Abstract
Background
Pituitary adenomas (PAs) are the second most common intracranial tumor. While current diagnostic practices rely primarily on histological testing, they often fail to capture the molecular complexities of pituitary adenomas, underscoring the need for a molecular-based classification to refine therapeutic strategies and prognostic assessments. This study aims to provide a molecularly unbiased classification of pituitary adenomas and explore their unique gene expression patterns and clinical features.
Methods
We performed unsupervised hierarchical clustering of the gene expression profiles of 117 PA samples to identify three distinct molecular subtypes. Subsequently, we analyzed the compiled transcriptomic profiles of each individual subtype for pathway enrichment. We also validated the new classification with a validation set containing 158 PAs and 24 pituitary adenoma stem cells (PASCs).
Results
Consensus clustering of transcriptomic data from 117 pituitary adenoma (PA) samples identified three distinct molecular subtypes, each showing unique gene expression patterns and associated biological processes: Group I is enriched in signaling pathways, such as the cAMP signaling pathway and the calcium signaling pathway. Group II is primarily related to metabolic processes, including nitrogen metabolism and arginine biosynthesis in cancer. Group III predominantly shows enrichment in immune responses and potential malignant transformation of the disease, especially through cancer-related pathways such as the JAK–STAT signaling pathway and the PI3K–Akt signaling pathway. The immune profiling revealed distinct patterns for each subtype: Group I had higher dendritic cells and fewer CD8+ T cells, Group II had more monocytes and macrophages, and Group III had elevated levels of T cells. Additionally, there were differences in clinical characteristics and prognosis among the subtypes, with Group III having a worse prognosis, despite the smaller tumor size compared to other groups. Notably, differences in PASCs correlated with the molecular subtypes, with Group III stem cells being enriched in tumorigenesis pathways, PI3K–Akt signaling pathway and Ras signaling pathway.
Conclusion
Our study introduces a novel molecular classification for pituitary adenomas, independent of traditional histological methods. Each subtype features distinct genetic, molecular, and immunological profiles. We have isolated pituitary adenoma stem-like cells (PASCs), pairing them with tumor tissues for detailed transcriptomic analysis. These PASCs exhibit diverse molecular traits consistent with the new classification.
Similar content being viewed by others
Background
Pituitary adenomas (PAs) represent the second most prevalent type of central nervous system tumors, comprising 15% of all primary brain tumors. They occur at an annual incidence rate of four cases per 100,000 individuals and have a prevalence of 37 cases per 100,000 [1, 2]. Clinically, these tumors are categorized into secretory adenomas and nonfunctional adenomas, while radiological classification distinguishes them as microadenomas (< 10 mm) and macroadenomas (≥ 10 mm). Recently, the term “adenomas” has been changed to “Pituitary neuroendocrine tumors (PitNETs)” to refer to these tumors. The most common PitNETs originate from the lactotroph lineage, followed in frequency by gonadotroph, somatotroph, corticotroph, and thyrotroph lineage tumors [3]. Although pituitary adenomas are usually benign, their clinical presentation is remarkably heterogeneous, ranging from asymptomatic lesions to mass effects leading to severe endocrine dysfunction and visual impairment, thus complicating treatment and prognosis [4,5,6]. Most pituitary adenomas (PAs) are noninvasive and grow slowly, first-line treatment typically managed with transsphenoidal surgery or medications like bromocriptine or cabergoline. However, 2.5% to 10% of PAs are aggressive, recurrent, and show resistance not only to these standard treatments but also to alternatives like Temozolomide [7,8,9]. The overall resection rate of PA is only 66% to 78% [10], with remission rates ranging from 40 to 70% for macroadenomas and 80% to 90% for microadenomas [11]. Currently, the diagnosis of pituitary adenomas is mainly based on histologic testing, which does not reflect pathological changes at the cellular and molecular levels in a timely and effective manner, and as a result, a subject of debate [12].
Transcriptomic studies have revealed that different subtypes of pituitary adenomas have different patterns of gene expression [3, 13, 14]. Upregulation of GNAS genes is frequently observed in somatotrophs, leading to activation of the cAMP signaling pathway, which promotes cell proliferation and hormone secretion [15]. Corticotrophs tend to exhibit involvement in the process of ubiquitylation of genes upregulated, such as USP8, which contributes to the pathogenesis of Cushing’s disease [16]. Although the pathogenesis of PAs has been extensively studied, identifying specific oncogenes and tumor suppressors [17,18,19], the molecular factors driving tumorigenesis, remission, and therapeutic response are still not well understood [20,21,22].
Cancer stem cells (CSCs) are regarded as a distinct subpopulation of cells possessing stem cell properties within certain types of solid tumors [23]. In gliomas, the presence of glioma stem cells has been associated with poor clinical prognosis, resistance to radiotherapy and high recurrence rates, and therapies targeting glioma stem cells are currently at the forefront. Furthermore, pituitary adenoma stem cells (PASCs) have emerged as a key component in understanding pituitary adenoma pathogenesis and recurrence. Initial studies identified PASCs using markers normally associated with neural stem cells such as CD133, Nestin, and SOX2, suggesting the presence of stem-like regions within these tumors [24]. Advanced methods such as single-cell RNA sequencing have likewise confirmed the presence of PASCs [25, 26]. These studies have shown that genes associated with cell proliferation, resistance to apoptosis and maintenance of stemness are significantly upregulated in adenomas from the pituitary gland, especially aggressive pituitary adenomas, highlighting the contribution of PASCs to tumor development and recurrence [27].
Materials and methods
Patients and tissue samples
A total of 117 patients were retrospectively identified for RNA-seq analysis which shown in Figure S1. All samples were snap-frozen immediately after collection and stored in liquid nitrogen to preserve RNA integrity for subsequent analysis. This method was chosen to ensure the highest quality of RNA for sequencing, minimizing degradation and ensuring reliable results. Additionally, from January 2020 to September 2020, 24 pituitary adenoma patients meeting the same criteria were enrolled at the Department of Neurosurgery, Beijing Tiantan Hospital. The subtypes of pituitary adenomas were determined based on pre-operative clinical features and post-operative pathological diagnoses, in accordance with the 2017 WHO Classification of Pituitary Tumors.
Clinical and molecular information was obtained from our database. All patients were evaluated preoperatively for tumor invasion using MRI. Postoperative MRI follow-up scans were performed to assess any residual mass and to evaluate tumor progression.
Culture and differentiation of PASCs
The pituitary adenoma specimen was resected and immediately divided into two portions during surgery. One portion was snap-frozen and stored in liquid nitrogen for subsequent pathological immunostaining and RNA sequencing analysis. The other portion was transported to the laboratory under sterile conditions for PASCs culture. Tumor specimens were thoroughly washed with 1× PBS and cut into small pieces. Washed in 10 ml DMEM/F-12 and centrifuged at 300×g for 2 min. The resultant cell pellet was lysed in 2 ml of 1× Accutase (Stemcell Technology, USA) and incubated at 37 °C for 5 min. Subsequent centrifugation was performed for 3 min. Erythrocytes were removed from the cell pellet using erythrocyte lysis buffer, with a 5-min treatment. After a washing step, the cells were resuspended and cultured in stem cell medium, which consisted of DMEM/F-12 supplemented with 1% penicillin–streptomycin, 1× B27 (50× concentration, Life Technologies, USA), 20 ng/ml of bFGF (Peprotech, USA), and 20 ng/ml of EGF (Peprotech, USA). The culture was maintained at 37 °C in a 5% CO2 incubator.
Immunohistochemical/immunofluorescence staining and analysis
Formalin-fixed and paraffin-embedded pituitary tumor specimens were cut into 5-μm thick sequential sections and processed for staining. The full slides were deparaffinized using xylene, followed by dehydration through a series of graded ethanol solutions. The slides were then rehydrated in PBS. For antigen retrieval, the sections were incubated in 10 mM citric acid (pH 6.0) at 97 °C for 20 min. To neutralize endogenous peroxidases, we treated the samples with 3% hydrogen peroxide for 15 min. The primary anti-bodies against CD4 (Abcam/ab133616; 1:250 dilution), CD8 (Abcam/ab17147; 1:100 dilution), CD20 (ab78237; 1:100 dilution), CD56 (ab75813; 1:100 dilution), CD68 (ab955; 1:100 dilution) were used.
PASCs were cultured in 6-well plates coated with poly-d-lysine, washed with 1× PBS, and fixed with 4% paraformaldehyde for 30 min at room temperature. Then incubated in 0.3% Triton X-100 (Sigma-Aldrich, USA) for 15 min, washed, and blocked with 5% BSA for 1 h. After overnight incubation at 4 °C with primary antibodies, cells were washed and incubated with secondary antibodies (Alexa-Fluor 488/647, Abcam, USA) at 1:1000 dilution. Images were captured at various magnifications using a Zeiss microimaging system equipped with a fluorescence emission system. The antibodies employed in the experiments included rabbit anti-Sox2 (Abcam/ab97959; 1:800 dilution), rabbit anti-Oct4 (Abcam/ab19857; 1:500 dilution), rabbit anti-Nestin (Abcam/ab134017; 1:200 dilution), and rabbit anti-CD133 (Abcam/ab19898; 1:500 dilution).
RNA extraction and synthesis of cDNA
Total RNA from PASCs or specimens was extracted using TRIzol reagent (Invitrogen, USA) and assessed with the RNA Nano 6000 Assay Kit on the Bioanalyzer 2100 system (Agilent Technologies, USA). RNA was stored at − 80 °C. RNA sequencing was performed by Novogene Co., Ltd. First-strand cDNA was synthesized using a random hexamer primer and RNase H, followed by second-strand synthesis with DNA Polymerase I and RNase H. After selecting 250–300 bp cDNA fragments, PCR was performed with High Fidelity DNA Polymerase. The PCR products were purified using the Agencourt AMPure XP, and the quality of the cDNA library was assessed with the Agilent Bioanalyzer 2100 system. Clustering of the index-coded samples was conducted on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina). The library preparations were sequenced on an Illumina Novaseq platform. We performed paired-end (PE) sequencing with a read length of 150 bp, generating a total of 10 Gb raw data per sample.
Identification of gene expression-based subtypes
We applied the unsupervised consensus clustering algorithm using the ConsensusClusterPlus package [28]. The clustering process was iterated 1000 times, with 80% of the data sampled in each iteration. The optimal number of clusters was determined based on the relative change in the consensus score and the area under the cumulative distribution function (CDF) curve of the consensus heatmap. SigClust analysis was performed to determine the significance of the clusters in a pairwise fashion (statistical significance of clustering for high-dimensional, low-sample size data).
Enrichment analysis of differentially expressed genes
After mapping the reads, the counts of differentially expressed genes from each library were adjusted and analyzed using the “Deseq2” package in R software (version 4.0.0). To account for multiple testing, P-values were adjusted via the Benjamini and Hochberg method. An adjusted P-value of 0.05 and log2 fold change (log2FC) threshold greater than 1 were established as cutoffs for significant differential expression. For gene ontology (GO) enrichment analysis, the ClusterProfiler and GOplot R packages were employed, considering GO terms with corrected p-values below 0.05 as significantly enriched. The ClusterProfiler R package also facilitated the statistical analysis of gene enrichment in various pathways.
Functional networks and gene connectivity
Gene connectivity and functional networks were analyzed using the STRING database, which supplied gene connectivity data with a high confidence level (interaction score of 0.9). These data were further analyzed in R using the iGraph package, employing techniques such as edge-betweenness and random walk methods to identify significant subnetworks or neighborhoods. Biological or pathway term enrichments for these neighborhoods were also conducted using STRING [29].
Identification of immune cell distribution patterns using ImmuCellAI
The ImmuCellAI algorithm was used to estimate the relative abundances of 24 immune cell types from RNA sequencing data, showing enhanced robustness and accuracy compared to other algorithms such as CIBERSORT and TIMER [30]. This accuracy was confirmed through validation using flow cytometry. The immune cells detected included six other types: B cells, NK cells, monocytes, macrophages, neutrophils, and dendritic cells, in addition to 18 subtypes of T cells.
Validation data set
Independent validation of class signatures was conducted using samples from 24 new patients and the E-MATB-7768 dataset [31] as a validation cohort, totaling 158 patients. Data normalization was performed using the “normalize Between Array” function within the SVA package in R software to mitigate batch effects and other variabilities.
Statistical analysis
Data analysis was conducted using IBM SPSS Statistical software (version 23.0). Differences in mean gene expression were assessed with unpaired two-sided Student’s t-tests after log2 transformation. Comparisons between normally distributed parameters were evaluated using the Mann–Whitney U-test. Pearson’s method was employed to analyze correlations between gene expression and immune cell levels. Patient prognosis was determined using Kaplan–Meier analysis.
Results
Consensus clustering identifies three subtypes of pituitary adenomas
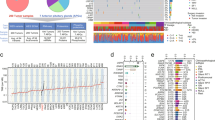
Initially, data from 117 pituitary adenoma samples were consolidated into a unified database. After excluding genes with extremely low expression, we selected 25,213 protein-coding genes for their consistent yet variable expression levels. We then employed unsupervised hierarchical clustering methods, specifically the K-means algorithm, to cluster the gene expression profiles of the 117 pituitary adenoma samples. This clustering resulted in three stable gene clusters, as shown in Fig. 1A. Three stable clusters were established, with cluster stability increasing from k = 2 to k = 3, but not for k > 3 (Fig. 1A, B). The statistical significance of the clustering was confirmed by SigClust, which showed that all class boundaries had statistical significance (Fig. 1C). The most representative samples of these clusters were determined based on their silhouette width, which describes the compactness of a class and the similarity of a sample to its own cluster versus other clusters. A positive silhouette coefficient indicates that the similarity of a sample to its own category is higher than to any other category (Fig. 1D). Based on this, we defined three molecular subtypes of pituitary adenomas: Group I, Group II, and Group III. Within each subtype, the gene expression patterns among samples were more similar to each other than to those in other subtypes.
Subsequently, we compared the histopathological classification with the molecular subtyping formed by the gene cluster clustering and found that this categorization was independent of the histological subtypes, detailed as follows:
-
Group I: 58 tumors (49.5%), including 40 nonfunctioning pituitary adenomas, 9 gonadotroph adenomas, 4 growth hormone adenomas, 4 prolactinomas, and 1 thyrotropin adenoma.
-
Group II: 22 tumors (18.8%), consisting of 10 nonfunctioning pituitary adenomas, 5 gonadotroph adenomas, 3 growth hormone adenomas, 3 prolactinomas, and 1 corticotroph adenoma.
-
Group III: 37 tumors (31.6%), with 16 growth hormone pituitary adenomas, 12 nonfunctioning pituitary adenomas, 7 prolactinomas, 1 thyrotropin adenoma, 1 corticotroph adenoma, and 1 gonadotroph adenoma.
To further explore genetic differences among the subtypes, we conducted t-tests to compare the differential genes between Group I and Group II and III, between Group II and Group I and III, and between Group III and Group I and II, respectively. We used a threshold of P < 0.05 and an absolute fold change greater than 1 as our criteria for selection. Each subtype exhibited a unique distribution pattern of protein-coding genes (Fig. 2A). We also conducted an analysis of transcription factors across the three groups, as shown in Figure S2. Our findings indicate that PIT1 is predominantly concentrated in Group III, while SF-1 is mainly found in Group I. The final results of the differential gene analysis are as follows (see Fig. 2B):
-
Group I: we identified 742 upregulated genes and 835 downregulated genes.
-
Group II: we discovered 477 upregulated genes and 1198 downregulated genes.
-
Group III: we found 932 upregulated genes and 955 downregulated genes.
Functional annotation of subtypes
To explore the biological functional differences behind the differentially expressed genes among the three subtypes, we performed Gene Ontology (GO) functional enrichment and KEGG pathway analyses of the above differential gene results, focusing on those with p-values below 0.05. In GO and KEGG analyses, several common terms appeared for each subtype, including cell adhesion, positive regulation of cell proliferation, positive regulation of gene expression, positive regulation of cell surface receptor signaling pathways, neuropeptide signaling pathways, and MAPK cascades. In addition, each subtype showed different functional enrichments. Group I primarily studies signaling pathways, including the cAMP and Calcium signaling pathways. Group II focuses on metabolic processes, specifically nitrogen metabolism and arginine biosynthesis in cancer. Group III predominantly showed enrichment in immune responses and potential malignant transformation of the disease, especially through cancer-related pathways such as the JAK–STAT signaling pathway and the PI3K–Akt signaling pathway (Fig. 3, Table S1).
Using the STRING database, we predicted associations between upregulated genes and their functional annotations, extracting sub-networks for each group (Fig. 4). Pathway enrichment analysis showed that the cAMP signaling pathway, followed by the cGMP–PKG signaling pathway and the Wnt signaling pathway, was predominantly affected in Group I, and the PI3K–Akt signaling pathway, the MAPK signaling pathway, and the Ras signaling pathway were predominantly affected in Group II. The PI3K–PKG signaling pathway and the Ras signaling pathway were mainly affected in Group II. In addition to the entries that appeared in Group I and Group II, Group III features enrichment in more pathways, including the JAK–STAT, mTOR, PI3K–Akt, and Hippo signaling pathways.
RNA-Seq analysis of up-regulated genes in each of the three groups. A–C Gene interaction network (confidence level = 0.9) of the differentially expressed genes in each of the three groups, using the STRING database. Subnetworks (neighborhoods) are colored and annotated with enriched functional categories. Gray lines, connections within a neighborhood; red lines, connections between neighborhoods. D GO enrichment for KEGG pathways
Immune landscape of the TME of PAs
The relative abundances of 24 immune cell types in the tumor microenvironment (TME) of PAs are depicted in Fig. 5A. Notably, immune cell proportions varied significantly not only between the groups but also within the same subtype, reflecting diverse immunological profiles.
Estimation of tumor-infiltrating immune cells (TIICs) in 117 pituitary adenomas based on ImmuCellAI. A Immune landscape of the TME, the relative abundances of the 24 types of TIICs are indicated by various colors. B Heatmap representing the distributions of 24 types of TIICs among the three groups. C Correlation analyses of the TIICs in three groups. The red color represents a positive correlation, and the blue color indicates a negative correlation
Analysis of immune cell infiltration revealed (Table 1):
-
Group I exhibited significantly higher levels of dendritic cells (DCs) and lower levels of various T cells, including CD8+ T cells, naïve CD8+ T cells, cytotoxic T cells, exhausted T cells, and mucosal-associated invariant T cells (MAIT), compared to the other groups.
-
Group II was characterized by elevated levels of B cells, monocytes, and macrophages, but significantly lower levels of CD4+ T cells and follicle-assisted T (Tfh) cells.
-
Group III displayed increased levels of natural killer (NK) cells, CD4+ T cells, gamma–delta (γ–δ) T cells, natural regulatory T (nTreg) cells, induced regulatory T (iTreg) cells, T-helper 1 (Th1) cells, T-helper 17 (Th17) cells, and central memory T cells, whereas natural killer T (NKT) cells were less prevalent.
To validate the effectiveness of our analysis, we utilized the available samples, which included a total of 49 cases: 21 from Group I, 10 from Group II, and 18 from Group III. We performed immunohistochemical (IHC) staining for tumor-infiltrating lymphocytes, targeting markers such as CD4, CD8, CD56, CD68, and CD20 (as shown in Fig. 6). Additionally, we quantified the number of positive cells and found that the results were consistent with our transcriptional data analysis (as shown in Fig. 6C). In correlation analyses, we observed distinct intercellular interactions between immune cells, with CD4+ T cell subsets (including CD4+ T cells in general as well as Tr1 cells, Th1 cells, and Tem cells showing correlation attenuated in all three groups. In addition, the CD8+ T cell subset (including CD8+ T cells in general as well as Tc cells, MAIT cells, and Tex cells) showed a strong positive correlation in all three groups. Group I showed strong positive correlations mainly between cytotoxic T cells, NK cells and B cells. Group II showed a complex pattern, showing positive correlations between macrophages and dendritic cells and negative correlations between T regulatory cells. Group III showed predominantly negative correlations (Fig. 5C).
Analysis of infiltrating immune cells within pituitary tumors. A Concordance between IHC-detected positive cell percentages and corresponding gene expression levels (log2 TPM) from RNA-seq. Pearson correlation (R) and p-values are indicated in the bottom right corner of each plot. B Representative pictures of CD4, CD8, CD56 and CD 20 IHC between three groups. C Quantification of IHC staining for CD4, CD8, CD56, and CD68 in pituitary adenomas across three patient groups. D PD-1/PD-L1, CTLA-4 expression in the data set according to the new molecular classification of PAs though RNA-seq. Unpaired two-sided Student’s t-tests were used. *P < 0.05. Scale bar 50 μm
We analyzed the expression profiles of 60 immune checkpoint molecules (ICMs) as reported by Rieder et al. [32], details of which are presented in Figure S3A. Group I predominantly showed high expression levels of CD40, CD276, C10orf54 and SIRPA, coupled with low expression levels of CD209 and TNFSF4. There were strong positive correlations between BTN3A1 and CD40, TNFRSF9 and CD40L, PVR and CD276, and negative correlations between PVR and HAVCR2, VTCN1 and CD27. Notably, most of the ICMs were significantly downregulated in Group II and negative correlations among nearly all HLA-related genes. In contrast, immunosuppressive genes such as CEACAM1, LAG3, CD209 and CD160 were upregulated. In Group III, a significant upregulation of immunostimulatory genes was observed, including BTN2A1, BTN3A1, various TNFRSF genes and several HLA genes (refer to Table S2). PD1 and PDL1, CTLA4 and CD80 demonstrated strong positive correlations in all groups. We specifically focused on analyzing the PD1/PDL1 and CTLA4 axis, as shown in Fig. 6D. Our results indicate that PD1 and PDL1 expression levels in Group III are significantly higher compared to the other two groups, while CTLA4 expression shows no significant difference across the three groups.
Subtypes and clinical correlations
To evaluate the clinical impact of our newly defined classification, we analyzed clinical and biological variables for all samples with data available at the time of sampling (Table 2). We observed differences in the age of diagnosis among the groups. Group I and Group II exhibited similar age distributions with mean ages of 49.6 ± 11.11 years and 49 ± 10.73 years, respectively, while Group III consisted of significantly younger patients, with a mean age of 40.67 ± 9.94 years. This younger age in Group III may be attributed to a higher prevalence of somatotroph PA patients, who typically exhibit more noticeable symptoms such as acromegaly and snoring.
Regarding gender distribution, Group I predominantly comprised male patients, whereas Group II was mostly female. Recurrent tumors were noted across all subtypes; however, Group III demonstrated a poorer prognosis with nearly half of the patient’s experiencing recurrence (Fig. 7). Notably, the tumors in Group III were comparable in size or even slightly smaller than those in the other groups. Additionally, there were no statistically significant differences in Knosp grading among the three groups.
External validation of the novel subtypes of PAs
To identify genes associated with each subtype, we employed the Random Forest method and designated genes with an importance score greater than 1 as core genes. A gene signature comprising 597 genes (243 in Group I, 190 in Group II, and 164 in Group III) was established based on differentially expressed genes, each demonstrating a distinct signature (Fig. 8). We collected samples from 24 new patients and used the E-MATB-7768 dataset [31] as the validation cohort, comprising a total of 158 patients to validate the reliability of our new molecular subtype classification. Additionally, we isolated and characterized pituitary tumor stem cells from these 24 patients in culture. The 158 pituitary adenomas in the validation set were similarly categorized into three distinct groups, achieving clear separation among them. Pituitary tumor stem cells from these patients were also classified into the same three groups.
-
Group I: 49 tumors (31.0%), including 36 nonfunctioning pituitary adenomas, 1 gonadotroph adenomas, 3 growth hormone adenomas.
-
Group II: 39 tumors (24.7%), consisting of 11 nonfunctioning pituitary adenomas, 1 thyrotrophin adenomas, and 27 corticotrophin adenomas.
-
Group III: 79 tumors (50.0%), with 44 somatotroph adenomas, 12 nonfunctioning pituitary adenomas, 18 prolactinomas, 5 thyrotropin adenomas.
Gene expression data identify three gene expression subtypes: A using the predictive 597 gene list, samples were ordered on the basis of subtype predictions. B Gene order from the E-MATB-7768 samples and 24 new samples were maintained in the validation data set (n = 158). C Visualizing the results of multi-cohort variance analysis of test and validation sets. D GO analysis of differentially expressed genes between three groups of the validation set. E KEGG enrichment of differentially expressed genes between three groups of the validation set
Each subtype exhibited a unique distribution pattern of protein-coding genes (Figure S3). The final results of the differential gene analysis (adjust p value < 0.05 and log2FC > 2) are as follows:
-
Group I: we identified 458 upregulated genes and 1177 downregulated genes.
-
Group II: we discovered 615 upregulated genes and 637 downregulated genes.
-
Group III: we found 601 upregulated genes and 865 downregulated genes.
Functional annotation and immune microenvironment in the validation set
In the validation set, the results of differential gene function annotation were similar to those observed in the training set. All three groups showed enrichment in functions such as cell adhesion, positive regulation of gene expression and neuropeptide signaling, cell–cell signaling, neuroactive ligand-receptor interactions, and the cAMP signaling pathway. Group I was also predominantly enriched in functions related to signaling, whereas Group III was predominantly related to potential malignant transformation. Notably, unlike the training set, group II in the validation set was enriched for PI3K–Akt, JAK–STAT, and Wnt signaling pathways, which may be due to the higher prevalence of Cushing’s disease patients in the validation cohort (Fig. 8, Table S3).
The results of the immune microenvironment for each group were also consistent with the test set results. Group I was characterized by high levels of B cells and macrophages, with strong correlations between B cells, macrophages and various T cell types. Group II had a marked monocyte infiltration, and showed a significant correlation between neutrophils, natural killer cells and monocytes; Group III had markedly elevated levels of T cells, with weaker correlations between dendritic cells and natural killer T cells and other immune cells (Figure S5, Table S4).
Pituitary adenoma stem cells and classification correlation
We isolated, cultured, and characterized pituitary adenoma stem cells. Their morphology under light microscopy is presented, along with immunofluorescence and confocal images highlighting markers such as Oct4, Sox2, CD133, and Nestin, as shown in Fig. 9. To examine the correlation between our new classification and pituitary adenoma stem cells, we isolated and characterized these cells from tumor tissues of 24 newly collected patients in the validation cohort; the specific details are provided in Table S5. Notable variations in stem cell characteristics were observed among the three groups.
The morphology of pituitary adenoma stem cells. A The morphology of human pituitary adenoma stem cells. A Representative morphological change of PASC on day 3, day 7, day 14 and day 21 by microscopic observation. B Immunohistochemistry and confocal analysis of Oct4, Sox2, CD133 and Nestin in PASCs cultured on day 14. All the markers were positively stained in the PASCs. Scale bar is 100um for all panels
Our analysis particularly focused on the differences between Group III and Groups I and II. We identified a total of 491 genes that were upregulated and 142 genes that were downregulated in Group III stem cells. Functionally, the upregulated genes in this group were predominantly enriched in processes such as cell adhesion, cell differentiation, positive regulation of gene expression, negative regulation of cell proliferation, and cell migration. Moreover, pathway enrichment analysis revealed significant involvement in tumorigenesis pathways in cancer, PI3K–Akt signaling, calcium signaling, cAMP signaling, and Ras signaling pathways (Fig. 10). Similar patterns of gene expression and pathway enrichment were observed in the tumor tissues as well.
Illustration of differentially expressed genes in PASC between Group III and Groups I and II. A Volcano plots illustrating differential gene expression in Group III, with 491 genes upregulated and 142 genes downregulated. B, D Pathway enrichment analysis of differentially expressed genes in Group III based on the KEGG database. C GO analysis of differentially expressed genes in Group III. E ID of GO term and KEGG pathway
Discussion
In this study, we identified three transcriptionally distinct subtypes of pituitary adenomas. These subtypes are independent of traditional histological classifications, each characterized by unique genetic and molecular features that correlate with various stages of tumor genesis, progression, and interactions within the immune microenvironment. The robustness of this novel classification was confirmed in an independent validation set, demonstrating that these subtypes are not artifacts of technical errors, chance, or biased sample selection. The identification of these subtypes is pivotal because it indicates that the clinical outcomes and therapeutic needs of patients vary significantly among the subtypes.
Additionally, we have isolated, cultured, and characterized PASCs, revealing widespread alignment with the new classification. This suggests that different subtypes may originate from distinct cells or are driven by divergent pathogenic mechanisms. Studying these subtypes and their corresponding PASCs could greatly enhance our understanding of pituitary adenoma pathology. Current classifications are largely empirical, based primarily on histological observations [33]. However, histo-prognostic classification is a topic of debate, which is not directly related to patient prognosis and does not reflect pathological changes at the cellular and molecular level. Our molecular classification, which is independent of traditional histological typing, closely correlates with clinical phenotypes and molecular distinctions. This approach provides a robust framework for enhancing our understanding of tumor biology and improving therapeutic strategies.
Our study delineates the unique clinical features and prognostic outcomes of each subtype, revealing significant biological diversity characterized by distinct gene expression profiles and clinical attributes. Group I primarily comprises nonfunctioning adenomas, typically presenting fewer symptoms and associated with a better prognosis. These adenomas are prevalent in older patients with a balanced gender distribution. The main molecular features are related to signal transduction mainly including the cAMP signaling pathway, followed by the cGMP–PKG signaling pathway, which significantly impacts cellular proliferation and hormone secretion [34]. Group II, characterized by a significant female predominance, includes a wide range of adenoma types; noteworthy, almost all corticotrophic adenomas fall within this group. Group II is notably affected by metabolic alterations, including nitrogen metabolism, arginine biosynthesis in cancer, and the PI3K–Akt pathway. The validation set showed enrichment for the Ras, Hippo, JAK–STAT, and Wnt signaling pathways, possibly due to the higher prevalence of Cushing’s disease patients in the cohort. There is a notable enrichment of males in Group I and females particularly in Group II. This is an intriguing observation, though we currently do not have a definitive explanation for the sex-related differences in these specific subtypes. Literature suggests that prolactinomas, which are typically benign prolactin-secreting adenomas derived from lactotrophs, account for approximately 50% of all pituitary adenomas in both women and men. Interestingly, in individuals aged 25–44 years, prolactinomas predominantly affect women, with a female-to-male ratio ranging from 5:1 to 10:1. However, this ratio tends to equalize after menopause [35]. Group III, consisting of younger patients and primarily harboring growth hormone adenomas, did not show a significant difference in tumor size compared to the other two groups. However, this group exhibited a higher recurrence rate, with nearly half of the patients experiencing recurrence. Group III showed upregulation in the JAK–STAT and PI3K–Akt signaling pathways. These pathways contribute to the subtype’s aggressive behaviors, promoting survival and proliferation and linking to the more severe tumor phenotypes observed in this group [26, 36]. In addition, recent studies have shown that transcription factors also play a key role in regulating pituitary adenoma behavior, which adds to the complexity of our understanding of pituitary adenomas, e.g. dysregulation of the transcription factor PIT1 may contribute to the pathogenesis of somatotroph and lactotroph adenomas by altering the network of genes involved in cell cycle control and apoptosis [18, 37].
In our analysis, the distribution of the 24 TIICs showed a high degree of heterogeneity across different PA histological subgroups. Also, a high degree of heterogeneity was shown in the new subtypes. Even within subgroups TIIC distribution differed significantly between cases. Our study reveals a complex immune microenvironment across the three subtypes of pituitary adenomas, emphasizing the significant heterogeneity in immune landscapes. This heterogeneous immune infiltration phenomenon in the tumor microenvironment (TME) has been observed in other tumors [50–53]. While previous studies have described generalized immune suppression in pituitary adenomas, researchers have not reached a consensus on the distributions of tumor-infiltrating immune cells (TIICs) in PAs. Andrew et al. reported that more Macrophages in larger adenomas [38] and Zhouwen et al. found that growth hormone adenomas had more T-cell infiltration [39].
Our findings provide a more nuanced understanding. Group I exhibited significantly higher levels of DCs and lower levels of various T cells. Group II was characterized by elevated levels of B cells, monocytes, and macrophages. Group III displayed increased levels of NK cells and most types of T cells including CD4+ T cells, γ–δ T cells, nTreg cells, iTreg cells, Th1 cells, Th17 cells, and Tcm cells. The expression of immune checkpoints across the subtypes varies, with Group III showing upregulation of both stimulatory and inhibitory checkpoints like PD-1, CTLA-4, and various HLA molecules, which is consistent with the findings of our previous studies [39]. This pattern could indicate a sophisticated strategy by the tumor to evade immune surveillance while maintaining an inflammatory state, making it a potential target for checkpoint blockade therapies. In contrast, Group I predominantly showed high expression levels of CD40, CD276, C10orf54 and SIRPA. Notably, most of the ICMs were significantly downregulated in group II and negative correlations among nearly all HLA-related genes, exhibits a distinctly “cold tumor” nature. In contrast to Group III, enhancing the overall immune response or modifying the inflammatory milieu in the first and second groups could improve the efficacy of existing therapies.
Studies targeting pituitary tumor PASC are still lacking, remaining at the stage of stem cell isolation, while genetic analysis of PASC is even more limited. We investigated the isolation, culture and characterization of pituitary adenoma stem cells (PASCs), performed transcriptomic analysis and matched tumor tissues to explore the characteristics of PASCs in molecular typing. Our analysis particularly focused on the third group with the worst prognosis. A recent study demonstrated that somatic mutations found in primary pituitary tumor samples were consistently detectable in the exomes of the pituitary glands corresponding to each tumor. Additionally, the analysis of tumor DNA revealed a relatively low count of somatic mutations [40,41,42]. Our RNA sequencing analysis showed that some pathways identified in tumor tissues were also present in matched PASCs. Functionally, the upregulated genes in this group predominantly showed enrichment in processes associated with anti-apoptotic behaviors and stemness characteristics, including cell adhesion, cell differentiation, positive regulation of gene expression, negative regulation of cell proliferation, and cell migration. Moreover, pathway enrichment analysis revealed significant involvement in tumorigenesis pathways in cancer, PI3K–Akt signaling, calcium signaling, cAMP signaling, and Ras signaling pathways. PASCs in aggressive pituitary tumors exhibit increased self-renewal capabilities and resistance to conventional therapies, likely driven by active signaling pathways such as PI3K–Akt and Wnt/β-catenin [43].
These insights enhance our understanding of the biological underpinnings of pituitary adenomas, advancing personalized treatment approaches. In response to the current problem of aggressive pituitary adenomas (PAs), which are often refractory to standard therapies and temozolomide (TMZ), targeted therapies against PI3K/Akt signaling and immunotherapies against PD1/PDL1 have been investigated [36, 44]. However, clinical trials are scarce, with only a few case reports confirming its efficacy, and most studies still in in vitro cellular validation or in animal studies [24]. Novel molecular classifications may enhance these personalized treatment strategies. For instance, patients in immunotherapy Group III might benefit more than those in Group II, suggesting targeted approaches based on tumor subtype. Further research is essential to translate these preliminary findings into effective clinical interventions.
Conclusion
In conclusion, our study proposes a new molecular classification of pituitary adenomas, which provides an important basis for understanding changes in tumor behavior and prognosis. This new molecular classification is independent of traditional histological classifications, with each subtype individually exhibiting unique genetic and molecular features, as well as a distinct immune microenvironment. The robustness of this novel classification has been confirmed through an independent validation set. We isolated and identified pituitary adenoma stem-like cells (PASCs) and paired them with tumor tissues for transcriptomic analysis.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available because it is used for further analysis, but are available from the corresponding author on reasonable request.
Abbreviations
- Pas:
-
Pituitary adenomas
- PASC:
-
Human pituitary adenoma stem cells
- PitNETs:
-
Pituitary neuroendocrine tumors
- CSC:
-
Cancer stem cells
- PBS:
-
Phosphate buffer saline
- DMEM:
-
Dulbecco’s modified eagle’s medium
- bFGF:
-
Basic fibroblast growth factor
- EGF:
-
Epidermal growth factor
- FBS:
-
Fetal bovine serum
- ELISA:
-
Enzyme-linked immunosorbent assay
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- PPI:
-
Protein–protein interaction
References
Molitch ME. Diagnosis and treatment of pituitary adenomas: a review. JAMA. 2017;317(5):516.
Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro Oncol. 2022;24(Supplement_5):v1–95.
Toader C, Dobrin N, Tataru CI, Covache-Busuioc RA, Bratu BG, Glavan LA, et al. From genes to therapy: pituitary adenomas in the era of precision medicine. Biomedicines. 2023;12(1):23.
Colao A, Grasso LFS, Giustina A, Melmed S, Chanson P, Pereira AM, et al. Acromegaly. Nat Rev Dis Prim. 2019;5(1):20.
Santos A, Resmini E, Martínez Momblán MA, Valassi E, Martel L, Webb SM. Quality of life in patients with cushing’s disease. Front Endocrinol. 2019;10:862.
Tampourlou M, Fountas A, Ntali G, Karavitaki N. Mortality in patients with non-functioning pituitary adenoma. Pituitary. 2018;21(2):203–7.
Dai C, Feng M, Liu X, Ma S, Sun B, Bao X, et al. Refractory pituitary adenoma: a novel classification for pituitary tumors. Oncotarget. 2016;7(50):83657–68.
Mehta GU, Lonser RR. Management of hormone-secreting pituitary adenomas. Neuro Oncol. 2017;19(6):762–73.
Raverot G, Burman P, McCormack A, Heaney A, Petersenn S, Popovic V, et al. European Society of Endocrinology Clinical Practice Guidelines for the management of aggressive pituitary tumours and carcinomas. Eur J Endocrinol. 2018;178(1):G1-24.
Almutairi RD, Muskens IS, Cote DJ, Dijkman MD, Kavouridis VK, Crocker E, et al. Gross total resection of pituitary adenomas after endoscopic vs. microscopic transsphenoidal surgery: a meta-analysis. Acta Neurochir. 2018;160(5):1005–21.
Ammirati M, Wei L, Ciric I. Short-term outcome of endoscopic versus microscopic pituitary adenoma surgery: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2013;84(8):843–9.
Rindi G, Klimstra DS, Abedi-Ardekani B, Asa SL, Bosman FT, Brambilla E, et al. A common classification framework for neuroendocrine neoplasms: an International Agency for Research on Cancer (IARC) and World Health Organization (WHO) expert consensus proposal. Mod Pathol. 2018;31(12):1770–86.
Jotanovic J, Tebani A, Hekmati N, Sivertsson Å, Lindskog C, Uhlèn M, et al. Transcriptome analysis reveals distinct patterns between the invasive and noninvasive pituitary neuroendocrine tumors. J Endocr Soc. 2024;8(5): bvae040.
Oh JY, Osorio RC, Jung J, Carrete L, Choudhary N, Lad M, et al. Transcriptomic profiles of normal pituitary cells and pituitary neuroendocrine tumor cells. Cancers. 2022;15(1):110.
Bi WL, Horowitz P, Greenwald NF, Abedalthagafi M, Agarwalla PK, Gibson WJ, et al. Landscape of genomic alterations in pituitary adenomas. Clin Cancer Res. 2017;23(7):1841–51.
Reincke M, Sbiera S, Hayakawa A, Theodoropoulou M, Osswald A, Beuschlein F, et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet. 2015;47(1):31–8.
Caimari F, Korbonits M. Novel genetic causes of pituitary adenomas. Clin Cancer Res. 2016;22(20):5030–42.
Vandeva S, Daly AF, Petrossians P, Zacharieva S, Beckers A. Somatic and germline mutations in the pathogenesis of pituitary adenomas. Eur J Endocrinol. 2019;181(6):R235–54.
Gruppetta M, Formosa R, Falzon S, Ariff Scicluna S, Falzon E, Degeatano J, et al. Expression of cell cycle regulators and biomarkers of proliferation and regrowth in human pituitary adenomas. Pituitary. 2017;20(3):358–71.
Paragliola RM, Corsello SM, Salvatori R. Somatostatin receptor ligands in acromegaly: clinical response and factors predicting resistance. Pituitary. 2017;20(1):109–15.
Colao A, Auriemma RS, Pivonello R. The effects of somatostatin analogue therapy on pituitary tumor volume in patients with acromegaly. Pituitary. 2016;19(2):210–21.
Gadelha MR, Wildemberg LE, Bronstein MD, Gatto F, Ferone D. Somatostatin receptor ligands in the treatment of acromegaly. Pituitary. 2017;20(1):100–8.
Wang JCY. Good cells gone bad: the cellular origins of cancer. Trends Mol Med. 2010;16(3):145–51.
Garcia-Lavandeira M, Quereda V, Flores I, Saez C, Diaz-Rodriguez E, Japon MA, et al. A GRFa2/Prop1/stem (GPS) cell niche in the pituitary. PLoS ONE. 2009;4(3): e4815.
Cui Y, Li C, Jiang Z, Zhang S, Li Q, Liu X, et al. Single-cell transcriptome and genome analyses of pituitary neuroendocrine tumors. Neuro Oncol. 2021;23(11):1859–71.
Yan N, Xie W, Wang D, Fang Q, Guo J, Chen Y, et al. Single-cell transcriptomic analysis reveals tumor cell heterogeneity and immune microenvironment features of pituitary neuroendocrine tumors. Genome Med. 2024;16(1):2.
Würth R, Thellung S, Corsaro A, Barbieri F, Florio T. Experimental evidence and clinical implications of pituitary adenoma stem cells. Front Endocrinol. 2020;11:54.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3.
Saini JS, Corneo B, Miller JD, Kiehl TR, Wang Q, Boles NC, et al. Nicotinamide ameliorates disease phenotypes in a human iPSC model of age-related macular degeneration. Cell Stem Cell. 2017;20(5):635-647.e7.
Miao Y, Zhang Q, Lei Q, Luo M, Xie G, Wang H, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci. 2020;7(7):1902880.
Neou M, Villa C, Armignacco R, Jouinot A, Raffin-Sanson ML, Septier A, et al. Pangenomic classification of pituitary neuroendocrine tumors. Cancer Cell. 2020;37(1):123-134.e5.
Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–62.
Lopes MBS. The 2017 World Health Organization classification of tumors of the pituitary gland: a summary. Acta Neuropathol. 2017;134(4):521–35.
Hernández-Ramírez LC, Trivellin G, Stratakis CA. Cyclic 3′,5′-adenosine monophosphate (cAMP) signaling in the anterior pituitary gland in health and disease. Mol Cell Endocrinol. 2018;463:72–86.
Petersenn S, Fleseriu M, Casanueva FF, Giustina A, Biermasz N, Biller BMK, et al. Diagnosis and management of prolactin-secreting pituitary adenomas: a Pituitary Society international Consensus Statement. Nat Rev Endocrinol. 2023;19(12):722–40.
Derwich A, Sykutera M, Bromińska B, Rubiś B, Ruchała M, Sawicka-Gutaj N. The role of activation of PI3K/AKT/mTOR and RAF/MEK/ERK pathways in aggressive pituitary adenomas—new potential therapeutic approach—a systematic review. IJMS. 2023;24(13):10952.
Drummond J, Roncaroli F, Grossman AB, Korbonits M. Clinical and pathological aspects of silent pituitary adenomas. J Clin Endocrinol Metab. 2019;104(7):2473–89.
Lu JQ, Adam B, Jack AS, Lam A, Broad RW, Chik CL. Immune cell infiltrates in pituitary adenomas: more macrophages in larger adenomas and more T cells in growth hormone adenomas. Endocr Pathol. 2015;26(3):263–72.
Zhou W. Comprehensive analysis of the immunological landscape of pituitary adenomas: implications of immunotherapy for pituitary adenomas. J Neurooncol. 2020;149(3):473–87.
Peculis R, Mandrika I, Petrovska R, Dortane R, Megnis K, Nazarovs J, et al. Pituispheres contain genetic variants characteristic to pituitary adenoma tumor tissue. Front Endocrinol. 2020;11:313.
Spada A, Mantovani G, Treppiedi D, Mangili F, Catalano R, Carosi G, et al. Pituitary tumors: genetic and molecular factors underlying pathogenesis and clinical behavior. Neuroendocrinology. 2022;112(1):15–33.
Peculis R, Niedra H, Rovite V. Large scale molecular studies of pituitary neuroendocrine tumors: novel markers, mechanisms and translational perspectives. Cancers. 2021;13(6):1395.
Bar-Hai N, Ishay-Ronen D. Engaging plasticity: differentiation therapy in solid tumors. Front Pharmacol. 2022;13: 944773.
Dai C, Zhang B, Liu X, Ma S, Yang Y, Yao Y, et al. Inhibition of PI3K/AKT/mTOR pathway enhances temozolomide-induced cytotoxicity in pituitary adenoma cell lines in vitro and xenografted pituitary adenoma in female nude mice. Endocrinology. 2013;154(3):1247–59.
Acknowledgements
We thank Dr. Qi Li and Dr. Wenyan He in helping the cell culture and molecular biological experiments in China National Clinical Research Center for Neurological Diseases, Beijing Tiantan Hospital, Capital Medical University.
Funding
This work was supported by the National Natural Science Foundation of China (82071996) and the Beijing Natural Science Foundation (7212008). Peng Kang is supported by Beijing Hospitals Authority Youth Programme (QML20180504) and Beijing Program of Excellent Young Investigator (2015000021469G221).
Author information
Authors and Affiliations
Contributions
Conception of the work: WJ, CBZ; acquisition of clinical samples: SCM, PK, GJJ, WJ; cell culture and molecular analysis: JYP, LHY, PK; bioinformatical analysis: JYP, CBZ, PK, LHY; data treatment and statistical analysis: JYP, CBZ, PK, LHY, SCM, SCJ, WJLZ; draft manuscript: JYP, CBZ, WJ; all authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The procedure was in accordance with the ethical standards and approved by the ethics committee of Beijing Tiantan Hospital (KY202215501).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peng, J., Yuan, L., Kang, P. et al. Comprehensive transcriptomic analysis identifies three distinct subtypes of pituitary adenomas: insights into tumor behavior, prognosis, and stem cell characteristics. J Transl Med 22, 892 (2024). https://doi.org/10.1186/s12967-024-05702-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-024-05702-w