Abstract
Background
Leukocyte immunoglobulin-like receptor B4 (LILRB4) plays a significant role in regulating immune responses. LILRB4 in microglia might influence the infiltration of peripheral T cells. However, whether and how LILRB4 expression aggravates brain damage after acute ischemic stroke remains unclear. This study investigates the role of LILRB4 in modulating the immune response and its potential protective effects against ischemic brain injury in mice.
Methods and results
Microglia-specific LILRB4 conditional knockout (LILRB4-KO) and overexpression transgenic (LILRB4-TG) mice were constructed by a Cre-loxP system. Then, they were used to investigate the role of LILRB4 after ischemic stroke using a transient middle cerebral artery occlusion (tMCAO) mouse model. Spatial transcriptomics analysis revealed increased LILRB4 expression in the ischemic hemisphere. Single-cell RNA sequencing (scRNA-seq) identified microglia-cluster3, an ischemia-associated microglia subcluster with elevated LILRB4 expression in the ischemic brain. Flow cytometry and immunofluorescence staining showed increased CD8+ T cell infiltration into the brain in LILRB4-KO-tMCAO mice. Behavioral tests, cortical perfusion maps, and infarct size measurements indicated that LILRB4-KO-tMCAO mice had more severe functional deficits and larger infarct sizes compared to Control-tMCAO and LILRB4-TG-tMCAO mice. T cell migration assays demonstrated that LILRB4-KD microglia promoted CD8+ T cell recruitment and activation in vitro, which was mitigated by CCL2 inhibition and recombinant arginase-1 addition. The scRNA-seq and spatial transcriptomics identified CCL2 was predominantly secreted from activated microglia/macrophage and increased CCL2 expression in LILRB4-KD microglia, suggesting a chemokine-mediated mechanism of LILRB4.
Conclusion
LILRB4 in microglia plays a crucial role in modulating the post-stroke immune response by regulating CD8+ T cell infiltration and activation. Knockout of LILRB4 exacerbates ischemic brain injury by promoting CD8+ T cell recruitment. Overexpression of LILRB4, conversely, offers neuroprotection. These findings highlight the therapeutic potential of targeting LILRB4 and its downstream pathways to mitigate immune-mediated damage in ischemic stroke.
Similar content being viewed by others
Introduction
Ischemic stroke, which constitutes 70% of all strokes and 50.3% of stroke-related deaths [1], presents a significant clinical challenge. A deeper understanding of the mechanisms underlying neuroinflammation post-ischemic stroke is crucial for developing effective therapies. Neurovascular damage following ischemic stroke is significantly driven by a cascade of inflammation response. These responses are triggered by the release of damage-associated molecular patterns and reactive oxygen species from ischemic and hypoxic brain tissues. And they play a critical role in mediating interactions between microglia and peripheral immune cells after a stroke, exacerbating brain edema and contributing to further neurovascular damage [2]. The intricate interplay between the brain and peripheral immune system is facilitated by complex communication networks. This interaction suggests that specifically modulating microglia-peripheral immune cell interactions could offer a potential therapeutic target for mitigating the inflammatory response post-ischemic stroke [3, 4]. Targeting these interactions may help to reduce neurovascular damage and improve outcomes for patients suffering from ischemic stroke.
Microglia, the primary immune cells of the central nervous system (CNS), are pivotal in the inflammatory response to ischemic stroke. Following an ischemic event, microglia rapidly activate and migrate to the injury site, releasing pro-inflammatory cytokines and chemokines that exacerbate tissue damage. Microglia also play a neuroprotective role by clearing cellular debris and promoting tissue repair, demonstrating their dual role in stroke pathology [5]. As ischemic inflammation progresses, resting microglia are typically activated and polarized into pro-inflammatory type M1 and anti-inflammatory type M2 [6]. However, this binary classification oversimplified the complexity and diversity of microglial subtypes observed after polarization. Recent studies using scRNA-seq, have revealed a more nuanced spectrum of microglial states that contribute to the inflammatory response post-stroke [7, 8]. These diverse subtypes exhibit varying functions and gene expression profiles, reflecting a dynamic range of activation states beyond the traditional M1/M2 dichotomy. Disease-associated microglia (DAM) subsets [7, 8] have been identified by scRNA-seq in various nervous system diseases. For example, DAM was firstly identified as a unique microglia cluster in Alzheimer’s model but not observed in the wild-type (WT) background [9]. This DAM associated with Alzheimer’s disease was characterized with down-regulated homeostasis microglial gene such as Cx3cr1, P2ry12 and Tmem119, while specially highly expression of Trem2, Apoe, Lpl and Ctsd, all of which were well known AD risk factors related genes. Moreover, DAM was enriched in the lipid metabolism pathways and phagocytic activity and conserved in human and other neurodegenerative diseases including amyotrophic lateral sclerosis [10]. In addition, a similar DAM gene signature was also demonstrated in a proliferative-region-associated microglia cluster in developing white matter of mouse brains [11], which could phagocytose newly formed oligodendrocytes. Utilizing scRNA-seq in stroke models, we also identified a DAM-like cluster that specific expressed the LILRB4 gene. This finding underscores the complexity and heterogeneity of microglial responses in ischemic stroke, highlighting the role of LILRB4 in modulating microglial function during neuroinflammation.
LILRB4, also known as ILT3, CD85k and LIR5, is a member of the leukocyte Ig-like receptor family, characterized by two C-type Ig-like extracellular domains D1 and D4, a transmembrane domain, and three cytoplasmic immunoreceptor tyrosine-based inhibitory motifs (ITIMs) [18]. ITIMs mainly exists in the intracellular part of some immunosuppressive receptors [12], and the tyrosine in ITIMs can bind to the SH2 domain on PTP molecules after phosphorylation, thereby recruiting and activating PTP, and playing a role in inhibiting the activation signal transduction pathway involved in PTK [13]. LILRB4 could bind to its ligands such as APOE [14], CD166 [15], fibronectin [16] and galectin-8 [17], then LILRB4’s ITIMs are phosphorylated, which in turn recruit the SH2 domain phosphatases such as SHP-1, SHP-2, and SHIP-1, resulting in negatively regulating or suppressing downstream NF-κB [18, 19], AKT, MAPK [15], TRAF6 [20] and other inflammatory signaling pathways [21]. LILRB4 is expressed on various immune cells, including microglia, monocytes, macrophages, DCs, T cells, neutrophils, plasma cells, and endothelial cells, where it mainly acts as immunosuppressive receptor via ITIMs involved in the inhibition of cytokine production, suppression of T cell activity and induction of tolerogenic DCs and Tregs [19, 20]. Therapeutic strategies targeting LILRB4 have demonstrated promise in models of solid tumors [22]. Moreover, LILRB4 is crucial in the pathological development of several inflammatory diseases, including atherosclerosis, non-alcoholic fatty liver disease, multiple sclerosis, and Alzheimer’s disease [20, 23]. The molecular mechanisms among the LILRB4 regulation of these inflammatory disorders demonstrated that the highly expression of LILRB4 on macrophages could suppress NF-κB-mediated inflammatory response in the development of atherosclerosis [24]. Additionally, LILRB4 expressed primarily by microglia in brain, and negatively regulated IFN-γ–induced microglia activation during meningoencephalitis [21]. In our research, we also identified a unique LILRB4+ microglia cluster which specifically only observed in a stroke model of MCAO, but not sham group. However, whether LILRB4 also involved in the regulation of the inflammatory responses during ischemic stroke, it remains unexplored.
In the present study, utilizing scRNA-seq analysis, we identified a DAM-like microglia subcluster post-ischemia, which characterized by highly expression of LILRB4. To further investigate the functional significance of this LILRB4+ microglia subset and its underlying mechanisms, we employed conditional gene knockout and overexpression techniques. Our findings reveal the dynamic expression of LILRB4 in microglia after ischemic stroke and its impact on CNS inflammation. Based on these observations, we propose that LILRB4 in microglia plays a crucial role in modulating inflammatory responses and tissue damage in the CNS following ischemia.
Materials and methods
Experimental design
To explore the expression level change of LILRB4 in murine models post-MCAO, C57BL/6J WT mice were randomly allocated into a sham group and a MCAO group. In addition, to investigate the influence and underlying mechanisms of LILRB4 on brain injury and neurological function in murine models post-MCAO, LILRB4-KO, LILRB4-TG and their littermate control transgenic floxed mice known as Control-KO and Control-TG were randomly distributed into a Sham group and a MCAO group. The animals were assigned a random number using a computer based random order generator method.
Animals
WT male C57BL/6 mice and mice carrying LILRB4-floxed alleles were obtained from the Shanghai Model Organisms Center, Inc. (Shanghai, China). Briefly, the floxed mice of LILRB4 knock in (overexpression), abbreviated as R26-LSL-LILRB4, was established as follows: the CAG-loxP-STOP-loxP-LILRB4-WPRE-pA vector was inserted into the Rosa26-Gt (ROSA)26Sor (104735) locus by the CRISPR/Cas9 genome editing technology. LILRB4 knock-in allele mouse strains were genotyped by PCR using the following primers: P1: 5’- AGGCTTCAGAAGGACACTGC-3’, P2: 5’- AAGGAAGGTCCGCTGGATTG − 3’. The PCR program started at 95 °C for 20 s, 60 °C for 20 s, 72 °C for 20 s for 35 cycles, and 72 °C for 5 min, generating a 626-bp fragment for LILRB4 knock in. The floxed mice of LILRB4 knock out was established as follows: the loxP sites were inserted around LILRB4 DNA sequence known as ‘floxing’ and were incorporated into the mouse genome by homologous recombination. LILRB4 knock-out allele mouse strains were genotyped by PCR using the following primers: P1:5’-TTCTTGTGTTGCTCTATGCTTTTA-3’, P2: 5’-GGAAGGAATTGCACCGAGATG-3’. The PCR program started at 94 °C for 30 s, 59 °C for 30 s, 72 °C for 1 min for 35 cycles, and 72 °C for 5 min, generating a 381-bp fragment for LILRB4 knock out. Cx3cr1-CreERT2 mice (strain #:021160) on the C57BL/6J background were obtained from the Jackson Laboratory. All mice were maintained at Tianjin Huanhu Hospital (Tianjin, China). Mice aged 8–12 weeks were randomly assigned to experiments. According to the Cre-LoxP system principle, Flox mice were bred with microglia-specific Cre (Cx3cr1-CreERT2)-expressing mice to generate transgenic mice with knockout or overexpression of LILRB4 in microglia (LILRB4-KO and LILRB4-TG), and their littermate transgenic floxed mice which were used as controls in the following experiments.
For mice above mentioned LILRB4-KO and LILRB4-TG, 4-5-week-old male were utilized for tamoxifen induction. Tamoxifen was administered intraperitoneally in corn oil at a dose of 100 mg/kg for five doses, with 48-hour intervals between doses. One month after the last tamoxifen dose, mice were subjected to following experiments. All animal procedures adhered to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Animal Experiments Ethical Committee of Tianjin Medical University. The experimental design adhered to the ARRIVE guidelines.
tMCAO procedure
Male C57BL/6 mice (8–10 weeks of age) were placed under controlled conditions (25 ± 2℃, 60-70% humidity, 12/12 h light-dark cycle) with access to food and water. Mice were anesthetized with 4% isoflurane in 70% N2O and 30% O2. During the procedure, the common carotid artery (CCA), internal carotid artery, external carotid artery and their main branches were exposed and separated, the external carotid artery and its main branches, along with the proximal end of the CCA were ligated first. The distal end of the internal carotid artery was clipped with an arterial clamp, and a spare silk thread was placed at the distal end of the CCA ligation to fix the embolus. A silicone-coated monofilament (Doccol, Sharon, MA, USA) was inserted through the CCA between the two ligature wires. The distal ligature wire was tightened, and the thread crossed the bifurcation of the internal and external carotid artery and entered the lumen of the internal carotid artery. When slight resistance was encountered, the monofilament had reached the origin of the middle cerebral artery, confirmed successfully occlusion. The ligature on the CCA was then tightened and secured. After 1 h of ischemia, the monofilament was removed. Cerebral blood flow was monitored during the whole procedure of tMCAO by laser Doppler flowmeter (PF5001, Perimed, Switzerland). In the sham-operated group, all procedures were identical except for the insertion of the monofilament. During the surgical procedure, the body temperature of the mice was controlled at 37.0 ± 0.5 °C. The exclusion criteria for successful tMCAO were as follows: the relative cerebral blood flow reduced less than 70% of preischemic levels for the mouse in the study.
scRNA-seq data collection and analysis
scRNA-seq data were collected and analyzed using the Gene Expression Omnibus database. scRNA-seq dataset, GSE174574, which includes expression profiling from the brain tissue of 3 control mice and 3 ischemic stroke mice at one day post-tMCAO or sham-operations, was obtained. We reanalyzed the dataset following the method as previously described [25]. This included data integration, dimensionality reduction, clustering, identification of differentially expressed genes (DEGs) and gene set enrichment analysis (GSEA). NCBI SRA database under Bioproject number PRJNA952594 were reanalyzed to examine spatial relationships pertaining to the expression of LILRB4 and CCL2.
Randomisation and blinding principle for the animal experiments
According to the ARRIVE guidelines, blinding was performed throughout the whole stages of the experiment, including random allocation, performing experiment, collecting data, outcome assessments and data analysis.
Neurological behavioral score assessment
The following neurological behavioral scores were assessed by investigators blinded to group allocation.
Rotarod test
The Rotarod test provides a convenient method for detecting motor function, coordination ability and anti-fatigue ability in rodents [26]. During the experiment, when the animal is placed on the rotating rod in the center of the roller, in order to avoid slipping, it needs to adjust the balance and run with the rotating rod. It was assessed by measuring the time the animals walked on the roller. Mice underwent a three-day training period. During the test, the rotarod speed increased from 4 rpm to 40 rpm over five minutes. The latency to fall or rotate three times was recorded, with a maximum duration of 120 s.
Novel object recognition
In this study, the cognitive memory ability of experimental animals was evaluated by the behavioral method of the length of exploration of familiar objects and new unfamiliar objects [27]. When the mice moved around the new and old objects, the number, time and distance of which were measured to estimate the cognitive performance of the mice. Conducted in an opaque acrylic box with a recording time of 200 s. On Day 1, mice acclimated to the box for five minutes. On Day 2, two identical objects (A) were placed 10 cm from the side wall, and mice explored for five minutes. On Day 3, one object A was replaced with object B, and the time spent exploring each object (TA and TB) was recorded. The recognition index (RI) was calculated as RI = TB/(TA + TB) × 100%.
Tail suspension test
It was first proposed by Steru et al. in 1985 to assess depression-like states in mice [28]. The emotional state of the mice was measured by looking at the hanging state from which they could not escape over a period of time. Conducted in a chamber with a white background (25 cm × 25 cm × 35 cm). Mice were suspended by the tail for six minutes, with the first two minutes as an adaptation period. Immobility time was recorded during the last four minutes to indicate depressive state, defined as cessation of movement and vertical suspension.
Foot-fault test
It also known as grid walking tests, are used to assess motor function and limb coordination in rodents [29]. Basically, the animals were placed on an elevated grid with square openings (1.69 cm2 for mice) and allowed to move across the entire grid. Assessed the proportion of missteps on irregularly spaced grids among 100 steps taken by both forelimbs.
Corner turning test
It assesses orientation patterns for sensorimotor dysfunction during a unilateral stroke model [30]. Mice were placed at acute angles formed by two glass plates, and the proportion of turns toward the paralyzed side was recorded. Normally, animals won’t prefer one direction over the other. However, stroke results in contralateral limb defects, animals tend to use the ipilateral limb to step back. The higher the inclination to one side, the more severe the outcome of the stroke. The formula used was [i/(i + C)] × 100, over 10 trials with 30-second intervals.
Modified neurological severity score (mNSS) test
The mNSS includes a comprehensive evaluation of motor, sensory, balance and reflexes of experimental animals to assess neurological deficits and the degree of neurological impairment of cerebral ischemia animals [31]. Evaluated neurological deficits on a scale of 0 to 18, the mild injury was 0 ~ 6, the moderate injury was 7 ~ 12, and the severe injury was 13 ~ 18. Higher scores indicated greater damage.
Laser speckle imaging
Laser speckle imaging assessed cortical cerebral blood flow (CBF) in mice following cerebral ischemia. Measurements were taken using a laser speckle imaging instrument (Reward, China) before tMCAO and at 60 min, 1 day, and 3 days post-occlusion. Mice were anesthetized with isoflurane and positioned under a laser speckle camera. The speckle contrast from laser light scattering was used to measure CBF, translating into correlation time values inversely related to blood flow velocity. Image contrast and color were adjusted for optimal visualization. Infarct volume and contralateral regional blood flow were analyzed using RFLSI software by an investigator blinded to the experimental groups.
Perfusion and tissue preparation
After tMCAO reperfusion, mice were anesthetized with 4% isoflurane in 70% N2O and 30% O2 at the appropriate time (1 and 3 days as per the experimental plan). A thoracotomy was performed, and the spleen was soaked in phosphate buffered saline (PBS) for flow cytometry. The animals were manually perfused with PBS through the left ventricle. The mice were then decapitated, and the left side of the brain was extracted.
Brain tissue for flow cytometry or 2,3,5-triphenyl tetrazolium chloride (TTC) staining was used directly in PBS, and tissue for frozen sections was immersed in 4% paraformaldehyde (PFA) (Solarbio, USA) and stored at 4℃. For other analyses, the left part of the brain tissue was isolated on the liquid nitrogen, then quickly frozen, dried and sealed in the − 80℃ environment for the subsequent extraction of protein and RNA.
TTC staining
Following PBS perfusion, and brain removal, The excised brains were washed in PBS and placed at -20℃ for 15 min. The brain were then cut into 1 mm thick coronal slices. Slices were inclubated in 2% TTC solution (Solarbio, USA), at 37℃ for 15–20 min in the dark. Normal brain tissue-stained bright red, while infarct area appeared white. Slices were fixed in 4% PFA solution and photographed 1 day later.
Immunofluorescence staining
Brain tissue was fixed in 4% PFA for 48 h, then soaked in 15% and 30% sucrose PBS solution for 2 days each. The tissue was cut into 1 mm coronal slices, embedded in OCT, and stored at -80℃. Section (6 μm thick) were cut and stored at -80℃. For immunofluorescence staining, frozen sections were fixed in 4% PFA for 15 min. The membrane was permeabilized with 1% tritonX-100 (Sigma, USA) for 15 min, followed by 5% bovine serum albumin blocking for 30 min. Sections were incubated with primary antibodies overnight at 4℃; Primary antibodies include: rabbit anti-Iba1 (1:500; 019-19741; Wako Pure Chemical Industries, Ltd., Japan); rabbit anti-NeuN (1:500; 94405, CST, USA); rabbit anti-GFAP (1:500; 3670; CST, USA); rabbit anti-LILRB4 (1:50, MBS 2018121; Mybiosource), Rabbit anti-CD3 (1:250; A12415; Abclonal, China). Then sections incubated with secondary fluorescent-conjugated antibodies at room temperature in the dark for 1 h. The secondary antibodies were AF 488-conjugated sheep anti-rabbit IgG, AF 594-conjugated sheep anti-mouse IgG (1:500; Thermo Fisher Scientific, USA); Sections were mounted with Fluoroshield Mounting Medium containing DAPI (Abcam) and imaged using a confocal microscope (LSM 800, Zeiss, Germany).
Protein extraction and western blotting
Tissues stored at -80℃ were ground into chips in liquid nitrogen and divided into two parts, with half used for total protein extraction. Tissue debris was lysed on ice for 30 min using a cell lysate (RIPA: PMSF = 100:1; Solarbio, USA), then centrifuged at 12,000 rpm to obtain the protein-containing supernatant. Protein were diluted to the same concentration and boiled for 5 min to denature them and stored at -80℃. The samples were separated by SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were incubated with primary antibodies overnight at 4℃: Rabbit anti-LILRB4 (orb1150417,1:1000, Biorbyt, UK); Rabbit anti-GAPDH (AC027,1:10000, Abclonal, China). After washing, membranes were incubated with HRP-labeled secondary antibodies (SA00001-2,1:10000, Proteintech, China) for 1 h at room temperature. The membrane was then treated with ECL reagent (Beyotime Biotechnology, USA) and imaged using a gel imager. Image analysis was performed using ImageJ software (NIH), and protein expression levels were normalized to the control group.
RNA extraction and quantitative polymerase chain reaction (qPCR)
Total RNA was isolated using Trizol reagent (OMEGA biotech, USA). and cDNA was synthesized using the PerfectStart Uni RT & PCR Kit (Transgene, China). qPCR was performed using SYBR Green (04913850001; Roche, Basel, Switzerland), and the relative expression of β-actin was calculated using the 2−ΔΔCT method. The specific primers used in this qPCR analysis are listed in Table 1.
Microglia isolation
Transcription levels of CCL2 were measured by sorting microglia from the brain tissues of Sham and tMCAO mice 1-day post-operation. Fresh left cerebral hemispheres were digested with 1 mg/ml collagenase D at 37 ° C for 30 min. Digestion was stopped with PBS, and the mixture was centrifuged at 2000 rpm for 5 min. The supernatant was discarded, and red blood cell lysis was used on ice for 15 min. After another centrifugation, cells were resuspended in 30% percoll (Percol stock: 10xPBS: 1xPBS = 27:3:70) and centrifuged at 700 g for 10 min to remove myelin sheaths, yielding a single-cell suspension. Microglia were isolated using the Mouse CD11b Selection Kit (480109, Biolegend, USA) and subsequently tested by PCR.
T cell isolation
T cells were isolated from the spleens of WT C57BL/6 mice using a spleen lymphocyte isolation kit (TBD, Tianjin, China). Spleens were homogenized and filtered through a 70-µm cell strainer. The homogenate was diluted with buffer and centrifuged with separation medium to isolate mononuclear cell in the interphase. Mononuclear cell were washed and transferred to fresh RPMI-1640 medium. CD3 and CD8 negative magnetic beads (480023/480007, Biolegend, USA) were used to select the desired T cell population. Purity of the sorted cells was verified by fluorescence-activated cell sorting (FACS) analysis using anti-CD3 and anti-CD8 antibodies (Biolegend). The isolated cells were resuspended in RPMI-1640 medium with 10% fetal bovine serum and seeded at a density of 1 × 106 cells/ml in 24-well plates. Cells were maintained at 37℃ and 5% CO2 for further experimentation.
Cell culture, lentiviral transfection, and treatment
The mouse microglia BV2 cells used in the experiment were purchased from Hanheng Biotechnology (Shanghai, China). BV2 were cultured in Dulbecco’s Modified Eagle’s Medium with 10% fetal bovine serum at 37℃ in an environment of 5% CO2. All cells were preserved and frozen at the Tianjin Key Laboratory of Cerebral Vascular and Neurodegenerative Diseases.
LILRB4 knockdown was achieved by transfecting lentiviral particles expressing a Flag epitope-tagged form of murine LILRB4 (LV-CMV-LILRB4a-3XFLAG-EF1-ZSGreen-T2A-PURO) into microglia 48 h before experiments. LV-CMV-EF1-ZSGreen-T2A-PURO was used as the control lentivirus. Lentiviral vectors were purchased from Hanheng Biotechnology (China). To achieve LILRB4 knockdown, the lentiviral vector was transfected into BV2 cells 48 h before the experiment.
Oxygen glucose deprivation/reoxygenation (OGD/R)
In the OGD/R experiment, BV2 cells in the logarithmic phase were selected. After 24 h, the culture medium was replaced with Dulbecco’s Modified Eagle’s Medium devoid of glucose and pyruvate, and the cells were transferred to a hypoxia chamber. A mixture of 95% N2 and 5% CO2 was introduced into the chamber at a rate of 15 L/h for 20 min to reduce the oxygen concentration. Following this, the chamber was transferred to an incubator maintained at 37℃ for 4 h. During reoxygenation, 1% pyruvate/glucose and 10% fetal bovine serum were added to the culture medium, and the cells were subsequently exposed to normoxic conditions.
Transwell co-culture system
BV2 cells (104 cells per well) were plated in 24-well plates and subjected to OGD/R modeling or control treatment. At the onset of reoxygenation, a 5-µm Transwell chamber (Corning, USA) was placed atop the well plates, and sorted T cells (105 cells per well) were seeded in the upper chamber. Anti-CCL2 neutralizing antibodies (BE0185, Bioxcell) were added to the lower chamber, with the isotype IgG as a control. Following 24 h of incubation, the cell suspensions from the 24-well plates were collected, and cell transmigration was assessed using flow cytometry.
In a separate experiment, BV2 cells (104 cells per well) were plated in 24-well plates, and arginase-1 (Arg-1) recombinant protein (0.25 mM, HY-P71833, MCE) was added to the lower chamber. CD8+ T cells (105 cells per well) were then plated onto the BV2 cells, following the same OGD/R modeling or control treatment protocol. Cells were collected after 24 h for further analysis.
FACS analysis
Cell suspensions from migration and co-culture experiments were collected were collected, washed and resuspended in PBS after 1% bovine serum albumin. Samples (100uL, 107/ml) were incubated with antibodies (1 µL/tube) for 30 min on ice in the dark. After re-washing, cells were analyzed with FACS Canto II Flow cytometry (BD Biosciences, USA). For intracellular antigen staining, 0.1% Cell Activation Cocktail (with Brefeldin A) was added 4 h before cell collection. Cells were stained for surface antigens, fixed with Cyto-Fast™ Fix/Perm Buffer Set (BioLegend, USA), permeabilized, and incubated with antibodies for 30 min on ice in the dark. Approximately 10,000 cells per sample were analyzed using FlowJo V10 software.
For In vivo analysis, spleen and the brain tissue were collected in PBS. Spleen tissue was centrifuged at 2000r for 5 min after grinding on ice, erythrocytes were lysed with erythroid lysate (Solarbio, USA). The cells were then resuspended in PBS and stained described above.
Brain tissue was digested within 1 mg/ml collagenase D at 37℃ for 30 min, and centrifuged at 2000 rpm for 5 min. The cells were resuspended in 30% percol solution, centrifugation at 700 g for 10 min, the upper myelin layer was removed. For intracellular antigen staining, cell suspensions were diluted to 106/ml, incubated with 0.1% Cell Activation Cocktail (with Brefeldin A) for 4 h at 5% CO2 and 37℃, and then processed as above.
Antibodies used for FACS: PECD85k (gp49 Receptor), PerCP-Cy5.5 CD11b, APC-Cy7 CD45, PerCP-Cy5.5 CD3, FITC CD4, PE CD8a, APC anti-mouse CD19, FITC NK-1.1, PE Ly-6G, FITC F4/80, APC IFN-γ, FITC CD69. All staining steps were performed in the dark. Flow cytometry and data analysis were conducted using a BD FACSCanto II (BD Bioscience, San Jose, CA, USA) and Flowjo software.
In vivo anti-CD8 monoclonal antibody (mAb) treatment and grouping
C57BL/6 mice were divided into 4 groups after tMCAO follow the principle of randomization allocation:
-
(1)
Control-tMCAO + IgG,
-
(2)
Control-tMCAO + anti-CD8 mAb,
-
(3)
LILRB4-KO-tMCAO + IgG,
-
(4)
LILRB4-KO-tMCAO + anti-CD8 mAb.
Anti-CD8 mAb (100746, Biolegend, USA) was stored at 4℃ in the dark. The stock solution was diluted to 1 mg/ml in PBS under sterile conditions. 1 day before establishing the tMCAO model, 0.1 ug of anti-CD8 mAb (100ul, 1 mg/ml) or equivalent dose of IgG control was slowly injected intraperitoneally.
Statistical analyses
During the data statistical analysis, the investigator will not know the specific information of group treatment. Data were presented as mean ± standard deviation (SD) for normal distribution group and medians with interquartile ranges for skewed distribution group. Group sizes were predetermined based on prior studies. Shapiro-Wilk test was used to assess the normality of the data. Levene test was used to assess the variance homogeneity among the group. Data analyzed via ordinary one-way ANOVA and Tukey’s multiple comparisons test for variance homogeneous data, or Welch’s ANOVA and Tamhane’s T2 multiple comparisons test for nonhomogeneous data. Welch t-test was used for comparing the means of two independent groups when the homogeneity of variance assumption is not met. A two-way ANOVA with Bonferroni’s post hoc test was utilized for comparisons across various time points. Statistical analyses and visualization were done with Prism 8.0 (GraphPad software, USA). All tests were two-tailed, with significance set at p < 0.05.
Results
scRNA-seq identified of a unique LILRB4+ microglia subtype associated with stroke
To study the spatial characteristics of LILRB4 after ischemic stroke, we firstly analyzed LILRB4 expression in the ischemic hemisphere of mice subjected to focal cortical ischemia using the single-cell spatial transcriptome sequencing. LILRB4 expression was predominantly found in the peri-infarct area (Fig. 1A). Figure 1B shows the five microglia subtypes identified by scRNA-seq. The tMCAO group exhibited significant changes in the ratio of MG3 cells compared to the Sham group (Fig. 1B and C). This MG3 cluster specifically expressed high levels of the LILRB4 gene (Fig. 1D, E). The MG3 cells also showed elevated expression of genes associated with neurodegenerative diseases, including LGALS3, LPL, and SPP1 (Fig. 1F). Gene enrichment analysis revealed that the LILRB4+ microglia subtype is involved in several key pathways, including Leukocyte transendothelial migration, Chemokine signaling pathway, Cytokine-cytokine receptor interaction, TNF signaling pathway and Arginine biosynthesis (Fig. 1G). These findings highlight the unique characteristics and potential functional roles of the LILRB4+ microglia subtype in ischemic stroke.
scRNA-seq identified of a unique LILRB4+ microglia subtype associated with stroke. (A) Spatial transcriptomics analysis of LILRB4 expression in the ischemic hemisphere in mice after stroke. (B) t-SNE plots showing the disease origin and the distribution of microglia. (C) Histograms of subcellular components in the control and deletion groups. (D) The t-SNE diagram showing that LILRB4 is expressed mainly in MG3. (E) Violin plot depicting the LILRB4 expression profile of microglia subtypes. (F) Heatmap of differentially expressed genes between LILRB4+ microglia subtype and WT steady state microglia. (G) Selected significantly enriched pathways by gene set enrichment analysis between LILRB4+ microglia and WT subtype
LILRB4 exhibits high expression in microglia during tMCAO in mice
To explore the correlation between LILRB4 and cerebral ischemia, a stroke model was established according to the flow chart (Fig. 2A). TTC staining and laser speckle photography were used to evaluate the modeling efficiency (Fig. 2B, C). The expression of LILRB4 was detected at 1 and 3 days after cerebral ischemia and reperfusion in mice. In the brains of stroke mice, LILRB4 expression was significantly increased compared to the Control group, showing an increasing trend from day 1to day 3 (Fig. 2D–F). Flow cytometry detected the dynamic change in LILRB4 expression levels in microglia (Fig. 2G, H). LILRB4 expression was significantly elevated in the infarct area and persistently increased from day 1 to day 3. Immunofluorescence staining confirmed the increase in LILRB4 expression at 1 and 3 days after infarction (Fig. 2K, L). The staining results also demonstrated that LILRB4 is indeed located in the microglia (Fig. 2M, N). These results suggest that LILRB4 is highly expressed in the infarcted area following ischemic stroke and indicate a correlation between LILRB4 and cerebral ischemia. The time-dependent increase in LILRB4 expression in microglia further supports its potential role in neuroimmune processes related to stroke pathology.
LILRB4 exhibits high expression in microglia during tMCAO in mice. (A) Overview of experimental design. (B) Representative laser speckle rheogram from WT tMCAO mice, IR: Ischaemia-Reperfusion. (C) Representative images of TTC staining of brain sections from tMCAO mice. (D, E) Increased expression of LILRB4 in stroke mice brains compared to controls, showing an increasing trend from day 1 to day 3. (n = 6; **p = 0.0082, ***p = 0.0090). (F) qPCR analysis of LILRB4 expression after tMCAO 1 h. (n = 6; ***p = 0.0003, **p = 0.0093). (G, H) FACS analysis of LILRB4 expression in microglia after tMCAO. (n = 6; ***p = 0.0008, **p = 0.0073). (I) Specimen sites of Immunofluorescent staining. (J) Representative photograph of Immunofluorescent staining at low magnification field of view. Scale bars, 100 μm. (K, L) Immunofluorescence staining showing increased LILRB4 expression at 1 and 3 days post-infarction. Scale bars, 50 μm. (n = 6; ****p<0.0001, **p = 0.0026). (M) LILRB4 and GFAP/NeuN/Iba − 1 immunofluorescence double staining, confirming LILRB4 localization in Iba-1+ microglia. Scale bar, 50/20µm. (N) Colocalization analysis of IBA-1 and LILRB4 in (M). ImageJ quantification
The characterization of LILRB4 transgenic mice have been evaluated under normal conditions after generation
To investigate the role of microglia LILRB4 in stroke, we constructed conditional microglia LILRB4 knockout (LILRB4-KO) and overexpression (LILRB4-TG) transgenic mice. The construction scheme is shown in Fig. 3A. We employed the inducible Cre-loxP system, establishing Cre mice by inserting the Cre-ERT2 element downstream of the promoter of CX3CR1. Flox mice with Lilrb4 knockout (Lilrb4-KO) were created by inserting the Lilrb4 gene flanked by loxP sequence, whereas Flox mice with Lilrb4 overexpression (Lilrb4-TG) were generated by knocking in a designed guide RNA target Lilrb4 sequence at the ROSA 26 site. The heterozygous LILRB4-TG mice were generated by crossing CX3CR1 CreER mice and Lilrb4-TG Flox mice. The homozygous LILRB4-KO mice were generated by crossing CX3CR1 CreER mice and Lilrb4-KO Flox mice. The above used CX3CR1 CreER mice and Lilrb4-TG Flox mice were heterozygous genotype, while the Lilrb4-KO Flox mice were homozygous genotype. Seven days after five intraperitoneal injections of tamoxifen, the expression level of LILRB4 mRNA in the brain were detected by PCR. The results indicated that LILRB4 expression was significantly decreased in the knockout model and increased in the overexpression model (Fig. 3B). Flow cytometry also confirmed that conditional LILRB4 knockout and overexpression in microglia were efficient (Fig. 3C, D).
We monitored the body weight and performed Rotarod test, Noval Object Recognition experiment and Tail suspension test on 10-week-old mice to assess the effects of LILRB4 transgenic technology on the basal status of mice. And there was no significant difference in body weight after knockout and overexpression of LILRB4 (Fig. 3E). Additionally, there were no differences among the four groups in cognitive, emotional and motor functions (Fig. 3F). We further used immunofluorescence to assess the effect of transgenic manipulation on the number of normal mouse brain cells. The results showed that, under normal conditions, knockout and overexpression of LILRB4 had no effect on the number of astrocytes, microglia, and neurons (Fig. 3G, H). Furthermore, H&E staining showed no obvious inflammatory cell infiltration in the brain sections (Fig. 3I), which indicated that transgenic manipulation did not increase the inflammatory response in the brain of normal mice.
The characterization of LILRB4 transgenic mice have been evaluated under normal conditions after generation. (A) Overview of the construction scheme for microglia LILRB4 knockout and overexpression mouse models. (B) qPCR was used to detect the level of LILRB4 transcription in mice brain. (n = 6; *p = 0.0127, **p = 0.0018). (C) FACS analysis of LILRB4 expression in microglia from LILRB4-KO, LILRB4-TG and Control mice. (n = 6; *p < 0.05). (D) Quantitative data plot in (C). (n = 6; ****p<0.0001). (E) Body weight of LILRB4-KO, LILRB4-TG and Control mice. (n = 6; p = 0.9973). (F) Quantitation and statistical evaluation of Rotarod test, Noval Object Recognition and Tail suspension test for the normal LILRB4-KO, LILRB4-TG and Control mice. (n = 6; p = 0.8803/0.4642/0.9887). (G) Representative immunohistochemical staining for neurons, astrocytes, microglia in brain sections from normal LILRB4-KO, LILRB4-TG and Control mice after tamoxifen injecton (7 days after the last dose). (H) Neurons (anti-NeuN, red), astrocytes (anti-GFAP, green) and microglia (anti-Iba-1, green) were quantified with DAPI counterstain (blue). (n = 6; p = 0.9479/0.3929/4557). (I) HE staining revealed no obvious inflammatory infiltration in brain sections of Control, LILRB4-KO or LILRB4-TG mice under normal conditions. (4x, Scale bar, 200 μm; 20x, Scale bar, 40 μm)
Microglial overexpression of LILRB4 protects against ischemia-induced brain damage
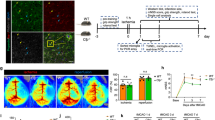
To assess whether LILRB4 could have an effect on neurological function, cerebral blood perfusion and brain injury post-ischemic stroke mice, we designed an experimental protocol plan as shown in Fig. 4A. After tMCAO, several behavioral experiments were conducted to examine neurological function in 4 groups of mice. Compared with Control group, LILRB4-KO mice developed more severe functional deficit, as evidenced by mNSS score, corner turning test, foot-fault test, and appearing from the first day after ischemia. In contrast, LILRB4-TG mice showed relatively lighter deficit in the mNSS, foot-fault test, and corner turning test (Fig. 4B). Cortical perfusion maps and statistics also supported the behavioral findings (Fig. 4C, D). The mean infarct size of LILRB4-KO mice was significantly larger than that of Control mice, whereas that of LILRB4-TG mice was smaller (Fig. 4E, F). In conclusion, selective overexpression of microglia LILRB4 plays a protective role after ischemic stroke.
Microglial overexpression of LILRB4 protects against ischemia-induced brain damage. (A) Experimental Protocol (Qod[Every Other Day], i.p.[intraperitoneal injection]). (B) MNSS score, Foot-Fault test, and corner turning test after tMCAO. (n = 8; *p = 0.0320/0.0320/0.0294/0.0126/0.0250/0.0272/0.0260, Control-KO/TG IR vs. LILRB4-KO/TG IR). (C) The representative laser speckle images of Control, LILRB4-KO and LILRB4-TG mice before operation and 1,3 days after tMCAO. (D) The infarct-side perfusion volume changed in mice Control, LILRB4-KO and LILRB4-TG mice at 1 and 3 days after tMCAO 1 h. (n = 8; *p = 0.0332, Control-KO/TG IR vs. LILRB4-KO/TG IR). (E) Representative images of TTC staining of brain sections from Control and LILRB4-KO, LILRB4-TG mice. (F) Quantitation and statistical evaluation of (E). (n = 5; **p = 0.0026, *p = 0.0447)
LILRB4 is associated with microglial inflammatory phenotypes and morphology after tMCAO
Since microglia are the primary immune cells in the brain, they can rapidly respond to the stimulation of ischemia and hypoxia. Therefore, we explored LILRB4-mediated microglia response after tMCAO. In order to investigate the effect of LILRB4 on microglia polarization and activation after ischemia-reperfusion, we used PCR to detect proinflammatory (M1 phenotype) and anti-inflammatory (M2 phenotype) markers in microglia of Control and LILRB4-KO mice in 1 day after tMCAO. As shown in Fig. 5A, the expression of M1-associated phenotype markers MCP-1, TNF-α, IL-1β, and CD32 increased after LILRB4 knockout, whereas the expression of the M2-associated phenotype markers Arg-1, TGF-β, and CD206 decreased. This indicates that LILRB4 knockout may lead to increased inflammation in the brain by promoting the conversion of microglial cells toward a proinflammatory phenotype.
As the morphology of microglia is closely related to its function, we further analyzed the morphology of Iba-1-positive microglia in the brains of transgenic and Control mice, focusing on the area near the infarct edge, where LILRB4 is highly expressed. Compared with the Control group, the branch (process) lengths of LILRB4-KO microglia were reduced. In contrast, LILRB4-TG microglia increased branch lengths. Sholl analysis revealed that the branches (processes) of microglia after overexpression of LILRB4 were more complex than that of Control microglia, while the degree of microglial process complexity was reduced after LILRB4-KO (Fig. 5B). In conclusion, LILRB4 expression levels affect the polarization and morphological plasticity of microglia and may ultimately modulate the immune response after ischemic stroke.
LILRB4 is associated with microglial inflammatory phenotypes and morphology after tMCAO. (A) Gene expression of M1-associated phenotype markers (MCP-1, TNF-α, IL-1β, and CD32) and M2-associated phenotype markers (Arg-1, TGF-β, and CD206). (n = 6; **p = 0.0061, ***p = 0.0007, ****p < 0.0001, *p = 0.0222; ns p>0.05). (B, C) Fluorescence imaging of microglia in the infarct border region in Control, LILRB4-KO, and LILRB4-TG mice. The lower shows Sholl analysis, where the cell body is the center, and the number of points intersecting several concentric circles is calculated. Shows the number, length of microglia processes (branches), scale bar 10 μm. (n = 10, *p = 0.0397/0.0306/0.0285)
LILRB4 deficiency in microglia aggravates cytotoxic CD8+ T cells accumulation in brain after cerebral ischemia
The damage of BBB after brain injury may cause the infiltration of peripheral immune cells to the CNS. To investigate the effect of microglia LILRB4 expression on the post-stroke immune response, we analyzed the numbers of immune cells in the brains of Control and LILRB4-KO mice after tMCAO, using flow cytometry to identify differences in immune cell populations. Figure 6A shows the flow gating strategy. One day after ischemic stroke, there were no differences in the numbers of neutrophil, natural killer cell, and macrophages in infarcted brain tissue between Control and LILRB4-KO mice. In contrast, the number of T cells in the brains of LILRB4-KO mice increased (Fig. 6B). Immunofluorescence also confirmed the above conclusion (Fig. 6H, I). Notably, the number of CD8+ T cell changed most significantly, suggesting that CD8+ T cell might be the most dramatically affected by LILRB4.
To further explore whether conditional microglia targeting of LILRB4 expression affects the function of CD8+ T cells, we examined the expression of CD69 and IFN-γ in CD8+ T cells after ischemic stroke, which are cell markers related to cytotoxicity. The expression of CD69 and IFN-γ were increased on CD8+ T cell in the brain of LILRB4-KO mice (Fig. 6D, E).
Interestingly, the numbers of T cells in spleen of LILRB4-KO mice decreased (Fig. 6C). There was no difference in the expression of CD69 and IFN-γ of T cells in spleen among the groups (Fig. 6F, G). These results suggest that LILRB4+ microglia may negatively affect the aggregation and activation of CD8+ T lymphocyte in the brain after ischemic stroke.
LILRB4 deficiency in microglia aggravates cytotoxic CD8+ T cells accumulation in brain after cerebral ischemia. (A) Flow-gated strategy after isolation of immune cells from brain and spleen 1 day after tMCAO in Control and LILRB4-KO mice. Fluorescent negative control was used. (B) Calculation of macrophages, neutrophil, CD4+ T cells, CD8+ T cells, NK cells, and B cells in the brains of Control, LILRB4-KO mice 1 day after tMCAO. The data show the absolute value of each brain. (n = 8; ***p = 0.0002, *p = 0.0102). (C) Quantification of macrophages, neutrophil, CD4+ T cells, CD8+ T cells, NK cells, and B cells in the spleens of Control, LILRB4-KO mice 1 day after tMCAO. The data show the absolute value of each spleen. (n = 8; **p = 0.0033, *p = 0.0192). (D–G) The expression of CD69 and IFN-γ on CD8+ T cells in the brain (D and F) and spleen (E and G) by FACS analysis for 1 day after tMCAO. (n = 5/6; *p = 0.0429, **p = 0.0017, ****p < 0.0001). (H) Representative immunofluorescence staining plots of T lymphocyte (anti-CD3, green; DAPI staining for nuclei, blue) at the infarct border site in Control, LILRB4-KO mice 1 day after tMCAO. Scale bar, 200 μm. (I) Quantification of data in (H). (n = 7; *p = 0.0376, ****p < 0.0001)
Depletion of CD8+ T cells rescue the aggravated brain injury in LILRB4-KO mice
We have found that LILRB4 can influence the inflammatory response in ischemic brain injury by regulating the infiltration of T cells into the brain, particularly CD8+ T cells. The suppressive effect of CD8+ T cells cytokine production by LILRB4 is consistent with previous studies [32,33,34], which found that LILRB4 inhibited T cells responses depended on the existence of CD8+ T cells. Depletion of CD8+ T cells eliminated the suppression effect of LILRB4 on T cell proliferation in autoimmune diseases [14, 23]. To further verify whether LILRB4 targeting specificity depends on CD8+ T cells in ischemic stroke, anti-CD8 mAb was used to deplete this subset of immune cells (Fig. 7A, B). Isotype IgG used as control was primary antibody that lack specificity to the target, but match the class and type of anti-CD8 mAb. The results showed that the depletion of CD8+ T cells by anti-CD8 mAb before tMCAO in LILRB4-KO mice compared with IgG control resulted in an increase in CBF and a decrease in neurologic function impairment degree and infarct size (Fig. 7C–G), whereas the above changes were similar in tMCAO mice treated with anti-CD8 mAb between Control and LILRB4-KO group. In addition, microglia morphology was also evaluated (Fig. 7H, I). Compared with IgG control group, CD8+ T cell depletion increased the branch length and complexity of microglia in both Control and LILRB4-KO mice. However, in tMCAO mice treated with anti-CD8 mAb group, the difference of microglia morphology between Control and LILRB4-KO mice was not significant. Theses above results indicated that the depletion of CD8+ T reversed the augmented brain injury and change of microglia morphology in LILRB4-KO mice. In conclusion, these data demonstrated LILRB4 conditional knockout aggravated ischemic brain injury in a CD8+ T cells‑dependent manner.
Depletion of CD8+ T cells rescue the aggravated brain injury in LILRB4-KO mice. (A) Gating strategy of brain and spleen CD8+ T cell in mice injected with IgG or anti-CD8 mAb. (B) Brain and spleen CD8+ T cell count in mice injected with IgG or anti-CD8 mAb. (n = 3, ****p < 0.0001). (C, D) Blood perfusion and representative laser speckle images in IgG treated Control/LILRB4-KO tMCAO mice and anti-CD8 mAb treated congtrol/LILRB4-KO tMCAO mice. (n = 6; *p = 0.0430). (E) mNSS score, Foot-Fault test and corner turning test in IgG/anti-CD8 mAb treated Control/LILRB4-KO tMCAO mice. (n = 6). (F, G) Representative images of TTC staining of brain sections from IgG/anti-CD8 mAb treated Control/LILRB4-KO tMCAO mice, with qantitation and statistical. (n = 5; *p = 0.0130). (H,) Fluorescence imaging of microglia in the infarct border region in IgG/anti-CD8 mAb treated Control/LILRB4-KO tMCAO mice. Scale bar, 10 μm. (I) Shows the number, length of microglia processes (branches). (n = 10; *p = 0.0323, **p = 0.0043)
Microglia LILRB4 deficiency increases the CCL2 production
We have shown that LILRB4 may regulate the infiltration of peripheral T cells, especially CD8+ T cell into the brain after stroke. Various cytokines and chemokines, such as CCL2, CCL3, CCL4, CCL12, CXCL10, CXCL16, acted on CD8+ T cells and cause them to enter the brain [35,36,37]. Firstly, we examined chemokines in the ischemic brain of LILRB4-KO mice 1 day after tMCAO and found that mRNA levels of CCL2 dramatically increased only in the LILRB4-KO mice after tMCAO (Fig. 8A). Targeted binding of CCL2 to CCR2 on the surface of the CD8+ T cell membrane promotes CD8+ T cell infiltration into brain tissue following ischemia and radiation brain injury, and CCR2 was found to be elevated on the T cell surface during this process [35]. Using scRNA-seq and the spatial transcriptomics analysis, we identified that CCL2 was mainly produced by microglia and macrophage clusters in the peri-infarct area after stroke (Fig. 8B, C). We further verified that the expression of CCL2 in microglia sorted from LILRB4-KO mice was also increased after tMCAO (Fig. 8D). Overall, these findings suggest that microglia LILRB4 can affect CCL2 expression in brain after ischemic stroke.
Microglia LILRB4 deficiency increases the CCL2 production. (A) qPCR analysis of CCL2, CCL5, CXCL1, CXCL5, CXCL10 in Control and LILRB4-KO mice 1 day after tMCAO. (n = 6; *p = 0.0109). (B) UMAP plots of 19 cell populations identified by single-cell spatial transcriptomics analysis and the expression level of CCL2 among each cell type in mice after stroke. (C) Spatially transcriptome heatmaps of expression patterns of CCL2 across tissue sections from sham or stroke mouse. (D) qPCR analysis of CCL2 in microglia of Control and LILRB4-KO mice 1 day after tMCAO. (n = 6; *p = 0.0229)
Blockade of CCL2 or addition of Arg-1 suppress CD8+ T cell activation and migration in co-culture with LILRB4-KD microglia
We found that LILRB4 may target and regulate peripheral T cell infiltration into the CNS. To further explore whether microglia recruit T cells directly by LILRB4, a LILRB4 knockdown model of mouse microglia BV2 were constructed and validated using viral transfection (Fig. 9A, B). The Control microglia were transfected with an empty virus that did not carry a fragment of the LILRB4 gene (ControlKD). These results of qPCR and flow-cytometric analyses suggested that the mRNA and protein expression of LILRB4 decreased significantly in LILRB4-KD microglia. T cells were extracted from the spleen of mice, and migration experiments were performed as shown in Fig. 9C, using Control and LILRB4-KD microglia. The microglia were exposed to an OGD environment for 4 h. To investigate the chemotactic effect of microglia derived CCL2 in the migration of T cells, CCL2 inhibitors was added as fellow. Prior to hypoxia, for LILRB4-KD microglia, one group did not add anything as a blank control, one group added CCL2 inhibitor to the medium 2 h earlier, and one group added IgG as a control. The Control microglia without LILRB4 deficiency would not been added anything. After hypoxia, they were placed in a Transwell system for 24 h, in which T cells (upper layer) and microglia were separated with Transwell membranes to verify T cell migration during microglia OGD-R in vitro. After reoxygenation, significant differences were observed in the proportion of T cells that migrated to the lower layers of the transwell device between the two groups. As shown in Fig. 9D, knockdown LILRB4 promoted the recruitment of T cells to microglia, and the recruited T cells were predominantly CD8+ T cells. When anti-CCL2 antibody was added to microglia medium, the migration-promoting effect of LILRB4-KD on T cells was decreased. This suggested that the recruitment of T cells by LILRB4-KD microglia may be mediated by chemokine CCL2.
To further assess whether LILRB4 influences the activation state of cytotoxic CD8+ T cells, we used flow cytometry to detect the expression of CD69 and IFN-γ in a vitro cocultured system (Fig. 9C). After 24 h OGD, we found the expression of CD69 and IFN-γ in CD8+ T cells increased when cocultured with LILRB4-KD microglia (Fig. 9E, F). Previous studies have proven that Arg-1 can inhibit T cell activity and can be upregulated by LILRB4 in leukaemia cells [14, 38]. In this study, we also found the secretion of Arg-1 in a LILRB4-depended on manner after stroke as shown in Fig. 5A. Hence, to investigate the impact of Arg-1 on CD8+ T cells, we added the recombinant Arg-1 into the co-culture system. Interestingly, Arg-1 also inhibited the effect of LILRB4-KD microglia on T cell proliferation by suppressing the expression levels of CD69 and IFN-γ on CD8+ T cells following OGD (Fig. 9E, F). Together, these results suggested that LILRB4 in microglia can inhibit CD8+ T cell activation depending on secretion of Arg-1.
Blockade of CCL2 or addition of Arg1 suppress CD8+ T cell activation and migration in co-culture with LILRB4-KD microglia. (A, B) Differential expression of LILRB4 in BV2 microglia transfected by knockdown and negative control lentiviral vectors. The expression of LILRB4 in BV2 was detected by PCR (A) and Flow cytometry assay (B) (n = 3; **p = 0.0025/0.0065/0.0057). (C) Experimental procedure. Transwell-placed, Control, and LILRB4-KD microglia (without or with CCL2 inhibitor or IgG) were cultured for 4 h under OGD conditions. During reoxygenation, the t-cell-containing Transwell device was placed on a 24-well plate and exposed to medium on its lower surface for 24 h, and the levels of t-cell migration to the lower layer were measured by Flow cytometry. In another experiment, microglia cells were co-cultured with CD8+ T cells. Control and LILRB4-KD microglia were collected and cultured under OGD conditions for 4 h, and CD8+ T cells were added directly to the medium during reoxygenation. One group was added recombinant Arg-1 and another group was not. After 24 h, CD69 and IFN-γ expression in CD8+ T cells were detected by flow cytometry. (D) T cell migration after exposure to OGD/R, measured by the number and type of T cells microglia into 24-well plates, with or without CCL2 inhibition. Flow cytometry tests for T cell migration and ratio of CD8+ T cell. (n = 4; *p = 0.0187/0.0383/0.0104, **p = 0.0029). (E) Control or LILRB4-KD microglia were exposed to OGD/R and co-cultured with CD8+ T cells with or without the addition of recombinant Arg-1. CD8+ T cells were collected for flow cytometry detection of CD69 and IFN-γ expression. (F) Quantitation and statistical evaluation of data in (E). (n = 6; *p = 0.0106/0.0427, ***p = 0.0006, **p = 0.0024/0.0024/0.0029). (G) Control or LILRB4-KD microglia were exposed to OGD/R and co-cultured with CD8+ T cells with or without the addition of recombinant Arg-1. Flow cytometry tests for T cell proliferation. (n = 5; *p = 0.0348/0.0487/0.0153)
Discussion
Microglia are now recognized as key modulator of inflammatory reactions following ischemic stroke, yet their cellar heterogeneity and functional diversity in the ischemic brain injury remains elusive. In this present study, using scRNA-seq, we identified a novel subcluster of microglia highly expressing LILRB4 specifically accumulated in the ischemic brain. We observed a neuroprotective effect of LILRB4 overexpression on cerebral infarction, while knockout of LILRB4 exacerbated the inflammatory response by regulating the infiltration of peripheral CD8+ T cells into the injured brain tissue. Microglia derived CCL2 and Arg-1 are important chemokine or cytokine involved in the mechanism of LILRB4 mediated inflammatory response as above. These findings suggest that targeting microglia LILRB4 may represent a viable therapeutic strategy to prevent the deleterious cerebral injury following ischemic stroke.
The most important biological significance of our study was the focus on the precise regulation of microglia gene expression in single cell level. Previous researches of microglia genes were based on bulk RNA-seq of whole brain or sorted all microglia [39, 40], which masked the gene expression on specific microglia subcluster. The reason for the failure of preclinical of clinical trials [41, 42] on targeting microglia may result from the ignoring of the heterogeneity of microglia subcluster in single cell level. Hence, in our study, we designed the research based on the application of scRNA-seq technology which identified a unique LILRB4+ microglia subset mainly occurred in the stroke tissue. Similarly, a recent study [43] also discovered a stroke-associated myeloid cell cluster which was specific highly in brain parenchyma using single-cell transcriptomic analysis on 24 h after MCAO. This cluster was a mixed macrophage and microglial phenotype also expressing LILRB4 exerting lipid phagocytosing and clearance function in vitro. However, whether LILRB4 in microglia could regulate lipid metabolism in stroke animal awaits to be verified in future researches.
Despite LILRB4 elevation in 24 h after MCAO was also identified [44], the dynamic expression at different time points, brain immune microenvironment alterations in LILRB4 upregulation and the resulting pathophysiological mechanisms remain further investigated. To better understand the role of LILRB4, we generated the transgenic mice strains with microglia-specific LILRB4 knockout and overexpression. In the early stage of acute cerebral infarction, LILRB4-KO mice suffered worst CBF reduction, while LILRB4-TG mice significantly improved CBF compared to Control and LILRB4-KO mice at 1–3 days after tMCAO. A recent study suggested that LILRB4 promoted myeloid-derived suppressor cells polarized to M2 phenotype expressing anti-inflammatory factors, such as TGF-β, IL-10 and stimulated tumor angiogenesis [45]. The expression of angiogenic biomarker VEGFA and CD31 was reduced in LILRB4−/− mice. Thus, LILRB4 might participate in anti-inflammatory function and contribute to the protection for neurovascular unit or the BBB structure in brain ischemia.
In our study, LILRB4-KO in the microglia after ischemic stroke increased the expression of pro-inflammatory factors and promoted the transformation of microglia into M1-like phenotype. Our findings align with other studies, the downregulation of LILRB4 markedly increased the secretion and synthesis of inflammatory factors, such as IL-1β, IL-6, and TNF-α [18, 19, 46] through NF-κB pathway in lung injury and heart disease, but decreased the anti-inflammatory mediators like IL-10 [20]. In our scRNA-seq analysis, we also discovered the LILRB4 in microglia enriched in TNF signaling pathway, however, whether the LILRB4 is also involved in the suppression function on microglia in stroke need for further study. Furthermore, morphological analysis of microglia showed a reduction in microglial process complexity in LILRB4 knockout mice, whereas LILRB4 overexpression recovered microglial morphology with more ramified and complex processes. Thus, LILRB4 was likely involved in enhancing the ramified processes of microglia, which might play a role in increasing their surveillance capabilities, allowing them to more rapidly sense and react to ischemic injury [47]. Similar phenomena were observed in bone marrow–derived macrophages, where LILRB4 expression was higher in M2-type macrophages, and LILRB4−/− macrophages switched toward a more inflammatory or M1-type phenotype with increased expression of IL-1β and iNOS2 cytokines [22]. Hence, LILRB4 in microglia/macrophages may play a protective role in resolution of neuroinflammation and help repair the damaged tissue.
In recent years, the infiltration of T cells into brain tissue have been recognized as a significant player in adaptive immunity in stroke [48, 49]. However, the mechanism and potential pathogenesis remain to be elucidated. In this study, LILRB4 knockout promoted T cell infiltration into ischemic brain tissue, particularly CD8+ T cells. Chih-Chao et al. [50] found that knockdown of LILRB4 in dendritic cells triggered the production of T cell chemokines including CXCL10 and CXCL11, which induced the transmigration of activated T cells in response to supernatants from LPS-treated LILRB4 knockdown-dendritic cells. In contrast, we detected evident CCL2 expression primarily in LILRB4-KD microglia, and then which promote the T cells migration in vitro OGD model. Although microglia and dendritic cells are all major antigen presenting cells in pathological conditions, whereas they might perform different pathways to process antigens and present them to T cells in different diseases. A recent study [35] consistent with our results also showed that microglia-derived CCL2 could trigger CD8+ T cell infiltration to the brain after radiation-induced or ischemic brain injury. Interference with the CCL2-CCR2 signaling pathway effectively inhibited CD8+ T cell migration and significantly reduced brain lesion. Our study will further continue to explore the molecular mechanisms between the elevated CCL2 after LILRB4 knockout and infiltration of CD8+ T cell in tMCAO model. Additionally, we observed the function state of cytotoxic CD8+ T cells was inhibited by LILRB4 in microglia, which was in line with the results of previous studies demonstrating that LILRB4 signaling inhibited CD8+ T cell cytotoxicity and infiltration in leukemia cells and cancer progression [14, 17]. It is well known that Arg-1 is primarily produced by myeloid-derived suppressor cells and has immunosuppressive effects on T cells proliferation, differentiation and cytokine production [14, 51]. Our study showed that Arg-1 transcription levels decreased significantly in the microglia LILRB4 knockout model after tMCAO. Moreover, addition of recombinant Arg-1 could also suppress the activation of CD8+ T cell in LILRB4-KD microglia in our OGD model. Thus, we proposed that LILRB4 may play a key role in suppressing CD8+ T cells function depending on Arg-1.
Our study has significant clinical guiding value for the selective microglia subset-specific genetic regulation program for inflammatory intervention after ischemic stroke. The clinical trial targeting LILRB4 receptor (IO-202) are ongoing with the aim to for the treatment of acute myeloid leukemia (NCT0437243) and advanced solid tumors (NCT05309187) [52]. Screening or designing small molecule compounds induce the upregulation of LILRB4 in microglia would be a powerful immunosuppressive strategy for treatment of stroke. Studies in humans support a role of IL-10, IFN-β, vitamin D analogues, niflumic acid, and tryptophan depletion could up-regulate the expression of LILRB4 among antigen-presenting cells in a variety of autoimmune diseases [33]. Further study of exploring suitable compounds or agonist for LILRB4 offers a new approach to clinical treatment of brain injury. An alternative strategy is to block the downstream factors of LILRB4 such as CCL2 or supplement Arg-1, which have been modified as Pegzilarginase [53] for treating patients with Arg-1 deficiency.
While our study offers valuable insights, it is not without limitations. The use of animal models necessitates caution in extrapolating results to human stroke. Although LILRB4 is highly conserved between human and mouse sharing 97% genetic homology [54], there are still likely to be functional differences within them. A recently published study established a new LILRB4 transgenic mouse line, known as ILT-Telo Tg mice [55], which structurally resembled human LILRB4 but was not its direct orthologue of mouse, to explore the function of human LILRB4 in Alzheimer’s disease. However, our research only designed the traditionally transgenic line of mouse LILRB4 gene. The LILRB4 gene in human might regulate the neuroinflammation through other signal pathways or mechanisms after stroke. Besides, the human LILRB4 might exhibit unique molecular receptor structure unlike mouse, which may bind to different ligands compared to mouse. Hence, our study ignored these species-specific differences, which might affect the translation of our results to humans. Furthermore, our study primarily focuses on the acute phase of ischemic stroke, and the long-term effects of LILRB4 modulation remain to be investigated. Future research should aim to confirm these findings in clinical settings and explore the long-term impact of LILRB4 on stroke recovery. It has been emergingly acknowledged that sex-specific lesion patterns exist in acute ischemic stroke and female suffer more widespread left hemisphere lesions than man [56]. Only male mice were used in our study and whether female mice share the same outcome in LILRB4 signaling after stroke warranted further investigation. Additionally, further elucidation of the molecular mechanisms underlying LILRB4’s protective effects is necessary. Exploring other potential chemokines and cytokines involved in LILRB4-mediated immune modulation could provide a more comprehensive understanding of its role in stroke pathology. These investigations will help to better understand the therapeutic potential of targeting LILRB4 in ischemic stroke.
Conclusion
In summary, microglial LILRB4 plays a crucial role in modulating the post-stroke immune response by regulating CD8+ T cell infiltration and activation. Knockout of microglial LILRB4 exacerbates CD8+ T cell transmigration in a CCL2‑dependent manner, and enhances CD8+ T cell responses which could be rescued by addition of Arg-1 in vitro OGD/R model. These findings underscore the therapeutic potential of targeting microglial LILRB4 and its downstream pathways to mitigate immune-mediated damage in ischemic stroke. Further research is warranted to translate these findings into clinical applications, potentially improving outcomes for stroke patients.
Data availability
The data that support the findings of this study are openly available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE174574.
Abbreviations
- Arg-1:
-
Arginase-1
- CBF:
-
Cerebral blood flow
- CCA:
-
Common carotid artery
- CNS:
-
Central nervous system
- DAM:
-
Disease-associated microglia
- FACS:
-
Fluorescence-activated cell sorting
- KO/KD/TG:
-
Knockout/Knockdown/Transgenic
- LILRB4:
-
Leukocyte immunoglobulin-like receptor B4
- mAb:
-
Monoclonal antibody
- mNSS:
-
Modified neurological severity score
- OGD/R:
-
Oxygen glucose deprivation/Reoxygenation
- PBS:
-
Phosphate buffered saline
- PFA:
-
Paraformaldehyde
- qPCR:
-
Quantitative polymerase chain reaction
- scRNA-seq:
-
Single-cell RNA sequencing
- tMCAO:
-
Transient middle cerebral artery occlusion
- TTC:
-
2,3,5-triphenyl tetrazolium chloride
- WT:
-
Wild type
References
Fan J et al. Global Burden, risk factor analysis, and Prediction Study of ischemic stroke, 1990–2030. Neurology. 2023;101(2).
Mun KT, Hinman JD. Inflammation and the link to vascular Brain Health: timing is brain. Stroke. 2022;53(2):427–36.
Lee J, McCullough LD. Revisiting regulatory T cells for stroke therapy. J Clin Invest. 2022;132(15).
Zheng P, et al. Group 2 innate lymphoid cells resolve neuroinflammation following cerebral ischaemia. Stroke Vasc Neurol. 2023.
Zhang G, et al. Spi1 regulates the microglial/macrophage inflammatory response via the PI3K/AKT/mTOR signaling pathway after intracerebral hemorrhage. Neural Regen Res. 2024;19(1):161–70.
Hu X, et al. Microglia/macrophage polarization dynamics reveal novel mechanism of injury expansion after focal cerebral ischemia. Stroke. 2012;43(11):3063–70.
Han, B., et al., Integrating spatial and single-cell transcriptomics to characterize the molecular and cellular architecture of the ischemic mouse brain. Sci Transl Med, 2024. 16(733): p. eadg1323.
Garcia-Bonilla L et al. Analysis of brain and blood single-cell transcriptomics in acute and subacute phases after experimental stroke. Nat Immunol. 2024.
Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s Disease. Cell. 2017;169(7):1276–e129017.
Mifflin L, et al. A RIPK1-regulated inflammatory microglial state in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A, 2021;118(13).
Li Q, et al. Developmental heterogeneity of Microglia and Brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101(2):207–e22310.
Daëron M, et al. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43.
Coxon CH, Geer MJ, Senis YA. ITIM receptors: more than just inhibitors of platelet activation. Blood. 2017;129(26):3407–18.
Deng M, et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature. 2018;562(7728):605–9.
Xu Z, et al. ILT3.Fc-CD166 Interaction induces inactivation of p70 S6 kinase and inhibits Tumor Cell Growth. J Immunol. 2018;200(3):1207–19.
Su MT, et al. Fibronectin-LILRB4/gp49B interaction negatively regulates osteoclastogenesis through inhibition of RANKL-induced TRAF6/TAK1/NF-kB/MAPK signaling. Int Immunol. 2023;35(3):135–45.
Wang Y et al. Discovery of galectin-8 as an LILRB4 ligand driving M-MDSCs defines a class of antibodies to fight solid tumors. Cell Rep Med, 2024. 5(1).
Zhou H, et al. Leukocyte immunoglobulin-like receptor B4 protects against cardiac hypertrophy via SHP-2-dependent inhibition of the NF-kappaB pathway. J Mol Med (Berl). 2020;98(5):691–705.
Qiu T et al. Leukocyte immunoglobulin-like receptor B4 deficiency exacerbates acute lung injury via NF-κB signaling in bone marrow-derived macrophages. Biosci Rep. 2019:39(6).
Lu Y, et al. Hepatic leukocyte immunoglobulin-like receptor B4 (LILRB4) attenuates nonalcoholic fatty liver disease via SHP1-TRAF6 pathway. Hepatology. 2018;67(4):1303–19.
Lee HN et al. NK cells require immune checkpoint receptor LILRB4/gp49B to control neurotropic Zika virus infections in mice. JCI Insight. 2022;7(3).
Sharma N et al. LILRB4 suppresses immunity in solid tumors and is a potential target for immunotherapy. J Exp Med. 2021;218(7).
Xu Z, et al. Suppression of experimental autoimmune encephalomyelitis by ILT3.Fc. J Immunol. 2021;206(3):554–65.
Jiang Z et al. LILRB4 deficiency aggravates the development of atherosclerosis and plaque instability by increasing the macrophage inflammatory response via NF-kappaB signaling. Clin Sci (Lond), 2017.
Butler A, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–20.
Kirschbaum KM, Hiemke C, Schmitt U. Rotarod impairment: catalepsy-like screening test for antipsychotic side effects. Int J Neurosci. 2009;119(10):1509–22.
Kim DH, et al. Assessment of cognitive phenotyping in inbred, genetically modified mice, and transgenic mouse models of Alzheimer’s Disease. Exp Neurobiol. 2019;28(2):146–57.
Rosa I et al. Comparison between tail suspension swing test and standard rotation test in revealing early motor behavioral changes and neurodegeneration in 6-OHDA hemiparkinsonian rats. Int J Mol Sci. 2020;21(8).
Horiquini Barbosa E, et al. Assessments of Motor abnormalities on the Grid-walking and Foot-Fault tests from Undernutrition in Wistar rats. J Mot Behav. 2016;48(1):5–12.
Ruan J, Yao Y. Behavioral tests in rodent models of stroke. Brain Hemorrhages. 2020;1(4):171–84.
Bieber M, et al. Validity and reliability of neurological scores in mice exposed to Middle cerebral artery occlusion. Stroke. 2019;50(10):2875–82.
Chang CC, et al. BCL6 is required for differentiation of Ig-like transcript 3-Fc-induced CD8+ T suppressor cells. J Immunol. 2010;185(10):5714–22.
Vlad G, et al. Membrane and soluble ILT3 are critical to the generation of T suppressor cells and induction of immunological tolerance. Int Rev Immunol. 2010;29(2):119–32.
Kim-Schulze S, et al. Recombinant Ig-like transcript 3-Fc modulates T cell responses via induction of Th anergy and differentiation of CD8+ T suppressor cells. J Immunol. 2006;176(5):2790–8.
Shi Z, et al. Microglia drive transient insult-induced brain injury by chemotactic recruitment of CD8(+) T lymphocytes. Neuron. 2023;111(5):696–e7109.
Jorfi M, et al. Infiltrating CD8+ T cells exacerbate Alzheimer’s disease pathology in a 3D human neuroimmune axis model. Nat Neurosci. 2023;26(9):1489–504.
Su W, et al. CXCR6 orchestrates brain CD8+ T cell residency and limits mouse Alzheimer’s disease pathology. Nat Immunol. 2023;24(10):1735–47.
Mussai F, et al. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood. 2013;122(5):749–58.
Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16(12):1896–905.
Ayata P, et al. Epigenetic regulation of brain region-specific microglia clearance activity. Nat Neurosci. 2018;21(8):1049–60.
Chamorro A, et al. The immunology of acute stroke. Nat Rev Neurol. 2012;8(7):401–10.
Butowski N, et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neurooncology. 2016;18(4):557–64.
Beuker C, et al. Stroke induces disease-specific myeloid cells in the brain parenchyma and pia. Nat Commun. 2022;13(1):945.
Zheng K et al. Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke. J Cereb Blood Flow Metab, 2021:271678X211026770.
Su MT, et al. LILRB4 promotes tumor metastasis by regulating MDSCs and inhibiting miR-1 family miRNAs. Oncoimmunology. 2022;11(1):2060907.
Li Q, Wei G, Tao T. Leukocyte immunoglobulin-like receptor B4 (LILRB4) negatively mediates the pathological cardiac hypertrophy by suppressing fibrosis, inflammation and apoptosis via the activation of NF-kappaB signaling. Biochem Biophys Res Commun. 2019;509(1):16–23.
Li D, et al. Upregulation of Microglial ZEB1 ameliorates brain damage after Acute ischemic stroke. Cell Rep. 2018;22(13):3574–86.
Cui P, et al. Microglia/macrophages require vitamin D signaling to restrain neuroinflammation and brain injury in a murine ischemic stroke model. J Neuroinflammation. 2023;20(1):63.
Li M, et al. Astrocyte-derived interleukin-15 exacerbates ischemic brain injury via propagation of cellular immunity. Proc Natl Acad Sci U S A. 2017;114(3):E396–405.
Chang CC, et al. Ig-like transcript 3 regulates expression of proinflammatory cytokines and migration of activated T cells. J Immunol. 2009;182(9):5208–16.
Czystowska-Kuzmicz M, et al. Small extracellular vesicles containing arginase-1 suppress T-cell responses and promote tumor growth in ovarian carcinoma. Nat Commun. 2019;10(1):3000.
Naing A et al. 747 A phase 1 trial of IO-202, an antagonist antibody targeting myeloid checkpoint LILRB4 (ILT3), as monotherapy and in combination with pembrolizumab in adult patients with advanced relapsed or refractory solid tumors. In Regular and Young Investigator Award Abstracts. 2022. pp. A780-A780.
Russo RS, et al. Efficacy and safety of pegzilarginase in arginase 1 deficiency (PEACE): a phase 3, randomized, double-blind, placebo-controlled, multi-centre trial. EClinicalMedicine. 2024;68:102405.
Arm JP, Nwankwo C, Austen KF. Molecular identification of a novel family of human ig superfamily members that possess immunoreceptor tyrosine-based inhibition motifs and homology to the mouse gp49B1 inhibitory receptor. J Immunol. 1997;159(5):2342–9.
Hou, J., et al., Antibody-mediated targeting of human microglial leukocyte Ig-like receptor B4 attenuates amyloid pathology in a mouse model. Sci Transl Med, 2024. 16(741): p. eadj9052.
Bonkhoff AK, et al. Outcome after acute ischemic stroke is linked to sex-specific lesion patterns. Nat Commun. 2021;12(1):3289.
Acknowledgements
We appreciated Xuebin Zhang for his guidance on the pathology experiments section of this manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82201442, 82371320 and 82071316), Tianjin Natural Science Foundation (23JCZXJC00110) and the Tianjin Key Medical Discipline (Specialty) Construction Project (No. TJYXZDXK-052B).
Author information
Authors and Affiliations
Contributions
Y.M. and K.Z. contributed equally to this manuscript as co-first authors, responsible for conducting the primary experiments, analyzing experimental data, and drafting and revising the manuscript. J.C., X.Y. provided guidance for the experimental design. C.Z., Y.Z., T.C. executed a portion of the in vivo experiments, while L.C. analyzed the experimental data. J.W and Y.C. contributed equally as corresponding authors, overseeing the entire research process and ensuring the accuracy of the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All of animal procedures were approved by the Animal Experiments Ethical Committee of Tianjin Medical University and Tianjin Huanhu Hospital, performed according to the Revised Guide for the Care and Use of Laboratory Animals. Studies were performed under Research ethics committee approvals HHLL-2023-003.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ma, Y., Zheng, K., Zhao, C. et al. Microglia LILRB4 upregulation reduces brain damage after acute ischemic stroke by limiting CD8+ T cell recruitment. J Neuroinflammation 21, 214 (2024). https://doi.org/10.1186/s12974-024-03206-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12974-024-03206-4