Abstract
Background
Fingolimod (FTY720), the first oral treatment for multiple sclerosis (MS), blocks immune cell trafficking and prevents disease relapses by downregulation of sphingosine-1-phosphate receptor. We determined the effect of FTY720 on human T cell activation and effector function.
Methods
T cells from MS patients and healthy controls were isolated to measure gene expression profiles in the presence or absence of FTY720 using nanostring and quantitative real-time polymerase chain reaction (qPCR). Cytokine protein expression was measured using luminex assay and flow cytometry analysis. Lentivirus vector carrying short hairpin RNA (shRNA) was used to knock down the expression of specific genes in CD4+ T cells. Chromatin immunoprecipitation was performed to assess T cell factor 1 (TCF-1) binding to promoter regions. Luciferase assays were performed to test the direct regulation of interferon gamma (IFN-γ) and granzyme B (GZMB) by TCF-1. Western blot analysis was used to assess the phosphorylation status of Akt and GSK3β.
Results
We showed that FTY720 treatment not only affects T cell trafficking but also T cell activation. Patients treated with FTY720 showed a significant reduction in circulating CD4 T cells. Activation of T cells in presence of FTY720 showed a less inflammatory phenotype with reduced production of IFN-γ and GZMB. This decreased effector phenotype of FTY720-treated T cells was dependent on the upregulation of TCF-1. FTY720-induced TCF-1 downregulated the pathogenic cytokines IFN-γ and GZMB by binding to their promoter/enhancer regions and mediating epigenetic modifications. Furthermore, we observed that TCF-1 expression was lower in T cells from multiple sclerosis patients than in those from healthy individuals, and FTY720 treatment increased TCF-1 expression in multiple sclerosis patients.
Conclusions
These results reveal a previously unknown mechanism of the effect of FTY720 on human CD4+ T cell modulation in multiple sclerosis and demonstrate the role of TCF-1 in human T cell activation and effector function.
Similar content being viewed by others
Background
Multiple sclerosis is a chronic inflammatory disorder of the central nervous system (CNS) driven by autoreactive lymphocytes that induce an inflammatory cascade leading to damage of myelin and axon, resulting in neurodegeneration [1]. In particular, T cells that produce pro-inflammatory cytokines such as interferon gamma (IFN-γ) and interleukin (IL)-17 play a critical role in multiple sclerosis pathogenesis [1–8], and reduced levels of these effector molecules are associated with better therapeutic responses [9–11]. Although the expression of granzyme B (GZMB) is linked to the pathogenic signature of T cells in experimental autoimmune encephalomyelitis (EAE) [12] and multiple sclerosis [13], its precise role in multiple sclerosis is still under investigation.
Fingolimod (FTY720), a sphingosine 1-phosphate (S1P) receptor modulator, was approved as the first oral treatment for multiple sclerosis based on results of three separate clinical trials among patients with relapsing-remitting multiple sclerosis (RRMS) [14–16]. S1P receptors are highly expressed on membranes of lymphocytes and are critical for T and B cell egress from secondary lymphoid organs. The phosphorylated form of FTY720 causes internalization and degradation of S1P receptors, resulting in the retention of lymphocytes in lymph nodes [17]. FTY720 primarily reduces the number of naïve T cells and central memory T cells in the circulation due to their expression of homing receptor CCR7 [18].
T cell factor 1 (TCF-1), also known as TCF7 (gene name), is a transcription factor present in hematopoietic T cells that has an important function in T cell development in the thymus. TCF-1 negatively regulates Th1 [19] and Th17 [20, 21] differentiation while promoting Th2 differentiation, via stimulation of GATA3 (a Th2-specific transcription factor) [19]. TCF7 knock-out mice are susceptible to EAE [20] and develop aggressive T cell deficiencies resembling human T cell acute lymphoblastic leukemia [22]. Interestingly, a computational re-analysis of multiple sclerosis-associated single nucleotide polymorphism data from 112 different cell types suggests that TCF7 is associated with multiple sclerosis [23], and a recent genome-wide association study identified the single nucleotide polymorphism rs756699 located on the TCF7 gene in multiple sclerosis patients [24]. However, the role of TCF-1 in the regulation of human CD4+ T cell effector function and its relevance to multiple sclerosis and treatment response are unknown.
In this study, we found that FTY720 modulates CD4+ T cell activation and effector function through TCF-1. FTY720-induced TCF-1 regulates the expression of IFN-γ and GZMB in T cells. Furthermore, T cells from multiple sclerosis patients exhibit lower TCF7 expression than those from healthy individuals, and FTY720 treatment upregulates TCF7 expression in T cells from both healthy controls and patients. Our findings establish that TCF-1 expression in human CD4+ T cells is linked to multiple sclerosis and that treatment with FTY720 increases TCF-1 expression, which regulates IFN-γ and GZMB production.
Methods
Subjects and blood samples
Peripheral venous blood was collected after obtaining informed consent from healthy individuals and multiple sclerosis patients. All patients were seen at the Partners Multiple Sclerosis Center at Brigham and Women’s Hospital. We included untreated RR multiple sclerosis patients and patients treated with FTY720 before and after 3 months of treatment. Patients were classified based upon their clinical characteristics as defined by 2010 Revisions to the McDonald Criteria [25] with the help of trained neurologists. Untreated multiple sclerosis patients had received no treatment with glatiramer acetate or interferons in the past 3 months, no treatment with other disease-modifying therapy in the past 6 months, and no steroids in the past month. Detailed characteristics of these patients are shown in Additional file 1: Table S1. Blood samples were collected under the Comprehensive Longitudinal Investigation of Multiple Sclerosis at Brigham and Women’s Hospital (CLIMB). This study was conducted in accordance with the WMA Declaration of Helsinki regarding ethical principles for medical research involving human subjects. The Partners Human Research Committee/Instutional Review Board approved the use of human material (IRB protocols 1999P010435/BWH and 2012P000394).
Naïve CD4+ T cell isolation, culture, and flow cytometry analysis
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation (Pharmacia LKB Biotechnology, Piscataway, NJ). Naïve T cells from PBMCs were isolated using a Miltenyi Biotec (Alburn, CA) negative selection kit. Purified naïve CD4+ T cells were activated with plate-bound anti-CD3 (5 μg/ml, BD Bioscience, San Jose CA), soluble anti-human CD28 (1 μg/ml, BD Bioscience), and IL-2 (20 ng/ml, R&D Systems) with or without FTY720 (100 ng/ml, Novartis). After 6 days, cell-free culture supernatants were collected for cytokine analysis by Luminex assay (Miltenyi Biotec), and cells were harvested for RNA extraction and intracellular staining. Naïve T cells were stimulated with PMA (Sigma), ionomycin (Sigma), and Golgistop for 4 h. Cells were stained for anti-human CD4 APC (BD Bioscience) and violet fluorescent reactive dye VVD (Life Technologies), and then cells were fixed and permeabilized with BD fixation and permeabilization buffer and stained for IFN-γ FITC and GZMB FITC (BD Bioscience). For surface staining, the following antibodies were used: anti-human CD4 pacific blue, anti-human CCR7 PE, and anti-human CD45RA APC, IgG2a PE isotype control, IgG2b, and k APC isotype control (all from BD Bioscience). All antibodies were titrated for flow cytometry, which was performed on a BD LSR II (BD Bioscience) and analyzed using Flowjo software.
Nanostring and quantification by real-time PCR
Total RNA was isolated from cultured T cells using RNA isolation kit (Norgen Biotek). RNA expression of 500 immune genes was detected by NanoString array (nCounter, Gene expression code set, Human Immunology Kit). Experiment was performed and analyzed following the manufacturers’ instructions. Data were normalized to spiked positive controls and housekeeping genes (nSolver Analysis system). Results from NanoString arrays were validated using Taqman PCR (Life Technologies). Briefly, first-strand cDNA synthesis was performed for each RNA sample from 0.5 to 1 μg of total RNA using Taqman reverse transcription reagents (Applied Biosystems). Quantitative real-time polymerase chain reaction (qPCR) was performed using primers for TCF-7 (Hs00175273_m1), IFNG (Hs99999041_m1), and GZMB (Hs001554355_m1) and the housekeeping genes GAPDH (Hs02758991_g1), G6PD (Hs00166169_m1), GUSB (Hs00939627_m1), TPB (Hs00427620_m1), TUBB (Hs00742828_s1), POLR1B (Hs00219263_m1), POLR2A (Hs00172187_m1), HPRT1 (Hs02800695_m1), PRL19 (Hs02338565_gH), EEFG1 (Hs01922638_u1), ALAS1 (Hs00963534_m1), and OAZ1 (Hs00427927_m1) with TaqMan Fast Universal qPCR Master Mix (No ampErase Uracil N-Glycosylase, Life Technologies). Samples were run on QuantStudio 7 (Applied Biosystems, Life Technologies). The mean of the 12 housekeeping genes was used as an endogenous control to normalize total cDNA for each sample, and all values are shown as relative expression.
In vitro knockdown with shRNA
The expression of TCF-1 was knocked down in primary naïve CD4+ T cells by lentivirus vector carrying short hairpin RNA (shRNA) against TCF-1 (mission shRNA bacterial glycerol stock TRCN0000281336, Sigma-Aldrich) or a non-targeting sequence (a gift from Dr. Thomas Pertel at BWH and Dr. David Root at Broad Institute) as a control. Primary naïve CD4+ T cells were cultured in a 24-well plate in the presence of anti-CD3, anti-CD28, and IL2 and incubated with lentivirus. After 3 days, the lentivirus was washed away, and puromycin was added (1.5 μg/ml) to select transduced T cells for 3 days. Live cells were sorted using intracellular fluorescence-activated cell sorting, and the expression of TCF-1 was measured using qPCR.
Chromatine immune precipitation assay
The binding of TCF-1, H3-Lys9, and H3-Lys27 on IFNG and GZMB were analyzed using chromatin immunoprecipitation (ChIP) assay as described previously [26, 27]. The antibodies used for ChIP assay were anti-TCF-1 (Cell Signaling), anti-trimethyl-histone H3-Lys9 (Millipore), and anti-trimethyl-histone H3-Lys27 antibodies (Millipore) or Rabbit IgG (Millipore). Input DNA and DNA recovered after immunoprecipitation were analyzed by real-time qPCR using primer pairs for IFNG and GZMB (IFNG: forward 5′-GAAGAGTCAACATTTTACCAGGGC and reverse 5′-GTGACAGATAGGCAGGGATGATAG; GMZB: forward 5′-GAACCTGGTGCAATTACCAGAAT and reverse 5′CTTTTCACAGGGATAAACTGCTGG) with SyBR green Fast Master mix (Applied Biosystems). Values for TCF-1 binding with IFNG promoter and GZMB enhancer regions were normalized to IgG.
Luciferase reporter assay
HEK293T cells were maintained in DMEM medium (Gibco) supplemented with 10 % fetal bovine serum (Gibco), 4 mM L-glutamine (Lonza), 1 mM sodium pyruvate (Lonza), 1 % non-essential amino acid (Lonza), and 10 mM HEPES (Lonza) at 37 °C/5 % CO2. Forty-eight hours prior to transfection, cells were seeded at 10,000 cells per well in a 96-well tissue culture plate (Perkin Elmer). Cells were transfected with the indicated amounts of each expression vector using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Renilla luciferase vector (12.5 ng) was used for normalization of transfection efficiency. Forty-eight hours after transfection, cells were assayed using the Dual-Luciferase Reporter Assay System kit (Promega) as per the manufacturer’s instructions. Firefly luciferase values were normalized to Renilla luciferase levels. The results were expressed as relative luciferase activity in relative light units (RLU). Transfections were carried out in triplicate in three independent experiments, and results are expressed as mean ± standard error.
Plasmids
The −3.6-kb human IFNG-luc in a pGL3 vector background was a gift from Howard Young (Addgene plasmid #17599) [28], pcDNA3-HA-TCF1 was a gift from Kai Ge (Addgene plasmid #40620) [29], plasmid encoding Tbet was a gift from Dr. Christine Campbell [30], 1436 pcDNA3 Flag HA was a gift from William Sellers (Addgene plasmid #10792), and pGL3 basic and pGL3 promoter vector were purchased from Promega. A 1.04-kb fragment of the human GZMB enhancer region was inserted as three repeats between KpnI and NheI sites into the pGL3 promoter vector to generate the GZMB-luc plasmid (GenScript).
Western blot
Naïve T cells were isolated from PBMCs, activated in the presence or absence of FTY720, and collected after 15, 30, 60, or 120 min. Cells were lysed with RIPA assay buffer (Thermo Scientific) supplemented with protease and phosphatase inhibitors (Thermo Scientific). Total protein concentration was determined by BCA assay (Thermo Scientific). Samples were prepared with 20 μg of protein, loading buffer (Life Technologies), and reducing agent (Life Technologies) and then heated for 10 min at 70 °C before use. Samples were run on a 10 % bis-tris gel (NOVO, Life Technologies), transferred to PVDF membrane, and detected by immunoblot. The following antibodies were used: p-AKT (Cell Signaling Technology), p-GSK3β (Cell Signaling Technology), pan-AKT (Cell Signaling Technology), pan-GSK3β (Cell Signaling Technology), beta actin (Cell Signaling Technology), and anti-rabbit IgG HRP (Cell Signaling Technology). The membrane was blocked with 5 % bovine serum albumin followed by primary and secondary antibody incubations as per the manufacturers’ instructions. Immunoblots were developed using ECL Prime (GE Healthcare).
Statistical analysis
All statistical analyses were performed using GraphPad Prism software. Paired and unpaired non-parametric t tests were used to test for group differences. Correlations were calculated using Spearman’s rank correlation coefficient.
Results
FTY720 treatment decreases circulating CD4+ T cells and increases TCF7 expression in CD4+ T cells.
FTY720 decreases relapses in multiple sclerosis, and its therapeutic effects are attributed to the sequestration of lymphocytes into the lymph nodes [18]. We characterized the frequency of total and naïve CD4+ T cells in multiple sclerosis patients before and 3 months after FTY720 treatment. Consistent with previous observation, [18] FTY720 treatment reduced the percentage and absolute number of circulating total CD4+ (Additional file 2: Figure S1 A and C) and naïve CD4+ T cells (Additional file 2: Figure S1 B and D), 3 months after treatment relative to baseline measurements. FTY720-treated patients continue to generate an immune response against foreign antigens indicating that the T cells still have the ability to egress from lymph nodes and potentially impact the CNS [31]. To further define the impact of FTY720 on immune cell function, we assessed the effect of FTY720 treatment on CD4+ T cell effector function.
Purified naïve T cells from healthy individuals were activated in the presence or absence of FTY720 at concentration of 100 ng/ml. Although the physiological concentration of FTY720 in blood is in a range of 5 to 10 ng/ml [18, 32], the concentration of the lymph nodes is 10 times higher [33]. Since we wanted to recapitulate the physiological effect of FTY720 on T cell activation within the lymph nodes, we chose to use the concentration of 100 ng/ml. We measured the effect of FTY720 on T cell-associated immune gene expression using nanostring-based immune arrays. We found that FTY720-treated T (T-FTY720) cells had a distinct immune gene signature, with 33 upregulated genes and 24 downregulated genes relative to untreated T (T-CTL) cells (Fig. 1a). We selected TCF7 based upon its relevance shown by several studies highlighting its role on T cell differentiation in mice and in EAE induction. In addition, recent observations in GWAS studies have correlated TCF7 with MS pathogenesis. These studies, however, did not address the functional role of TCF7 in MS patients, thus, we focused our attention on TCF7. We found that T-FTY720 cells showed greater TCF7 expression than T-CTL cells (Fig. 1b). These results were further confirmed by qPCR, which also showed increase in the expression of TCF7 in T-FTY720 compared to T-CTL (Fig. 1c). To determine whether this increase in TCF7 expression was specifically brought about by FTY720, we activated T cells in the presence of other approved drugs for multiple sclerosis treatment, namely interferon beta and natalizumab. We found increased expression of TCF7 only in T-FTY720 cells but not T cells treated with interferon beta (T-IFN beta) or natalizumab (T-Ty) (Additional file 3: Figure S2), suggesting that expression of TCF7 is specifically enhanced upon FTY720 treatment.
FTY720 treatment increases TCF7 expression in T cells. a Heatmap showing significantly changing mRNA expression upon CD4 naïve T cell activation in the presence or absence of FTY720 as measured by a nanostring array of 500 immune genes. b Expression of TCF7 in in vitro activated T cells from healthy control (HC) individuals in the presence (T-FTY720) or absence (T-CTL) of FTY720 analyzed using nanostring (n = 6). c Expression of TCF7 in in vitro activated T cells from HC individuals in the presence or absence of FTY720 analyzed using qPCR (n = 10). **p < 0.01, paired non-parametric t tests
FTY720 treatment decreases effector T cell function
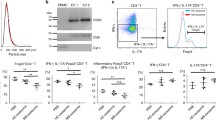
To investigate whether FTY720 modulates effector function of CD4+ T cells, we measured cytokine expression in naïve CD4+ T cells treated with or without FTY720. We observed that, compared to T-CTL cells, T-FTY720 cells showed lower messenger RNA (mRNA) expression of IFNG and GZMB as measured by nanostring array (Fig. 2a) and qPCR (Fig. 2b) and lower protein expression of IFN-γ and GZMB as measured by Luminex-based assay (Fig. 2c) and intracellular fluorescence-activated cell sorting analysis (Fig. 2d).
FTY720 treatment decreases effector T cell function. a Expression of IFNG and GZMB in in vitro activated T cells from healthy individuals in the presence or absence of FTY720 as determined by nanostring (n = 6). b Expression of IFNG and GZMB in in vitro activated T cells from healthy individuals in the presence or absence of FTY720 as determined by qPCR (n = 6). c Expression of IFN-γ and GZMB in in vitro activated T cells from healthy individuals in the presence or absence of FTY720 as determined by Luminex-based assay (n = 6). d Expression of IFN-γ and GZMB in in vitro activated T cells from healthy individuals in the presence or absence of FTY720 using flow cytometry (n = 6). *p < 0.05, **p < 0.01, paired non-parametric t tests
In addition to IFN-γ and GZMB expressions, we measured the expression of pro-inflammatory cytokines IL-17, granulocyte macrophage colony-stimulating factor (GMCSF), and tumor necrosis factor alpha (TNF-α) in supernatant collected from T-FTY720 and T-CTL cells using Luminex-based assay. We found decreased expression of IL-17, GMCSF, and TNF-α in T-FTY720 cells compared to T-CTL cells (Additional file 4: Figure S3 A). To determine whether this decreased cytokine production was related to excessive cell death, we measured the percentage of dead cells in T cell cultures in the presence or absence of FTY720. We found no difference in the percentage of dead T cells between groups (Additional file 4: Figure S3 B), suggesting that the effect of FTY720 is not mediated by increased cell death.
TCF-1 regulates IFN-γ and GZMB expression
TCF-1 regulates T cell development and differentiation in mice [19–21, 34–36]. However, the role of TCF-1 in the regulation of CD4+ T cell function in humans remains unclear. Therefore, we analyzed the transcriptional regulation of IFNG and GZMB in CD4+ T cells by TCF-1 upon FTY720 treatment. We performed an in silico analysis and found that TCF-1 has binding sites in the IFNG promoter (−321 to −85 bp) and GZMB enhancer (+1606 to +1841 bp) regions (Fig. 3a). Using chromatin immunoprecipitation (ChIP) assay, we found increased binding of TCF-1 on both IFNG promoter and GZMB enhancer regions in FTY720-treated cells (Fig. 3b).
TCF-1 regulates IFNG and GZMB expression. a Binding region of TCF-1 in the promoter of IFNG and enhancer of GZMB. Blue arrows represent transcription start site. b qPCR analysis of chromatin-immunoprecipitated T cells showing interaction of TCF-1 with the IFNG promoter and GZMB enhancer region in primary T cells treated with or without FTY720 (n = 7). Data are shown as mean ± SEM. *p < 0.05, paired non-parametric t tests. c qPCR analysis of TCF7 expression in T cells treated with TCF7 shRNA (sh-TCF7) or negative control (sh-CTL). **p < 0.01, paired non-parametric t tests. d qPCR analysis of IFNG and GZMB expression in T cells treated with TCF7 shRNA or negative control. **p < 0.01, paired non-parametric t tests. e Luciferase activity in HEK293T cells transfected with plasmids encoding firefly luciferase downstream of the IFNG promoter region (IFNG-luc), Tbet, TCF-1, or empty vector control. f Luciferase activity in HEK293T cells transfected with plasmids encoding firefly luciferase downstream of the GZMB enhancer region (GZMB-luc), TCF-1 or empty vector control. RLU relative light units. Data are shown as mean ± SEM. * **p < 0.01, ***p < 0.001, unpaired non-parametric t tests
Regulation of IFN-γ and GZMB expression by TCF-1 was investigated by knockdown of TCF7 expression using lentivirus particles carrying shRNA against TCF7 or negative control. We found that shRNA-mediated knockdown of TCF7 (Fig. 3c) resulted in an increased IFNG and GZMB expression in T cells suggesting that TCF-1 might directly affect IFNG and GZMB expression (Fig. 3d).
Next, we performed a dual-luciferase reporter assay in HEK293T cells to determine whether TCF-1 directly represses IFN-γ and GZMB expression by binding to their promoter and enhancer regions, respectively. We used a plasmid encoding firefly luciferase downstream of the IFNG promoter region (IFNG-luc) that included the putative TCF-1 binding site determined by the ChIP assay as well as a binding site for Tbet, a transcription factor that reportedly activates IFN-γ expression [30, 37, 38]. We found increased relative firefly luciferase levels in cells co-transfected with IFNG-luc and Tbet expression plasmid compared with cells transfected with an empty vector. However, cells co-transfected with TCF-1 expression plasmid and IFNG-luc did not induce luciferase reporter activity; however, when both Tbet and TCF-1 expression plasmids were co-transfected with IFNG-luc, Tbet-induced firefly luciferase activity was reduced in a TCF-1 dose-dependent manner (Fig. 3e), suggesting that TCF-1 represses transcription at the IFNG promoter. We used a reporter plasmid containing the 1.04-kb GZMB enhancer sequence identified from our ChIP assay at a site upstream of the SV40 promoter that regulates firefly luciferase (GZMB-luc). We observed that firefly luciferase activity decreased significantly when HEK293T cells were co-transfected with GZMB-luc and increasing amounts of the TCF-1 expression plasmid (Fig. 3f). Firefly luciferase activity was unchanged in cells co-transfected with GZMB-luc reporter plasmid and an empty vector suggesting that repression of luciferase activity was TCF-1 dependent.
Using in silico analysis, we observed that TCF-1 also has putative binding sites in TNF-α, IL-17, and CSF2 promoters (Additional file 4: Figure S3 C). However, we did not observe the binding of TCF-1 to CSF2 and TNF-α promoters in ChIP assay (data not shown). We did not observe IL-17 production in more than 50 % of healthy individuals. Together, these results suggest that TCF-1 binds to IFN-γ promoter and GZMB enhancer regions and regulates their expression in FTY720-activated T cells, and the effect of FTY720 on TNF-α and GMCSF expression does not depend on TCF-1.
FTY720 downregulates Akt and GSK3 β phosphorylation and induces epigenetic modification
Increased expression of TCF-1 upon FTY720 treatment suggests that perhaps FTY720 affects events downstream of S1P and Wnt signaling pathways. Phosphorylation of Akt, which is a fundamental step of the sphingosine pathway [39], induces phosphorylation of other proteins and enzymes such as GSK3β [34, 40]. Akt inhibits GSK3 kinase activity via phosphorylation of serine 9 (Ser9) in GSK3β, resulting in its inactivation [41, 42]. GSK3β also plays an important role in signaling downstream of the Wnt pathway. Inactivation of GSK3β allows unphosphorylated β-catenin to accumulate, translocate into the nucleus, and activate gene transcription by replacing co-repressor molecules bound to TCF-1 [43, 44]. On the other hand, decreased levels of phosphorylated/inactive GS3Kβ result in phosphorylation of β-catenin and its subsequent proteasomal degradation [43]. We performed western blot analysis to assess the phosphorylation status of Akt at Ser473 and of GSK3β at Ser9 in FTY720-treated and FTY720-untreated T cells at different time points. We found decreased phosphorylation of Akt at Ser473 (Fig. 4a) and GS3Kβ at Ser9 (Fig. 4b) in T-FTY720 cells compared with T-CTL cells at all time points. This reduced phosphorylated GS3Kβ associated with lower levels of β-catenin in T-FTY720 cells than in T-CTL cells (Fig. 4c). These results indicate that FTY720 inhibits β-catenin accumulation by reducing Akt-mediated inactivation/phosphorylation of GSK3β.
FTY720 downregulates Akt and GSK3β phosphorylation and induces epigenetic modification. a Western blot analysis of cell extracts from FTY720-treated (T-FTY720) and control T (T-CTL) cells at the indicated time points and analyzed for phosphorylated Akt (p-Akt) and total Akt (tot-Akt). β-actin served as a loading control. A representative image from one experiment is shown. b Immunoblot of phosphorylated GSK3β (pGSK3β) and total GSK3β (tot-GSK3β) in cell extracts from T-FTY720 and T-CTL cells at the indicated time points. β-actin served as a loading control. A representative image from one experiment is shown. c Immunoblot of β-catenin in cell extracts from FTY720 and T-CTL cells at the indicated time points. β-actin served as a loading control. A representative image from one experiment is shown. d qPCR analysis of chromatin-immunoprecipitated T cells showing methylation status of H3K9 and H3K27 on the IFNG promoter region (n = 7). Data are shown as mean ± SEM. *p < 0.05, paired non-parametric t tests. e qPCR analysis of chromatin-immunoprecipitated T cells showing the methylation status of H3K9 and H3K27 on the GZMB promoter region (n = 7). Data are shown as mean ± SEM. *p < 0.05, paired non-parametric t tests
In the absence of β-catenin, target genes are silenced by TCF-1-mediated recruitment of TLE/Groucho proteins [45]. qPCR analysis showed no differences in the expression of TLE1, TLE2, and TLE4 but a modest increase in TLE3 expression in T-FTY720 cells compared with T-CTL cells (Additional file 5: Figure S4). The recruitment of TLE/Groucho proteins by TCF-1 results in transcriptional suppression of target genes by inducing epigenetic modifications [46, 47]. To gain further insight into the molecular mechanism by which FTY720-induced TCF-1 regulates IFN-γ and GZMB expression, we measured methylation status of histones H3K9 and H3K27 on the IFNG promoter and GZMB enhancer regions after FTY720 treatment using ChIP assay. We observed increased levels of tri-methylation at H3K9 and H3K27 in the IFNG promoter (Fig. 4d) and GZMB enhancer (Fig. 4e) regions in T-FTY720 cells compared with T-CTL cells. These results suggest that FTY720-induced TCF-1 regulates IFN-γ and GZMB expression by directly binding and inducing repressive chromatin modifications such as tri-methylation of Lys9 and Lys27 of histone H3.
Expression of TCF7 is decreased in CD4+ T cells from multiple sclerosis patients
To determine the relevance of TCF-1 in multiple sclerosis pathogenesis, we first assessed its expression in CD4+ T cells from multiple sclerosis patients. We found that TCF7 expression was decreased in T cells from untreated RRMS patients (T-RRMS) compared with T cells from healthy controls (T-CTL) in both ex vivo isolated (Fig. 5a) and in vitro-activated (b) T cells. To assess whether FTY720 upregulates TCF7 expression in T cells from RRMS patients, we activated T-RRMS cells in the presence or absence of FTY720 and found higher TCF7 expression in FTY720-treated T-RRMS cells (Fig. 5c).
RRMS T cells show decreased TCF7 expression. a Expression of TCF7 in ex vivo T cells from RRMS patients (T-RRMS, n = 26) and healthy individuals (T-CTL, n = 21) measured using qPCR. b Expression of TCF7 in activated T-RRMS cells (n = 18) and T-CTL cells (n = 19) measured using qPCR. c TCF7 expression measured by qPCR in in vitro activated T cells from RRMS patients (n = 10) in the presence or absence of FTY720. d Correlation between TCF7 expression and EDSS score. *p < 0.05, **p < 0.01, ***p < 0.001, unpaired non-parametric t tests and Spearman correlation
In order to determine whether the expression of TCF7 is related to multiple sclerosis disease activity, we computed correlations between TCF7 expression in T cells and clinical parameters such as Expanded Disability Status Scale (EDSS) score, disease duration, and number of relapses 2 years before the date of blood draw. We found a significant negative correlation (r 2 = −0.45, p < 0.05) of TCF7 expression with EDSS score (Fig. 5d). We did not find any correlations between TCF7 expression and disease duration or number of relapses (data not shown). These results suggest that TCF7 is involved in multiple sclerosis pathogenesis.
Discussion
FTY720, the first oral drug approved for multiple sclerosis treatment, is an S1P analog that induces internalization of S1P receptors on T cells, resulting in lymphocyte sequestration inside lymph nodes [33]. In addition, FTY720 exhibits immunomodulatory effects in various immune cell populations [17, 31, 48–51], including T cells [52, 53], B cells [54], dendritic cells [55, 56], regulatory T cells [9, 57–60], and monocytes [56]. FTY720 also regulates the dichotomy between Th1 and iTregs through S1P1- and mTOR-dependent pathways [60]. Here, we examined the effect of FTY720 in human T cells and studied its role in the regulation of pathogenic effector T cell function in MS.
We found that FTY720 treatment increased TCF7 expression and downregulated IFNG and GZMB expression in CD4+ T cells. These results are in line with a previous report which states that FTY720 treatment ameliorates EAE in mice and decreases IFN-γ and GZMB production in splenic CD8+ T cells [61].
In mice, TCF-1 expression is linked to regulation of inflammatory Th1 and Th17 differentiation and development of EAE [21, 36, 62]. In this study, we found that shRNA-mediated knockdown of TCF-1 increases IFNG and GZMB expressions, indicating that TCF-1 may play a direct role in IFNG and GZMB expressions. Indeed, ChIP assay and reporter assay showed that TCF-1 not only directly binds to IFNG promoter and GZMB enhancer regions in FTY720-treated T cells but also regulates their expression. Although we found decreased expression of IL-17, GMCSF, and TNF-α upon FTY720 treatment, these cytokines were not followed further in the current study because IL-17 expression and production was not observed in more than 50 % of healthy individuals and ChIP assay did not detect binding of TCF-1 to GMCSF or TNF-α promoter regions (data not shown).
Tbet is a master regulator of Th1 differentiation that regulates TCF-1 expression through direct interaction with Bcl-6 [63]. However, we found no differences in the expression of TBX21 or BCL6 in FTY720-treated T cells (data not shown), suggesting that a different mechanism was involved in TCF-1 induction in this context. TCF-1 has also been shown to regulate Th2 differentiation through GATA3 in mice [19]. However, we found no changes in the expression of GATA3 in human CD4+ T cells treated with FTY720 (data not shown).
Crosstalk between the sphingosine and Wnt signaling pathways has been previously shown in osteoblast-like cells through activation of Akt, GSK3β, and nuclear translocation of β-catenin followed by its association with TCF-1 [34]. In our study, we found decreased phosphorylation of Akt and GSK3β in FTY720-treated T cells and a reduction in β-catenin levels upon FTY720 treatment. Therefore, we speculate that in the presence of FTY720, there is a decrease in S1P1-mediated phosphorylation of Akt, which in turn decreases GSK3β phosphorylation. The active unphosphorylated GSK3β phosphorylates β-catenin, resulting in its proteasomal degradation. In the absence of β-catenin, TCF-1 binds to co-repressors such as TLE/Groucho family proteins and suppresses the transcription of target genes such as IFNG and GZMB (Additional file 6: Figure S5).
A recent mouse study shows that selective targeting of GSK3β in T cells leads to less severe EAE [63]. As the effect of GSK3β on TCF-1 expression was not investigated in this study, it is possible that therapeutic targeting of GSK3β might be linked to increased TCF-1 expression, which could reduce pathogenic T cell function and alleviate EAE symptoms.
In the absence of Wnt signaling, TCF-1 interacts with transcriptional repressor TLE/Groucho family members [62, 64], which repress transcription of target genes by inducing epigenetic modification [46, 47]. Acetylation and methylation on specific lysine residues are important epigenetic modifications for gene regulation. Transcription sites of actively transcribed genes are characterized by the presence of methylation at H3K4 or acetylation at H3K27, whereas gene repression is mediated by tri-methylation of H3K9 or H3K27 [65]. In mice, TCF-1 regulates Th17 differentiation through epigenetic changes [20, 21]. Here, we observed higher levels of histone H3K9 and H3K27 tri-methylation in the promoter region of IFNG and enhancer region of GZMB in T-FTY720 cells compared with T-CTL cells, supporting the hypothesis that FTY720 induces TCF-1-mediated regulation of IFN-γ and GZMB expression by chromatin modification. Although the mechanism of TCF-1 mediated epigenetic modifications was not addressed in this particular study, it is conceivable that TCF-1 brings about epigenetic changes by recruiting histone methyltransferases and other co-repressors such as TLE in the presence of FTY720.
Multiple sclerosis is the primary cause of non-traumatic neurologic disability in young adults. IFN-γ producing and GZMB secreting inflammatory T cells are implicated in its pathogenesis [3–6, 10, 66–68]. However, the regulatory mechanisms controlling pathogenic effector T cell function in multiple sclerosis are not completely understood. We found significantly decreased TCF7 expression in T cells from relapsing-remitting multiple sclerosis patients compared with those from healthy individuals and the treatment with FTY720 increased TCF-1 expression in both groups. Moreover, TCF7 expression in T cells is inversely correlated to disease disability as measured by EDSS.
Conclusions
We observed that TCF-1 expression is reduced in T cells from relapsing-remitting patients. FTY720 treatment increases TCF-1 expression in CD4+ T cells from multiple sclerosis patients as well as from healthy individuals. The modulation of the sphingosine pathway is able to interfere with the downstream signaling of the Wnt pathway by reducing the availability of β-catenin and subsequently repressing the transcription of target genes. FTY720-induced TCF-1 expression in CD4+ T cells downregulates the pathogenic cytokines IFN-γ and GZMB by binding to their promoter/enhancer region and mediating epigenetic modifications. We also showed that TCF7 expression negatively correlates with EDSS score in multiple sclerosis patients, suggesting a possible role of TCF-1 in the pathogenesis of multiple sclerosis. Together, our results reveal a previously unknown mechanism by which FTY720 functions in multiple sclerosis, and suggests that targeting TCF-1 signaling in CD4+ T cells could be a potential treatment strategy for multiple sclerosis.
Abbreviations
- CNS:
-
central nervous system
- EAE:
-
experimental autoimmune encephalomyelitis
- EDSS:
-
expanded disability status scale
- FTY720:
-
fingolimod
- GZMB:
-
granzyme B
- IFN-γ:
-
interferon gamma
- PBMCs:
-
peripheral blood mononuclear cells
- RLU:
-
relative light units
- RRMS:
-
relapsing remitting multiple sclerosis
- TCF-1:
-
T cell factor 1
- TNF-α:
-
tumor necrosis factor alpha
References
Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372:1502–17.
Brucklacher-Waldert V, Stuerner K, Kolster M, Wolthausen J, Tolosa E. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain. 2009;132:3329–41.
Jager A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol. 2009;183:7169–77.
Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747.
Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–45.
Neumann H, Medana IM, Bauer J, Lassmann H. Cytotoxic T lymphocytes in autoimmune and degenerative CNS diseases. Trends Neurosci. 2002;25:313–9.
Dhib-Jalbut S, Arnold DL, Cleveland DW, Fisher M, Friedlander RM, Mouradian MM, et al. Neurodegeneration and neuroprotection in multiple sclerosis and other neurodegenerative diseases. J Neuroimmunol. 2006;176:198–215.
Kurschus FC. T cell mediated pathogenesis in EAE: molecular mechanisms. Biomed J. 2015;38:183–93.
Serpero LD, Filaci G, Parodi A, Battaglia F, Kalli F, Brogi D, et al. Fingolimod modulates peripheral effector and regulatory T cells in MS patients. J Neuroimmune Pharmacol. 2013;8:1106–13.
Durelli L, Conti L, Clerico M, Boselli D, Contessa G, Ripellino P, et al. T-helper 17 cells expand in multiple sclerosis and are inhibited by interferon-beta. Ann Neurol. 2009;65:499–509.
Mehling M, Lindberg R, Raulf F, Kuhle J, Hess C, Kappos L, et al. Th17 central memory T cells are reduced by FTY720 in patients with multiple sclerosis. Neurology. 2010;75:403–10.
Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–9.
Cao Y, Goods BA, Raddassi K, Nepom GT, Kwok WW, Love JC, et al. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci Transl Med. 2015;7:287ra74.
Kappos L, Antel J, Comi G, Montalban X, O’Connor P, Polman CH, et al. Oral fingolimod (FTY720) for relapsing multiple sclerosis. N Engl J Med. 2006;355:1124–40.
Kappos L, Radue EW, O’Connor P, Polman C, Hohlfeld R, Calabresi P, et al. A placebo-controlled trial of oral fingolimod in relapsing multiple sclerosis. N Engl J Med. 2010;362:387–401.
Cohen JA, Barkhof F, Comi G, Hartung HP, Khatri BO, Montalban X, et al. Oral fingolimod or intramuscular interferon for relapsing multiple sclerosis. N Engl J Med. 2010;362:402–15.
Cohen JA, Chun J. Mechanisms of fingolimod’s efficacy and adverse effects in multiple sclerosis. Ann Neurol. 2011;69:759–77.
Mehling M, Brinkmann V, Antel J, Bar-Or A, Goebels N, Vedrine C, et al. FTY720 therapy exerts differential effects on T cell subsets in multiple sclerosis. Neurology. 2008;71:1261–7.
Yu Q, Sharma A, Oh SY, Moon HG, Hossain MZ, Salay TM, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat Immunol. 2009;10:992–9.
Ma J, Wang R, Fang X, Ding Y, Sun Z. Critical role of TCF-1 in repression of the IL-17 gene. PLoS One. 2011;6, e24768.
Yu Q, Sharma A, Ghosh A, Sen JM. T cell factor-1 negatively regulates expression of IL-17 family of cytokines and protects mice from experimental autoimmune encephalomyelitis. J Immunol. 2011;186:3946–52.
Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–26.
Disanto G, Kjetil Sandve G, Ricigliano VA, Pakpoor J, Berlanga-Taylor AJ, Handel AE, et al. DNase hypersensitive sites and association with multiple sclerosis. Hum Mol Genet. 2014;23:942–8.
Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60.
Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302.
Mascanfroni ID, Takenaka MC, Yeste A, Patel B, Wu Y, Kenison JE, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nat Med. 2015;21:638–46.
Gandhi R, Kumar D, Burns EJ, Nadeau M, Dake B, Laroni A, et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol. 2010;11:846–53.
Gonsky R, Deem RL, Bream JH, Lee DH, Young HA, Targan SR. Mucosa-specific targets for regulation of IFN-gamma expression: lamina propria T cells use different cis-elements than peripheral blood T cells to regulate transactivation of IFN-gamma expression. J Immunol. 2000;164:1399–407.
Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A. 2012;109:15324–9.
Butz NV, Gronostajski RM, Campbell CE. T-box proteins differentially activate the expression of the endogenous interferon gamma gene versus transfected reporter genes in non-immune cells. Gene. 2006;377:130–9.
Mehling M, Hilbert P, Fritz S, Durovic B, Eichin D, Gasser O, et al. Antigen-specific adaptive immune responses in fingolimod-treated multiple sclerosis patients. Ann Neurol. 2011;69:408–13.
David OJ, Kovarik JM, Schmouder RL. Clinical pharmacokinetics of fingolimod. Clin Pharmacokinet. 2012;51:15–28.
Sensken SC, Bode C, Graler MH. Accumulation of fingolimod (FTY720) in lymphoid tissues contributes to prolonged efficacy. J Pharmacol Exp Ther. 2009;328:963–9.
Matsuzaki E, Hiratsuka S, Hamachi T, Takahashi-Yanaga F, Hashimoto Y, Higashi K, et al. Sphingosine-1-phosphate promotes the nuclear translocation of beta-catenin and thereby induces osteoprotegerin gene expression in osteoblast-like cell lines. Bone. 2013;55:315–24.
Schilham MW, Wilson A, Moerer P, Benaissa-Trouw BJ, Cumano A, Clevers HC. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–91.
Yu Q, Sharma A, Sen JM. TCF1 and beta-catenin regulate T cell development and function. Immunol Res. 2010;47:45–55.
Lugo-Villarino G, Maldonado-Lopez R, Possemato R, Penaranda C, Glimcher LH. T-bet is required for optimal production of IFN-gamma and antigen-specific T cell activation by dendritic cells. Proc Natl Acad Sci U S A. 2003;100:7749–54.
Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–42.
Jo E, Sanna MG, Gonzalez-Cabrera PJ, Thangada S, Tigyi G, Osborne DA, et al. S1P1-selective in vivo-active agonists from high-throughput screening: off-the-shelf chemical probes of receptor interactions, signaling, and fate. Chem Biol. 2005;12:703–15.
Means CK, Brown JH. Sphingosine-1-phosphate receptor signalling in the heart. Cardiovasc Res. 2009;82:193–200.
Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, et al. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–32.
Wu D, Pan W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci. 2010;35:161–8.
Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–79.
Staal FJ, Clevers HC. WNT signalling and haematopoiesis: a WNT-WNT situation. Nat Rev Immunol. 2005;5:21–30.
Xue HH, Zhao DM. Regulation of mature T cell responses by the Wnt signaling pathway. Ann N Y Acad Sci. 2012;1247:16–33.
Chodaparambil JV, Pate KT, Hepler MR, Tsai BP, Muthurajan UM, Luger K, et al. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 2014;33:719–31.
Patel SR, Bhumbra SS, Paknikar RS, Dressler GR. Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol Cell. 2012;45:185–95.
Ingwersen J, Aktas O, Kuery P, Kieseier B, Boyko A, Hartung HP. Fingolimod in multiple sclerosis: mechanisms of action and clinical efficacy. Clin Immunol. 2012;142:15–24.
Mehling M, Kappos L, Derfuss T. Fingolimod for multiple sclerosis: mechanism of action, clinical outcomes, and future directions. Curr Neurol Neurosci Rep. 2011;11:492–7.
Mehling M, Johnson TA, Antel J, Kappos L, Bar-Or A. Clinical immunology of the sphingosine 1-phosphate receptor modulator fingolimod (FTY720) in multiple sclerosis. Neurology. 2011;76:S20–7.
Johnson TA, Shames I, Keezer M, Lapierre Y, Haegert DG, Bar-Or A, et al. Reconstitution of circulating lymphocyte counts in FTY720-treated MS patients. Clin Immunol. 2010;137:15–20.
Yokoseki A, Saji E, Arakawa M, Hokari M, Ishiguro T, Yanagimura F, et al. Relapse of multiple sclerosis in a patient retaining CCR7-expressing T cells in CSF under fingolimod therapy. Mult Scler. 2013;19:1230–3.
Johnson TA, Lapierre Y, Bar-Or A, Antel JP. Distinct properties of circulating CD8+ T cells in FTY720-treated patients with multiple sclerosis. Arch Neurol. 2010;67:1449–55.
Miyazaki Y, Niino M, Fukazawa T, Takahashi E, Nonaka T, Amino I, et al. Suppressed pro-inflammatory properties of circulating B cells in patients with multiple sclerosis treated with fingolimod, based on altered proportions of B-cell subpopulations. Clin Immunol. 2014;151:127–35.
Luessi F, Kraus S, Trinschek B, Lerch S, Ploen R, Paterka M, et al. FTY720 (fingolimod) treatment tips the balance towards less immunogenic antigen-presenting cells in patients with multiple sclerosis. Mult Scler. 2015.
Durafourt BA, Lambert C, Johnson TA, Blain M, Bar-Or A, Antel JP. Differential responses of human microglia and blood-derived myeloid cells to FTY720. J Neuroimmunol. 2011;230:10–6.
Muls N, Dang HA, Sindic CJ, van Pesch V. Fingolimod increases CD39-expressing regulatory T cells in multiple sclerosis patients. PLoS One. 2014;9, e113025.
Wu T, Zhang L, Xu K, Sun C, Lei T, Peng J, et al. Immunosuppressive drugs on inducing Ag-specific CD4(+)CD25(+)Foxp3(+) Treg cells during immune response in vivo. Transpl Immunol. 2012;27:30–8.
Liu Y, Jiang J, Xiao H, Wang X, Li Y, Gong Y, et al. The sphingosine-1-phosphate receptor agonist FTY720 and its phosphorylated form affect the function of CD4 + CD25+ T cells in vitro. Int J Mol Med. 2012;30:211–9.
Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat Immunol. 2010;11:1047–56.
Ntranos A, Hall O, Robinson DP, Grishkan IV, Schott JT, Tosi DM, et al. FTY720 impairs CD8 T-cell function independently of the sphingosine-1-phosphate pathway. J Neuroimmunol. 2014;270:13–21.
Ma J, Wang R, Fang X, Sun Z. beta-catenin/TCF-1 pathway in T cell development and differentiation. J Neuroimmune Pharmacol. 2012;7:750–62.
Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–13.
Kokabu S, Sato T, Ohte S, Enoki Y, Okubo M, Hayashi N, et al. Expression of TLE3 by bone marrow stromal cells is regulated by canonical Wnt signaling. FEBS Lett. 2014;588:614–9.
Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–45.
Bhela S, Kempsell C, Manohar M, Dominguez-Villar M, Griffin R, Bhatt P, et al. Nonapoptotic and extracellular activity of granzyme B mediates resistance to regulatory T cell (Treg) suppression by HLA-DR-CD25hiCD127lo Tregs in multiple sclerosis and in response to IL-6. J Immunol. 2015;194:2180–9.
Furlan R, Rovaris M, Martinelli Boneschi F, Khademi M, Bergami A, Gironi M, et al. Immunological patterns identifying disease course and evolution in multiple sclerosis patients. J Neuroimmunol. 2005;165:192–200.
Haile Y, Simmen KC, Pasichnyk D, Touret N, Simmen T, Lu JQ, et al. Granule-derived granzyme B mediates the vulnerability of human neurons to T cell-induced neurotoxicity. J Immunol. 2011;187:4861–72.
Acknowledgements
We thank all subjects for their blood donation. We thank Ivan Mascanfroni, Sushrut Jangi, and Felipe von Glehn for their valuable assistance with experiments and comments on the manuscript.
Study support
This study was supported by funding from Novartis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MAM designed, performed, and analyzed the experiments and wrote the manuscript. RR and MG designed performed the experiments and provided comments on the manuscript. HR performed the experiments. DK designed the experiments and provided comments on manuscript. TP provided suggestions and material for the lentivirus experiments. KR and PB analyzed nanostring and clinical data. RG and LA performed the experiments. PN performed the experiments. NG and HH performed the experiments. PK and BG assisted in the patient sample collection. TC and HLW provided patient samples. RG designed the experiments, supervised the study, and edited the manuscript. All authors read and approved the final manuscript.
Additional files
Additional file 1: Table S1.
Multiple sclerosis patient characteristics. All multiple sclerosis patients had relapsing-remitting disease courses. s.d. = standard deviation.
Additional file 2: Figure S1.
Flow cytometry analysis of CD4+ and naïve CD4+ T cells from multiple sclerosis patients before and after 3 months of treatment with fingolimod. BL = baseline, RV = revisit (n = 8). *p < 0.05, **p < 0.01, paired non-parametric t tests.
Additional file 3: Figure S2.
FTY720 increases TCF7 expression in T cells. Expression of TCF7 in in vitro activated T cells from healthy individuals in the presence or absence of FTY720, natalizumab (T-Ty), or interferon beta (T-IFN beta). Data are shown as mean ± SEM. *p < 0.05, ns = not significant, paired non-parametric t tests.
Additional file 4: Figure S3.
FTY720 reduces expression of pro-inflammatory cytokines. A) Expression of IL-17 (n = 9), GMCSF (n = 24), and TNFα (n = 24) in in vitro activated T cells from healthy individuals in the presence or absence of FTY720 using Luminex-based assay. (Note: IL-17 expression could not be measured in 15 healthy individuals.) B) Viabity staining of in vitro-activated T cells in the presence or absence of FTY720. Data are shown as mean ± SEM. *p < 0.05, ***p < 0.001, ns = not significant, paired non-parametric t tests. C) Binding region of TCF-1 in the promoter of IL17, CSF2, and TNF-α. Blue arrows represent transcription start site.
Additional file 5: Figure S4.
Effect of FTY720 on expression of TLE family proteins. Expression of TLE1, TLE2, TLE3, and TLE4 using qPCR (n = 8). Data are shown as mean ± SEM. *p < 0.05, ns = not significant, paired non-parametric t tests.
Additional file 6: Figure S5.
Proposed model of interaction between S1P1 and Wnt signaling pathways. A) S1P1 receptor activation induces Akt phosphorylation, which in turn induces phosphorylation of GSK3β. Phosphorylation of GSK3β at Ser9 leads to its inactivation, which allows unphosphorylated β-catenin to accumulate and translocate into the nucleus, where it binds to TCF-1 and induces transcription of target genes. B) In the presence of FTY720, the S1P1 receptor is downregulated, and less Akt is phosphorylated. Inactive Akt cannot phosphorylate GSK3β, which leads to the ability of GSK3β to phosphorylate β-catenin and thus, induce its degradation. In the absence of β-catenin, TCF-1 binds to co-repressors, such as TLE/Groucho family proteins, and represses the transcription of target genes.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mazzola, M.A., Raheja, R., Murugaiyan, G. et al. Identification of a novel mechanism of action of fingolimod (FTY720) on human effector T cell function through TCF-1 upregulation. J Neuroinflammation 12, 245 (2015). https://doi.org/10.1186/s12974-015-0460-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12974-015-0460-z