (http://www.homozygositymapper.org/) was used to determine the existence of candidate homozygous variants in the proband [14].
Sanger sequencing
The targeted variant in PADI6 was amplified by polymerase chain reaction (PCR) using the ProFlex™ Base (Applied Biosystems, Singapore). Specific primers flanking the mutation sites were listed in Additional file 1: Table S1. The mutation variant was then confirmed by Sanger sequencing in the patients and controls.
Evolutionary conservation analysis and molecular modelling
Evolutionary conservation was assessed using Clustal Omega software (https://www.ebi.ac.uk/Tools/msa/clustalo/). DNA structures was drawn using the Gene Structure Display Server (http://gsds.gao-lab.org/) [15]. The Pfam database (http://pfam.xfam.org/) was used to analyze the protein domains. In addition, wild-type (wt) PADI6 was built using SWISS-MODEL software (https://swissmodel.expasy.org/) in the automated model and mutated PADI6 was mapped using SWISS-Pdb Viewer.
Plasmid construction and mutagenesis
Plasmids were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). Briefly, the full-length coding sequences of wild-type and mutant PADI6 (NM_207421.4) were synthesized and inserted into the p.cDNA3.1(+) vector with a C-terminal Flag tag. Sanger sequencing confirmed the wild-type and mutant clones.
Cell culture and transfection
Human embryonic kidney 293T (HEK293T) cells were kindly provided by the Center for Medical Genetics & Hunan Key Laboratory of Medical Genetics, School of Life Sciences, Central South University. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Biological Industries) supplemented with 10% fetal bovine serum (Biological Industries) and 1% penicillin/streptomycin (Gibco) in a humidified incubator with a 5% CO2 atmosphere at 37 °C. Transient transfections were performed using the Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions. DNase and RNase water of equal volumes were added into the blank groups as controls.
Western blotting
The cells were washed thrice with cold PBS after transfection for 48 h. Cells were harvested and lysed with RIPA buffer (Beyotime Biotechnology) containing with 1× protease inhibitor cocktail (Beyotime Biotechnology). After incubating on the ice for 20 min and centrifuging at 12,000 rpm at 4 °C for 20 min, protein lysates were collected in a new centrifuge tube. After quantification using a bicinchoninic acid kit (Beyotime Biotechnology), equal amounts of samples were denatured in protein loading buffer (NCM Biotech) and heated at 100 °C for 10 min. Protein samples were separated by 9% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes (Millipore). The membranes were blocked in 5% skim milk diluted in 1× Tris-buffered saline with Tween 20 (TBST) for 1 h and incubated at 4 °C overnight with primary antibodies against Flag (1:1000, 8146S, Cell Signaling Technology) or GAPDH (1:10,000, 60004-1-Ig, Proteintech). After 1 h incubation with the corresponding secondary antibodies (1:10,000, 7076S, Cell Signaling Technology) for 1 h at room temperature on the secondary day, the membranes were visualized using Amersham ImageQuant 800 (Cytiva, United States) after detection with BeyoECL Plus (Beyotime).
Immunofluorescence
After transfection for 48 h, cells on slides were fixed with 4% paraformaldehyde for 20 min, and then incubated in permeabilizing solution (0.02% Triton X-100 in PBS) for 15 min and blocking buffer (5% goat serum in PBS) for 1 h. Anti-Flag antibody (1:400, 8146S, Cell Signaling Technology) was added at 4 °C overnight. On the secondary day, the cells were incubated with CoraLite488-conjugated goat anti-mouse IgG(H+L) (1:400, SA00013-1, Proteintech) for 1 h to visualize PADI6-Flag staining. DAPI Fluoromount-G (SouthernBiotech) was used to label DNA. Finally, the cells were imaged on a fluorescence microscope (Leica, Germany).
RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HEK293T cells 48 h after transfection using the Total RNA Purification Kit (SimGen). Reverse transcription and amplification were performed using the Evo M-MLV RT Mix Kit (Accurate Biotechnology), and RT-qPCR was performed using the SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology) according to the manufacturer’s instructions. All reactions were performed with at least three technical replicates. The relative expression level was calculated using 2−△△Ct normalized to endogenous GAPDH expression. Primers used in the experiments are listed in Additional file 1: Table S1.
Statistical analysis
All experiments were independently conducted in triplicate. RT-qPCR analysis was performed using Student’s t-tests when comparing experimental groups. Statistical analyses were performed using GraphPad Prism 8.0. P < 0.05 was considered as statistically significant.
Results
Clinical characteristics of the affected individual
A novel homozygous PADI6 mutation was identified in an infertile patient with recurrent IVF/ICSI failure. The proband had been diagnosed with primary infertility for several years, and had normal menstruation and hormone levels (Additional file 1: Table S2). The family pedigree is shown in Fig. 2a.
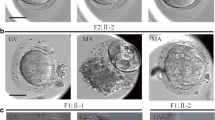
The proband was 34 years old with a 15-year infertile history at examination. Initially, it was found that the two sides of the fallopian tubes were not completely obstructed. She had tried four cycles of ovulation monitoring, but had been not pregnant. The couple then accepted an ICSI attempt at another hospital, but it failed because of the lack of available embryos. At our hospital, an antagonist protocol with ICSI treatment was administered. Fifteen oocytes were retrieved, including three germinal vesicles (GV) and 12 MII oocytes. A total of 11 oocytes were fertilized and ten of them formed cleaved embryos, although all of them were of poor quality (Table 1). Two 3cells/II embryos were transferred on the Day 2. In the end, no viable blastocysts were obtained in prolonged cultivation, and no pregnancy was established, indicating early embryonic arrest (Table 1, Fig. 1).
Identification of novel mutations in PADI6
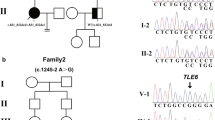
In the proband, a homozygous frameshift insertion mutation was identified in exon 6 of PADI6 (NM_207421.4: c.558dupA: p.Thr187Asnfs*48) (Fig. 2a). Sanger sequencing was performed to confirm that the mutation was homozygous (Fig. 2b). Homozygosity mapping detected a strong signal in patient (II-2) in the region of chromosome 1 that contained PADI6 (Fig. 2c). Because the DNA samples of their families were unavailable, the mutation inheritance was unknown.
Identification of PADI6 mutation in one affected family. a Pedigree of a family who carry the PADI6 mutation. The affected individual shows homozygous mutation. Squares indicate male individuals, and circles indicate female individuals. The black circle with an arrow indicates the affected individual, and blank symbols indicate unaffected individuals. The equal sign represents infertility. b Sanger sequencing confirmation of the proband and control. c Homozygosity mapping of the proband. Homozygous regions harboring the strongest signal are indicated in red, and the asterisk (*) denotes where PADI6 is located
Conservative and in silico analysis of the variant effects on protein function
The mutation site was located in exon 6 of the PADI6 gene (Fig. 3a). The amino acid at position p.Thr187 of PADI6 was not conserved among the eight mammalian orthologs (Fig. 3b). The mutation was absent in the 1000G, ExAC, and ExAC_EAS databases (Table 2). The variant that we identified was not predicted by MutationTaster (Table 2). According to the three-dimensional (3D) structure of PADI6, the frameshift insertion mutation caused the replacement of threonine with asparagine at position 187, resulting in a truncated protein (Fig. 3c). The mutation site was in the α-helix, which might influence the formation and stability of the α-helix. The mutation was pathogenic according to the ACMG guidelines.
The locations and conservation of mutated residues position in PADI6. a The localization of the mutations is indicated in the genomic and protein structure of PADI6. b Conservation of amino acids in PADI6 among different 8 mammalian orthologs. c Protein conformation prediction of the effect of mutation in PADI6. The view shows the structure comparison of structure of the frameshift insertion mutation in PADI6 protein with wild-type PADI6 protein
Effects of the variant on protein and mRNA expression in HEK293T cells
To evaluate the functional influences of the PADI variant in vitro, PADI6 wild-type and mutant plasmids identified in this study were constructed and transiently transfected into HEK293T cells. Western blotting and immunofluorescence analysis demonstrated that compared with the wild-type group, the p.Thr197Asnfs*48 protein could not be expressed (Fig. 4a and c). RT-qPCR showed a significant reduction in the mRNA levels of the p.Thr197Asnfs*48 variant (Fig. 4b).
Effects of the variant on PADI6 protein and mRNA expression in vitro. a Western blot of PADI6 protein expression in HEK-293T cells transfected with Flag-tagged wild-type and mutant plasmids. GAPDH was used as the internal control. b RT-qPCR analysis of wild-type and mutant PADI6 mRNA level. The bars showed the mean of three separate measurements, and the error denoted the SEM. c Immunofluorescence of wild-type and mutant PADI6. The experiment was performed with three independent biological replicates yielding similar results. **P < 0.01
Discussion
In the present study, a novel homozygous mutation (NM_207421.4: c.558dupA: p.Thr187Asnfs*48) in gene PADI6 was discovered in a patient diagnosed with primary infertility due to early embryonic arrest. We constructed wild-type and mutant PADI6 plasmids and transfected them into HEK293T cells to investigate the effects of the variant on protein and mRNA expression in vitro. The results showed that p.Thr187Asnfs*48 could cause a significant reduction in mRNA levels and did not induce protein expression. This discovery expanded the genetic spectrum of early embryonic arrest diagnosis.
The PADI family is comprised of five members, including PADI1, PADI2, PADI3, PADI4, and PADI6. Among these, PADI6 is exclusively expressed in oocytes and early embryos [9]. PADI6 is one of SCMC components that localizes to other components in oocytes and early embryos. It encodes a peptidylalarginine deiminase. Peptidylarginine deiminase belongs to a class of Ca2+-dependent enzyme that convert arginine residues to citrulline, which is called citrullination and is crucial for the formation of rigid structures such as hair, skin, and myelin sheath [16, 17]. It is highly conserved in their amino acid sequence of mammalian peptidylarginine deiminases [18]. PADI6 knockout mouse studies confirm that PADI6 null oocytes lack a specialized cytoskeletal structure called as cytoplasmic lattices and consequently have abnormal organelle positioning and redistribution [19]. Furthermore, the stored PADI6 mRNAs are largely degraded in the zygote [20, 21], altering spindle microtubule assembly and movement to mediate proper cleavage in early embryo development [20, 22]. Therefore, PADI6 null embryos were arrested between the two-cell and four-cell stage [20].
In 2016, Xu et al. first reported that PADI6 variants were associated with embryonic arrest due to embryonic genome activation failure in humans [23]. To date, studies have reported six homozygous mutations or four compound-heterozygous mutations that caused early embryo arrest (Additional file 1: Table S3). Other studies confirm that PADI6 does not only cause (early) pregnancy losses and recurrent hydatidiform moles, but maternal effect variants in this gene cause chromosomal aberrations and disturbed imprinting, producing children with Beckwith–Wiedemann syndrome or Silver–Russell syndrome (Additional file 1: Table S3). These studies indicated that different mutations in PADI6 cause phenotypic variability in patients.
In this study, a homozygous mutation c.558dupA (p.T187Nfs*48) caused early embryonic arrest in the proband (II-2), resulting in 3- or 5-cell arrested embryos on Day 3 and no viable blastocytes on Day 5, which was congruent with previous reports [8, 9, 23,24,25]. Due to the limitation of the PADI6 antibody for immunofluorescence, we did not investigate the in vivo expression of PADI6 in the oocytes of the patient. However, bioinformatics predicted that the mutation could result in a truncated protein because of a premature stop codon. Although most reported mutations are localized in the protein arginine deiminase (PAD) domain, this novel homozygous mutation is in the PAD middle domain. In addition, the variant displayed a significant reduction in mRNA levels and no protein expression in vitro experiments. It was possibly caused by nonsense-mediated mRNA and protein decay. Therefore, we speculate that the variant could result in no expression of PADI6 in the patient’s oocytes, finally leading to unstable SCMC and embryo developmental arrest.
For patients diagnosed with infertility caused by PADI6 mutations, genetic counselling is recommended. Oocyte donation may be a feasible option for preventing an adverse pregnancy and unnecessary IVF treatment. Several studies have shown that complementary RNAs (cRNAs) injection can rescue abnormal oocyte development, resulting in blastocytes with normal copy number variations [26, 27]. Although no rescue study has been performed to treat oocyte/embryo with PADI6 mutations, it may be a promising gene therapy for these patients.
Conclusions
A novel homozygous mutation in PADI6 was identified in the present study, which can account for early embryo arrest caused by genetic abnormalities and expand the spectrum of genetic causes and phenotypes of human infertility. These findings also provide an additional possible diagnostic marker for patients with recurrent IVF/ICSI failure.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- HEK:
-
Human embryonic kidney
- IVF:
-
In vitro fertilization
- ICSI:
-
Intracytoplasmic sperm injection
- PATL1 :
-
PAT1 Homolog2
- TUBB8 :
-
Tubulin beta 8 class VIII
- TLE6 :
-
TLE Family member
- CDC20 :
-
Cell division cycle 20
- PADI6 :
-
Peptidyl arginine deiminase 6
- NLRP2 :
-
NLR Family pyrin domain containing 2
- NLRP5 :
-
NLR Family pyrin domain containing 5
- KHDC3L :
-
KH Domain containing 3 like
- REC114 :
-
REC Meiotic recombination protein
- MEI1 :
-
Meiotic double-stranded break formation protein 1
- SCMC:
-
Subcortical maternal complex
- OOEP :
-
Oocyte permitting embryonic development
- ZBED3 :
-
Zinc finger BED-type containing 3
- WES:
-
Whole-exome sequencing
- hg19:
-
Human genome 19
- SNP:
-
Single nucleotide polymorphism
- InDel:
-
Insertion and deletion
- 1000G:
-
1000 Genomes Project
- ExAC:
-
Exome Aggregation Consortium
- ExAC_EAS:
-
Exome Aggregation Consortium_East Asian
- ACMG:
-
American College of Medical Genetics and Genomics
- PCR:
-
Polymerase chain reaction
- GV:
-
Germinal vesicles
- 3D:
-
Three dimension
- PAD:
-
Protein arginine deiminase
- cRNAs:
-
Complementary RNAs
References
Nikshad A, Aghlmandi A, Safaralizadeh R, Aghebati-Maleki L, Warkiani ME, Khiavi FM, et al. Advances of microfluidic technology in reproductive biology. Life Sci. 2021;265:118767. https://doi.org/10.1016/j.lfs.2020.118767.
Wang W, Wang W, Xu Y, Shi J, Fu J, Chen B, et al. FBXO43 variants in patients with female infertility characterized by early embryonic arrest. Hum Reprod. 2021. https://doi.org/10.1093/humrep/deab131.
Gardner DK, Lane M. Culture and selection of viable blastocysts: a feasible proposition for human IVF? Hum Reprod Update. 1997;3:367–82. https://doi.org/10.1093/humupd/3.4.367.
Wang W, Dong J, Chen B, Du J, Kuang Y, Sun X, et al. Homozygous mutations in REC114 cause female infertility characterised by multiple pronuclei formation and early embryonic arrest. J Med Genet. 2020;57:187–94. https://doi.org/10.1136/jmedgenet-2019-106379.
Dong J, Zhang H, Mao X, Zhu J, Li D, Fu J, et al. Novel biallelic mutations in MEI1: expanding the phenotypic spectrum to human embryonic arrest and recurrent implantation failure. Hum Reprod. 2021. https://doi.org/10.1093/humrep/deab118.
Sang Q, Zhou Z, Mu J, Wang L. Genetic factors as potential molecular markers of human oocyte and embryo quality. J Assist Reprod Gen. 2021. https://doi.org/10.1007/s10815-021-02196-z.
Begemann M, Rezwan FI, Beygo J, Docherty LE, Kolarova J, Schroeder C, et al. Maternal variants in NLRP and other maternal effect proteins are associated with multilocus imprinting disturbance in offspring. J Med Genet. 2018;55:497–504. https://doi.org/10.1136/jmedgenet-2017-105190.
Wang X, Song D, Mykytenko D, Kuang Y, Lv Q, Li B, et al. Novel mutations in genes encoding subcortical maternal complex proteins may cause human embryonic developmental arrest. Reprod Biomed Online. 2018;36:698–704. https://doi.org/10.1016/j.rbmo.2018.03.009.
Liu J, Tan Z, He J, Jin T, Han Y, Hu L, et al. Two novel mutations in PADI6 and TLE6 genes cause female infertility due to arrest in embryonic development. J Assist Reprod Gen. 2021. https://doi.org/10.1007/s10815-021-02194-1.
Monk D, Sanchez-Delgado M, Fisher R. NLRPs, the subcortical maternal complex and genomic imprinting. Reproduction. 2017;154:R161–70. https://doi.org/10.1530/REP-17-0465.
Parry DA, Logan CV, Hayward BE, Shires M, Landolsi H, Diggle C, et al. Mutations causing familial biparental hydatidiform mole implicate C6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011;89:451–8. https://doi.org/10.1016/j.ajhg.2011.08.002.
Alazami AM, Awad SM, Coskun S, Al-Hassan S, Hijazi H, Abdulwahab FM, et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015. https://doi.org/10.1186/s13059-015-0792-0.
Lin J, Xu H, Chen B, Wang W, Wang L, Sun X, et al. Expanding the genetic and phenotypic spectrum of female infertility caused by TLE6 mutations. J Assist Reprod Gen. 2020;37:437–42. https://doi.org/10.1007/s10815-019-01653-0.
Seelow D, Schuelke M, Hildebrandt F, Nurnberg P. HomozygosityMapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–9. https://doi.org/10.1093/nar/gkp369.
Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–7. https://doi.org/10.1093/bioinformatics/btu817.
Cubellis MV, Pignata L, Verma A, Sparago A, Del Prete R, Monticelli M, et al. Loss-of-function maternal-effect mutations of PADI6 are associated with familial and sporadic Beckwith-Wiedemann syndrome with multi-locus imprinting disturbance. Clin Epigenet. 2020. https://doi.org/10.1186/s13148-020-00925-2.
Qian J, Nguyen NMP, Rezaei M, Huang B, Tao Y, Zhang X, et al. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur J Hum Genet. 2018;26:1007–13. https://doi.org/10.1038/s41431-018-0141-3.
Lu X, Gao Z, Qin D, Li L. A maternal functional module in the mammalian oocyte-to-embryo transition. Trends Mol Med. 2017;23:1014–23. https://doi.org/10.1016/j.molmed.2017.09.004.
Liu X, Morency E, Li T, Qin H, Zhang X, Zhang X, et al. Role for PADI6 in securing the mRNA-MSY2 complex to the oocyte cytoplasmic lattices. Cell Cycle. 2017;16:360–6. https://doi.org/10.1080/15384101.2016.1261225.
Vitale AM, Yurttas P, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, et al. Role for PADI6 and the CPLs in ribosomal storage in oocytes and translation in the early embryo. Development (Cambridge). 2008;135:2627–36. https://doi.org/10.1242/dev.016329.
Li L, Baibakov B, Dean J. A subcortical maternal complex essential for pre-implantation mouse embryogenesis. Dev Cell. 2008;15:416–25. https://doi.org/10.1016/j.devcel.2008.07.010.
Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, et al. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol. 2011;350:311–22. https://doi.org/10.1016/j.ydbio.2010.11.033.
Xu Y, Shi Y, Fu J, Yu M, Feng R, Sang Q, et al. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet. 2016;99:744–52. https://doi.org/10.1016/j.ajhg.2016.06.024.
Zheng W, Hu H, Dai J, Zhang S, Gu Y, Dai C, et al. Expanding the genetic and phenotypic spectrum of the subcortical maternal complex genes in recurrent preimplantation embryonic arrest. Clin Genet. 2021;99:286–91. https://doi.org/10.1111/cge.13858.
Zheng W, Chen L, Dai J, Dai C, Guo J, Lu C, et al. New biallelic mutations in PADI6 cause recurrent preimplantation embryonic arrest characterized by direct cleavage. J Assist Reprod Gen. 2020;37:205–12. https://doi.org/10.1007/s10815-019-01606-7.
Zhao L, Xue S, Yao Z, Shi J, Chen B, Wu L, et al. Biallelic mutations in CDC20 cause female infertility characterized by abnormalities in oocyte maturation and early embryonic development. Protein Cell. 2020;11:921–7. https://doi.org/10.1007/s13238-020-00756-0.
Yoon SY, Eum JH, Lee JE, Lee HC, Kim YS, Han JE, et al. Recombinant human phospholipase C zeta 1 induces intracellular calcium oscillations and oocyte activation in mouse and human oocytes. Hum Reprod. 2012;27:1768–80. https://doi.org/10.1093/humrep/des092.
Acknowledgements
The authors thank the staff in the Department of Reproductive Medicine, Xiangya Hospital, Central South University, without whom the study could not be carried out.
Funding
This study was supported by the Technology Innovation Guidance Program of Hunan Province (2017SK50103).
Author information
Authors and Affiliations
Contributions
Conceptualization: YL, ZY, XW and HZ; Methodology: YH; Formal analysis and investigation: HZ and JuZ, JiZ and QX collected the samples; Writing-original draft preparation: XW. Writing-review and editing: ZY, YL, HZ and LW. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by Institute Review Board of Department of Reproductive Medicine, Central South University. Informed consent was obtained from all individual participants included in the study.
Consent for publication
Patients signed informed consent regarding publishing their data and photographs.
Competing interests
Authors declared that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
PADI6 primer sequences. Table S2. Sex hormone characteristics of the proband during the controlled ovarian stimulation. Table S3. Overview of mutation sites and associated phenotypes of PADI6 in previous studies.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, X., Zhu, H., He, Y. et al. A novel homozygous mutation in the PADI6 gene causes early embryo arrest. Reprod Health 19, 190 (2022). https://doi.org/10.1186/s12978-022-01495-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12978-022-01495-7