Abstract
Background
Whether cytochrome P450 24A1 (CYP24A1) polymorphism is associated with cancer susceptibility, the individual study results are still controversial. Therefore, we performed a comprehensive study to identify the association of CYP24A1 polymorphisms (rs4809960, rs6068816, rs2296241, rs4809957, rs2762939) with cancer susceptibility.
Methods
Electronic databases including Cochrane Library, PubMed, and Embase were systematically retrieved for relevant publications. Fixed or random-effect model was selected to calculate odds ratios (ORs) with their 95% confidence intervals (95%CI).
Results
Eighteen published articles were identified. The results indicated that rs4809960 polymorphism was associated with a decreased cancer risk in Caucasian (TT vs. TC+CC: P=0.035; C vs. T: P=0.016) and Asian population (CC vs. TC+TT: OR P=0.044; TT vs. TC+CC: P=0.021; CC vs. TT: P=0.020; C vs. T: P=0.008) and breast cancer risk (TT vs. TC+CC: P = 0.007; TC vs. TT: P=0.004; C vs. T: P=0.033). A significant association was found between rs2296241 polymorphism and esophageal squamous cell carcinoma risk (AA vs. GG+AG: P = 0.023) and prostate cancer susceptibility (A vs. G: P=0.022). Furthermore, rs4809957 polymorphism was associated with prostate cancer susceptibility in Caucasian (GG vs. GA+AA: P=0.029; GA vs. GG: P=0.022) and breast cancer susceptibility (AA vs. GG+GA: P=0.012; AA vs. GG, P=0.010; A vs. G: P=0.024). Additionally, rs6068816 polymorphism significantly decreased the lung cancer (CC vs. CT+TT: P = 0.016; TT vs. CC: P = 0.044; CT vs. CC: P = 0.036; T vs. C: P = 0.016) and breast cancer risk (TT vs. CC+CT: P = 0.043; TT vs. CC: P = 0.039). No association was found for rs2762939 polymorphism with overall cancer risk. However, for rs2296241, rs4809957, and rs6068816 polymorphisms, there were no significant differences after the Bonferroni correction.
Conclusion
The meta-analysis suggested that rs4809960 was associated with cancer risk and might be a genetic marker for predicting cancer risk. More large-scale and large-sample studies are necessary to further confirm these results.
Similar content being viewed by others
Introduction
Cancer is a global public health problem, and incidence and mortality are rapidly growing worldwide. According to the data of the International Agency for Research on Cancer, GLOBOCAN 2020 investigation results showed 19.3 million new cancer cases and 10.0 million cancer deaths in 2020 [1]. In 2023, it is estimated that there will be 1,958,310 new cancer cases and 609,820 cancer-related deaths in the USA [2]. With the rapid growth and aging of the world population, the predominance of cancer is a leading cause of death. Current evidence suggests that factors, such as irregular lifestyles, smoking, alcohol intake, environmental factors, and genetic factors, are closely associated with the occurrence of cancer [1, 2]. Accumulative evidence has demonstrated that genetic factors may be associated with the etiology of cancer and the individual’s risk of cancer development, especially whole-genome association studies (GWAS) have identified various genes that may be involved in cancer development [3, 4].
Vitamin D, an essential fat-soluble vitamin, is mainly come from ultraviolet exposure and diet metabolism [5]. Meanwhile, it plays critical roles in cellular growth and anti-proliferative activities [6]. Clinical studies have indicated that vitamin D deficiency contributed to cancer risk, including prostate cancer, breast cancer, and thyroid carcinoma [7]. 25 hydroxy vitamin D (25(OH)D) is the main circulating form of vitamin D. In addition, 1,25 dihydroxy vitamin D (1,25(OH)2D3), an active form of vitamin D, which is associated with cell functions and gene expression. In the process of vitamin D metabolism, 25(OH)D and 1,25(OH)2D3 are converted to 24,25 dihydroxy vitamin D (24,25(OH)2D3) and 1,24,25 trihydroxy vitamin D (1,24,25(OH)3D3), respectively, which are degraded by 25-hydroxyvitamin D 24-hydrolase (encoded by CYP24A1 gene) [8]. Mutation of CYP24A1 may influence the metabolism of Vitamin D and anti-proliferative effects [9, 10].
Single-nucleotide polymorphisms (SNPs) are the most common form of variation in the human genome, which can alter the expression level or function of genes or their encoded products and thus determine the phenotype of the organism [11, 12]. Therefore, it is increasingly recognized that SNPs play a crucial role in the mechanisms of cancer [13]. Epidemiological studies have demonstrated that several common SNPs of CYP24A1 are involved in the concentration of circulating 25(OH)D [14]. To date, five common SNPs (rs4809960, rs6068816, rs2296241, rs4809957, rs2762939) were found to be associated with cancer risk, including esophageal squamous cell carcinoma prostate cancer, breast cancer, and lung cancer [5, 14, 15]. However, controversial results were reported and the association was not yet well established. Therefore, a comprehensive meta-analysis was performed to better explore the associations of CYP24A1 polymorphisms with cancer risk.
Materials and methods
This study was performed under the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) and was registered in PROSPERO (CRD42023446451).
Search strategy
The relevant paper was identified (published until Feb. 2023) through Embase, PubMed, and Cochrane Library using the following strategy: (CYP24A1 or rs2296241 or rs4809957 or rs2762939 or rs4809960 or rs6068816) and (polymorphism or SNP or variant or variation or mutation or genotype) and (cancer or carcinoma or tumor or neoplasm). In addition, other potential publications were also searched by scanning the reference list. The details of the search strategy can be found in Supplementary Table 1.
Inclusion and exclusion criteria
Relevant studies were included according to the following criteria: (1) case-control studies, (2) evaluated the association between CYP24A1 polymorphism and cancer risk, (3) provided sufficient data to calculate the OR with 95%CI, and (4) control group conform to the Hardy–Weinberg equilibrium (HWE). The exclusion criteria were (1) review, abstract, comment, or letter; (2) duplication publications; and (3) relevant data not reported. In addition, for studies with repeat data, the study with the largest sample size was included. Each ethnicity was regarded as a separate study when different ethnicities were reported in a study.
Data extraction and quality assessment
Two authors independently extracted relevant data from the included studies. The extraction parameters included the first author, publication year, country, ethnicity, sample size, cancer type, genotype, and allele distribution in cases and controls, and the P value of HWE in the control group, methodology quality of each study was assessed according to the Newcastle–Ottawa Scale (NOS).
Statistical analyses
HWE was assessed by the chi-square test. The ORs and 95%CIs were calculated to evaluate the strength under allelic, recessive, dominant, homozygous, and heterozygous models. The P value of < 0.05 was considered as statistically significant. The chi-square test and I2 statistics were calculated to evaluate the heterogeneity across studies. If heterogeneity was found (P<0.10 or I2> 50%), the random-effect model was adopted. Otherwise, the fixed-effect model was adopted. Bonferroni correction was performed to adjust multiple-test P value [16]. Sensitivity analyses were performed to evaluate the stability of the results. Stratified analyses were performed by cancer type and ethnicity. Begg’s and Egger’s test was used to assess publication bias. Statistical analyses were completed using Stata 12.0 software (StataCorp, College Station, TX).
Results
Characteristics of the included studies
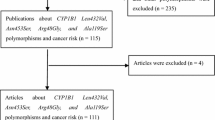
A total of 258 articles were retrieved in the initial search. Finally, a total of 18 articles [14, 15, 17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32] (19,017 cancer patients and 21,623 controls) were identified (Fig. 1). Among these 18 articles, nine publications about rs2296241 polymorphism, four publications focused on rs4809957 polymorphism, four on rs2762939 polymorphism, six on rs4809960 polymorphism, and six on rs6068816 polymorphism. In addition, four studies focused on prostate cancer, three on lung cancer, five on breast cancer, one on thyroid carcinoma, three on colorectal cancer, one on esophageal squamous cell carcinoma, and one on pancreas cancer. The characteristics of the included studies were described in Table 1.
Meta-analysis of rs4809960
Six publications [21, 26, 30,31,32,33] including seven studies (4509 cancer patients and 5210 controls) examined rs4809960 polymorphism. As shown in Table 2, no significant association between rs4809960 polymorphism and overall cancer susceptibility (Table 2). Subgroup analyses by ethnicity indicated that rs4809960 polymorphism was related to Caucasian population (TT vs. TC+CC: OR 1.18, 95%CI 1.01~1.37, P=0.035; C vs. T: OR 0.88, 95%CI 0.79~0.98, P=0.016) and Asian population (CC vs. TC+TT: OR 1.52, 95%CI 1.01~2.28, P=0.044; TT vs. TC+CC: OR 0.79, 95%CI 0.65~0.97, P=0.021; CC vs. TT: OR 1.52, 95%CI 1.08~2.48, P=0.020; C vs. T: OR 1.24, 95%CI 1.06~1.46, P=0.008). Subgroup analyses by cancer type revealed that rs4809960 polymorphism decreased breast cancer risk (TT vs. TC+CC: OR 1.19, 95%CI 1.05~1.36, P = 0.007; TC vs. TT: OR 0.82, 95%CI 0.72~0.94, P=0.004; C vs. T: OR 0.89, 95%CI 0.80~0.99, P=0.033). However, we only observed that rs4809960 polymorphism was significantly associated with the risk of breast cancer after Bonferroni correction.
Significant heterogeneity was found in all genetic models. Sensitivity analysis suggested that a significant association between rs4809960 polymorphism and overall cancer susceptibility was found (TT vs. TC+CC: OR 1.20, 95%CI 1.03~1.39, P=0.020, I2 = 53.1%; TC vs. TT: OR 0.84, 95%CI 0.70~0.99, P=0.043, I2 = 60.2%; C vs. T: OR 0.88, 95%CI 0.81~0.95, P=0.001, I2 = 37.2%) when after removed Yi et al. (Fig. 2). No visual publication bias was detected under the allelic genetic model. In addition, Egger’s test showed that there was no publication bias under the allelic genetic model (P=0.347).
Meta-analysis of rs2296241
Nine publications [14, 15, 17, 19, 21,22,23,24,25] including 5831 cancer patients and 6179 controls were used to calculate pooled ORs and 95%CIs. As shown in Table 3, there was no significant association between rs2296241 polymorphism and overall cancer susceptibility in all genetic models. Subgroup analysis was performed according to ethnicity and cancer type. Stratification by ethnicity indicated that rs2296241 polymorphism was not related to ethnicity. In addition, subgroup analyses by cancer type revealed that rs2296241 polymorphism increased the risk of esophageal squamous cell carcinoma (AA vs. GG+AG: OR 1.34, 95%CI 1.04~1.74, P = 0.023) and decreased risk in prostate cancer (A vs. G: OR 0.91, 95%CI 0.84~0.99, P=0.022) (Fig. 3) (Table 3). However, these associations were no longer significant after the Bonferroni correction.
Significant heterogeneity was found under the recessive, homozygous, and allelic models. Sensitivity analysis showed that the initial result was not changed by removing each study respectively. No visual publication bias was detected under the allelic genetic model (Fig. 4). In addition, Egger’s test showed that there was no publication bias under the allelic genetic model (P=0.066).
Meta-analysis of rs4809957, rs2762939 and rs6068816
Four publications [18, 20, 27, 28] (1851 cancer patients and 2570 controls) about rs4809957 polymorphism, four publications including five studies [15, 21, 22, 26] (2731 cancer patients and 2736 controls) about rs2762939 polymorphism and six publications [21, 26, 29,30,31, 33] (4095 cancer patients and 4829 controls) about rs6068816 polymorphism. As shown in Table 4, subgroup analyses revealed that rs4809957 polymorphism was significantly associated with Caucasian, especially pancreas cancer patients (GG vs. GA+AA: OR 0.80, 95%CI 0.66, 0.98, P=0.029; GA vs. GG: OR 1.27, 95%CI 1.04, 1.56, P=0.022). Furthermore, a significant association was found in breast cancer (AA vs. GG+GA: OR 1.70, 95%CI 1.22~2.58, P=0.012; AA vs. GG, OR 1.80, 95%CI 1.15~2.82, P=0.010; A vs. G: OR 1.27, 95%CI 1.03~1.55, P=0.024) (Fig. 5). In addition, there was no association between rs2762939 polymorphism with cancer risk (Table 5). For rs6068816, we found that rs6068816 polymorphism significantly decreased lung cancer (CC vs. CT+TT: OR 1.45, 95%CI 1.07~1.97, P = 0.016; TT vs. CC: OR 0.58, 95%CI 0.35~0.99, P = 0.044; CT vs. CC: OR 0.71, 95%CI 0.52~0.98, P = 0.036; T vs. C: OR 0.76, 95%CI 0.61~0.95, P = 0.016) and breast cancer risk (TT vs. CC+CT: OR 0.52, 95%CI 0.27~0.98, P = 0.043; TT vs. CC: OR 0.52, 95%CI 0.25~0.97, P = 0.039) (Fig. 6) (Table 6). However, these associations were no longer significant after the Bonferroni correction.
Sensitivity analysis showed that removing each study respectively from the meta-analysis did not change the initial result. No publication bias was detected in the studies about rs4809957 and rs2762939 polymorphism meta-analysis.
Discussion
CYP24A1, a member of the cytochrome P450 enzyme family, is located on the long arm of chromosome 20 (20q13.2). It is a key gene that converted 1,25(OH)2D3 to 1,24,25(OH)2D3 by 24-hydroxylation25-hydroxyvitamin D 24-hydrolase [34]. Albertson et al. [35] first identified the 20q13 gene amplification in breast cancer and identified the CYP24A1 gene as a candidate oncogene using array comparative genomic hybridization. CYP24A1 has been identified as a potential biomarker for cancer [36]. Numerous studies have suggested the expression level of the CYP24A1 was abnormally increased in several cancers, such as breast cancer, ovarian cancer, cervix carcinoma, lung cancer, and colon cancer [7, 37, 38]. Kong et al. [39] revealed that the rs6068816 and rs4809957 polymorphisms were associated with NSCLC risk. For breast cancer, Wei et al. [27] reported a significant association between the rs4809957 and breast cancer risk. Anderson et al. [18] revealed no significant correlation between rs4809957 with pancreas cancer. Among these publications reported the associations of CYP24A1 polymorphisms with cancer susceptibility, while the results remain controversial. The previous meta-analysis was performed by Zhu et al. [40], but they had not controlled the type I error rate through Bonferroni correction and had a smaller sample size. Therefore, the present meta-analysis aimed to re-evaluate the associations of CYP24A1 polymorphisms with cancer risk.
The present study indicated that there was no association between CYP24A1 polymorphisms (rs4809960, rs2296241, rs4809957, rs2762939, rs6068816) and overall cancer risk. For rs4809960 polymorphism, it was related to the Caucasian and Asian populations and decreased breast cancer risk. Moreover, our results suggested that rs2296241 polymorphism increased esophageal squamous cell carcinoma risk and decreased prostate cancer risk. For rs4809957 polymorphism, it was associated with pancreas cancer and breast cancer risk. In addition, we found that rs6068816 polymorphism significantly decreased lung cancer and breast cancer risk. However, rs4809960 polymorphism was associated with a decreased breast cancer risk after Bonferroni correction. A previous meta-analysis also reported CYP24A1 rs2296241 polymorphism was associated with prostate cancer risk [41]. Although our work found rs2296241 polymorphism was associated with an increased esophageal squamous cell carcinoma risk and decreased prostate cancer risk, these results could not withstand the Bonferroni correction.
The improvements of our meta-analysis are as follows: Firstly, more case-control studies about rs4809960, rs6068816, and rs2296241 polymorphism were included in the meta-analysis. Secondly, this is the first meta-analysis to assess the relationship between CYP24A1 (rs4809957, rs2762939) polymorphism and cancer risk. Thirdly, all included studies conform to the HWE, which may improve the reliability and stability of our study. In addition, all CYP24A1 polymorphisms were considered at the beginning. Ultimately, due to a lack of eligible articles and overlapping studies, our further evaluation of other CYP24A1 polymorphisms was limited. Therefore, in this meta-analysis, we only focused on five polymorphisms.
There are several limitations should be noted in the present study. First, the sample size of the included studies was relatively small, which might weaken the strength of the results. Second, the number of included studies in the subgroup analysis was also relatively small, which might lead to statistical bias. Third, not sufficient data to analyze whether environmental factors may influence the statistical result. Four, the association of CYP24A1 polymorphism with different types or stages, drinking, smoking, age, gender, exposure factors, or other risk factors was not considered in this study. Five, the patients of included studies mainly come from Caucasians. The African and Asian populations were relatively small.
In conclusion, this meta-analysis indicated that rs4809960 polymorphism was associated with a decreased breast cancer risk. No association between rs4809957, rs2296241, rs2762939, rs4809957 polymorphism, and overall cancer risk was found after Bonferroni correction. Considering the above limitations, more large-scale and large-sample studies are necessary to confirm these results.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CYP24A1:
-
Cytochrome P450 24A1
- CI:
-
Confidence interval
- GWAS:
-
Genome-wide association studies
- HWE:
-
Hardy-Weinberg equilibrium
- NOS:
-
Newcastle-Ottawa Scale
- OR:
-
Odds ratio
- SNPs:
-
Single-nucleotide polymorphisms
References
Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48.
Decker B, Allen J, Luccarini C, et al. Targeted resequencing of the coding sequence of 38 genes near breast cancer GWAS loci in a large case-control study. Cancer Epidemiol Biomarkers Prev. 2019;28(4):822–5.
Houlston RS, Cheadle J, Dobbins SE, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42(11):973–7.
Holick MF. Vitamin D: its role in cancer prevention and treatment. Prog Biophys Mol Biol. 2006;92(1):49–59.
Masuda S, Byford V, Arabian A, et al. Altered pharmacokinetics of 1alpha,25-dihydroxyvitamin D3 and 25-hydroxyvitamin D3 in the blood and tissues of the 25-hydroxyvitamin D-24-hydroxylase (Cyp24a1) null mouse. Endocrinology. 2005;146(2):825–34.
Sakaki T, Yasuda K, Kittaka A, Yamamoto K, Chen TC. CYP24A1 as a potential target for cancer therapy. Anticancer Agents Med Chem. 2014;14(1):97–108.
Jones G, Prosser DE, Kaufmann M. 25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): its important role in the degradation of vitamin D. Arch Biochem Biophys. 2012;523(1):9–18.
Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014;14(5):342–57.
Sun H, Wang C, Hao M, et al. CYP24A1 is a potential biomarker for the progression and prognosis of human colorectal cancer. Hum Pathol. 2016;50:101–8.
Koberle B, Koch B, Fischer BM, Hartwig A. Single nucleotide polymorphisms in DNA repair genes and putative cancer risk. Arch Toxicol. 2016;90(10):2369–88.
Yang W, Zhang T, Song X, Dong G, Xu L, Jiang F. SNP-target genes interaction perturbing the cancer risk in the post-GWAS. Cancers (Basel). 2022;14(22):5636.
Yao L, Tak YG, Berman BP, Farnham PJ. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114.
Yang J, Wang H, Ji A, et al. Vitamin D signaling pathways confer the susceptibility of esophageal squamous cell carcinoma in a Northern Chinese population. Nutr Cancer. 2017;69(4):593–600.
Wu X, Cheng J, Yang K. Vitamin D-related gene polymorphisms, plasma 25-hydroxy-vitamin D, cigarette smoke and Non-Small Cell Lung Cancer (NSCLC) risk. Int J Mol Sci. 2016;17(10):1597.
Armstrong RA. When to use the Bonferroni correction. Ophthalmic Physiol Opt. 2014;34(5):502–8.
Anderson LN, Cotterchio M, Cole DE, Knight JA. Vitamin D-related genetic variants, interactions with vitamin D exposure, and breast cancer risk among Caucasian women in Ontario. Cancer Epidemiol Biomarkers Prev. 2011;20(8):1708–17.
Anderson LN, Cotterchio M, Knight JA, Borgida A, Gallinger S, Cleary SP. Genetic variants in vitamin d pathway genes and risk of pancreas cancer; results from a population-based case-control study in ontario, Canada. PLoS One. 2013;8(6):e66768.
Beuten J, Gelfond JA, Franke JL, et al. Single and multigenic analysis of the association between variants in 12 steroid hormone metabolism genes and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1869–80.
Gong C, Long Z, Yu Y, et al. Dietary factors and polymorphisms in vitamin D metabolism genes: the risk and prognosis of colorectal cancer in northeast China. Sci Rep. 2017;7(1):8827.
Holick CN, Stanford JL, Kwon EM, Ostrander EA, Nejentsev S, Peters U. Comprehensive association analysis of the vitamin D pathway genes, VDR, CYP27B1, and CYP24A1, in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(10):1990–9.
Holt SK, Kwon EM, Peters U, Ostrander EA, Stanford JL. Vitamin D pathway gene variants and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1929–33.
McCullough ML, Stevens VL, Diver WR, et al. Vitamin D pathway gene polymorphisms, diet, and risk of postmenopausal breast cancer: a nested case-control study. Breast Cancer Res. 2007;9(1):R9.
Oh JJ, Byun SS, Lee SE, et al. Genetic variants in the CYP24A1 gene are associated with prostate cancer risk and aggressiveness in a Korean study population. Prostate Cancer Prostatic Dis. 2014;17(2):149–56.
Penna-Martinez M, Ramos-Lopez E, Stern J, et al. Impaired vitamin D activation and association with CYP24A1 haplotypes in differentiated thyroid carcinoma. Thyroid. 2012;22(7):709–16.
Reimers LL, Crew KD, Bradshaw PT, et al. Vitamin D-related gene polymorphisms, plasma 25-hydroxyvitamin D, and breast cancer risk. Cancer Causes Control. 2015;26(2):187–203.
Wei Y, Wang X, Zhang Z, et al. Role of Polymorphisms of FAM13A, PHLDB1, and CYP24A1 in Breast Cancer Risk. Curr Mol Med. 2019;19(8):579–88.
Zhuo M, Zhuang X, Tang W, et al. The impact of IL-16 3’UTR polymorphism rs859 on lung carcinoma susceptibility among Chinese Han individuals. Biomed Res Int. 2018;2018:8305745.
Qu R, Li X, Quan X, et al. Polymorphism in CYP24A1 is associated with lung cancer risk: a case-control study in Chinese female nonsmokers. DNA Cell Biol. 2019;38(3):243–9.
Yi C, Huang C, Wang H, et al. Association study between CYP24A1 gene polymorphisms and cancer risk. Pathol Res Pract. 2020;216(1): 152735.
Clendenen TV, Ge W, Koenig KL, et al. Genetic polymorphisms in vitamin D metabolism and signaling genes and risk of breast cancer: a nested case-control study. PLoS One. 2015;10(10): e0140478.
Sadeghi H, Nazemalhosseini-Mojarad E, Piltan S, et al. A candidate intronic CYP24A1 gene variant affects the risk of colorectal cancer. Biomark Med. 2020;14(1):23–9.
Holt SK, Kwon EM, Koopmeiners JS, et al. Vitamin D pathway gene variants and prostate cancer prognosis. Prostate. 2010;70(13):1448–60.
Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007;7(9):684–700.
Albertson DG, Ylstra B, Segraves R, et al. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25(2):144–6.
Shiratsuchi H, Wang Z, Chen G, et al. Oncogenic potential of CYP24A1 in lung adenocarcinoma. J Thorac Oncol. 2017;12(2):269–80.
Chen XQ, Mao JY, Li WB, et al. Association between CYP24A1 polymorphisms and the risk of colonic polyps and colon cancer in a Chinese population. World J Gastroenterol. 2017;23(28):5179–86.
Parise RA, Egorin MJ, Kanterewicz B, et al. CYP24, the enzyme that catabolizes the antiproliferative agent vitamin D, is increased in lung cancer. Int J Cancer. 2006;119(8):1819–28.
Kong J, Xu F, Qu J, et al. Genetic polymorphisms in the vitamin D pathway in relation to lung cancer risk and survival. Oncotarget. 2015;6(4):2573–82.
Zhu M, Qiu S, Zhang X, et al. The associations between CYP24A1 polymorphisms and cancer susceptibility: a meta-analysis and trial sequential analysis. Pathol Res Pract. 2018;214(1):53–63.
Wang P, Zhang H, Zhang Z, Qin L, Li B. Association of the CYP24A1-rs2296241 polymorphism of the vitamin D catabolism enzyme with hormone-related cancer risk: a meta-analysis. Onco Targets Ther. 2015;8:1175–83.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
YBW, SFY, and XTL contributed to the conception of the study. YBW and RWW performed the data collection and data analysis. YBW, RWW, and XTL wrote the manuscript. The authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable since our study is a meta-analysis.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1.
Search strategy.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, Y., Wang, R., Yuan, S. et al. Genetic polymorphisms of CYP24A1 gene and cancer susceptibility: a meta-analysis including 40640 subjects. World J Surg Onc 21, 279 (2023). https://doi.org/10.1186/s12957-023-03156-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-023-03156-w