Abstract
Objective
Regulation of single nucleotide polymorphisms (SNP) in micro-RNA (miRNA) on the host cells may be one of the most important factors influencing the occurrence of cervical cancer based on the prevalence of HPV infection and the development of cervical cancer. In order to explore the contribution of miRNA polymorphism to the occurrence and development of cervical cancer, we conducted an analytical study.
Methods
We selected the polymorphisms of three widely studied miRNAs (miRNA-146a rs2910164, miRNA-499 rs3746444, and miRNA-196a2 rs11614913). Then, we conducted a meta-analysis (for the first time) to investigate their susceptibility to cervical cancer. Case control studies on the correlation between these three miRNAs and cervical cancer susceptibility were investigated by searching on from Pubmed, The Cochrane Library, Embase, CBM, CNKI, Wanfang database, and VIP database. Basic characteristics were recorded and meta-analysis of the case studies was performed using the STATA 15.1 software.
Results
The miRNA-146a rs2910164 mutation significantly reduced the risk of cervical cancer in both recessive model (OR = 0.804, 95% CI = 0.652-0.992, P = 0.042; CC vs. CG+GG) and allelic model (OR = 0.845, 95% CI = 0.721-0.991, P = 0.038; C vs. G). There was no significant correlation between miRNA-499 rs3746444 and the risk of cervical cancer. The miRNA-196a2 rs11614913 mutation was significantly associated with a reduced risk of cervical cancer in homozygous model (OR = 0.641, 95% CI = 0.447-0.919, P = 0.016; TT vs. CC), dominant model (OR = 0.795, 95% CI = 0.636-0.994, P = 0.045; CT+TT vs. CC), recessive model (OR = 0.698, 95% CI = 0.532-0.917, P = 0.01; TT vs. CC+CT), and allelic models (OR = 0.783, 95% CI = 0.643-0.954, P = 0.015, T vs. C).
Conclusion
In summary, this meta-analysis shows that the mutant genotypes of miRNA-146a rs2910164 and miRNA-196a2 rs11614913 are associated with a reduced risk of cervical cancer. Therefore, they may be two gene regulatory points for the prevention of cervical cancer.
Systematic review registration
PROSPERO registration number CRD42021270079.
Similar content being viewed by others
Introduction
Cervical cancer is the second most common malignancy in women, whose incidence is second only to breast cancer [1]. About 500,000 new cases of cervical cancer occur globally each year, accounting for 5% of all new cancer cases. Every year, more than 260,000 women die of cervical cancer [2], accounting for 7.5% of all female cancer deaths. The number is increasing year by year, and the patient population is getting younger. Therefore, cervical cancer is an important disease endangering women’s health and life [3, 4]. Focusing on women’s health is an important step in human reproduction and development. At present, in the face of cervical cancer, we still focus on prevention. Hence, the factors affecting the occurrence and development of cervical cancer are worthy of our research and discussion.
It is well-known that human papillomavirus (HPV) is associated with the occurrence of cervical cancer. HPV is the major risk factor for cervical cancer [5]. Women infected with HPV can get rid of it by their own immune system, but there are still some women who develop cervical cancer due to persistent infection with high-risk HPV [6]. In the process of continuous infection with the high-risk HPV until the development of cervical cancer, these women are also affected by environmental, genetic, and other factors [7].
Many diseases have been found to be related to genes. Nowadays, the genetic changes of patients and their influence on the occurrence and development of diseases are the focus of research. MicroRNAs (miRNAs) are a class of non-coding single-stranded RNA molecules with a length of about 22 nucleotides that regulate various intracellular activities such as cell proliferation, apoptosis, and carcinogenesis [8,9,10]. Each miRNA can have multiple target genes, and several miRNAs can regulate the same gene. MicroRNAs have been reported to regulate about one-third of human genes [11]. The length of miRNA is usually shorter than the length of the encoding gene. Its small genetic changes may alter human miRNA expression and target selection [12]. MicroRNAs have been shown to be associated with cervical cancer [13]. Therefore, single nucleotide polymorphisms (SNPs) in miRNA genes may lead to abnormal gene expression by changing miRNA maturation and expression [14, 15]. This may be related to cervical cancer susceptibility. In the past few years, several studies have investigated the association between some miRNA polymorphisms and cervical cancer risk, but the results were different [16,17,18,19,20,21,22,23]. Therefore, three miRNA polymorphisms (miRNA-146a rs2910164, miRNA-499 rs3746444, and miRNA-196a2 rs11614913) associated with cervical cancer risk were selected for meta-analysis. The polymorphism sites and base pair locations of the above three miRNAs are shown in Table 1.
Materials and methods
Research program
This protocol of systematic review and meta-analysis has been drafted under the guidance of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocols (PRISMA-P). The meta-analysis was based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) report (Table S1) [24]. It has been shared on open science framework (OSF) (registration number: DOI 10.17605/OSF.IO/P38H2). Moreover, it has been registered on PROSPERO (registration number: CRD42021270079).
Literature search and selection
We searched Pubmed, The Cochrane Library, Embase, CBM, CNKI, Wanfang Database, and VIP database; all of which were established until May 2021. The retrieval method adopted the combination of subject words and free words. The retrieval strategy took PubMed as an example, as shown in Table S2. We searched all miRNA associated with cervical cancer susceptibility, the retrieved articles were screened by inclusion and exclusion criteria. Finally, three miRNA were included in this study (including miRNA-146a, miRNA-499, and miRNA-196a2). Where necessary, we contacted the original study author by email or phone for the undetermined but important information required in this study.
Inclusion criteria: (1) The association between miRNA and the cervical cancer risk was assessed; (2) studies with relevant genotype OR allele frequency, information to calculate OR and 95% CI; (3) the subjects were humans; (4) the study was designed as a case control.
Exclusion criteria: (1) Specific information on genotype or allele frequency of miRNA was not provided in the literature; (2) the genotype frequency of the control group was not in Hardy-Weinberg equilibrium; (3) the relationship with cervical cancer is unclear or the number of studies does not meet the miRNA for systematic evaluation; (4) cervical abscess cell pathology in the control group showed cervical intraepithelial neoplasia (CIN); (5) literatures with less than 6 stars in the Newcastle-Ottawa Scale [25].
Data extraction and quality evaluation
The final included literature recorded the following information: first author name, year of publication, patient nationality, genotyping method, genotype, and allele frequency, and tabulated the basic characteristics. The selected literature was evaluated using the Newcastle-Ottawa Scale.
Statistical analysis
Meta-analysis was used to assess the relationship between three miRNA polymorphisms and cervical cancer susceptibility. The genotype frequency of the control group was tested by the Hardy-Weinberg equilibrium (HWE) test using the chi-square test. If P > 0.05, it was regarded to be in the Hardy-Weinberg equilibrium. The odds ratio (OR) and 95% confidence interval (95%CI) were calculated to evaluate the relationship between the six gene models (homozygous, heterozygous, dominant, recessive, over dominant, and allelic genetic models) of the polymorphisms of miRNA-146a rs2910164, miRNA-499 rs3746444, and miRNA-196a2 rs11614913 and the risk of cervical cancer. The strength of association between each polymorphism and the risk of cervical cancer was evaluated by the combined odds ratio (OR) and its 95% confidence interval (95% CI). The statistical significance of combined OR was tested by z test; and P < 0.05 showed a significant statistical difference. We assessed heterogeneity between studies by chi-square test and I2: If P ≥ 0.1 and I2 < 50%, we used a fixed-effect model to assess OR and 95% CI. If P < 0.1 and I2 > 50%, we evaluated OR and 95% CI using a random effect model.
This meta-analysis quantitatively assessed publication bias using the Begg’s test and the Egger’s test. Sensitivity analysis assessed the stability of the results by excluding each study in turn. All analyses were performed using the STATA 15.1 software (STATA Corporation, College Station, TX, USA).
Results
Literature screening results
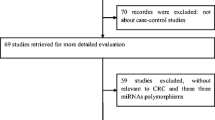
In total, 8 articles were included in this meta-analysis [16,17,18,19,20,21,22,23]. From the studies, 14 data sets, comprising 3142 patients and 3971 controls were retrieved. Among these, five studies related to the correlation between miRNA-146a rs2910164 and cervical cancer; three related to the correlation between miRNA-499 rs3746444 and cervical cancer; and six related to the correlation between miRNA-196a2 rs11614913 and cervical cancer. The flowchart of literature screening is as shown in Fig. 1.
Basic characteristics of the included literature and results of quality assessment
The basic characteristics of the literature are described in Table 2. Eight literatures were evaluated by Newcastle-Ottawa Scale, and the scores were all above 7☆ (full score was 9☆). The quality evaluation results of the included literature are as shown in Table 3.
Meta-analysis of the relationship between three miRNAs and cervical cancer
Relationship between miRNA-146a rs2910164 and cervical cancer
The combined analysis showed that the mutant C allele significantly reduced the risk of cervical cancer in both the recessive genetic model (OR = 0.804, 95% CI = 0.652-0.992, P = 0.042; CC vs. CG+GG) and the allele genetic model (OR = 0.845, 95% CI = 0.721-0.991, P = 0.038; C vs. G), and the difference was statistically significant (Table 4 and Fig. 2).
Subgroup analysis was performed according to the analysis methods used for genotyping and countries. In the study involving PCR-RFLP analysis, the polymorphism of miRNA-146a rs2910164 reduced the risk of cervical cancer in the homozygote (OR = 0.626, 95% CI = 0.453-0.865, P = 0.004; CC vs. GG), heterozygote (OR = 0.787, 95% CI = 0.637-0.972, P = 0.026; CG vs. GG), dominant (OR = 0.726, 95% CI = 0.590-0.893, P = 0.002; CC+CG vs. GG), recessive (OR = 0.753, 95% CI = 0.581-0.976, P = 0.032; CC vs. CG+GG), allele (OR = 0.795, 95% CI = 0.681-0.928, P = 0.004; C vs. G) genetic models, and the difference was statistically significant. The difference was not statistically significant in the analysis of China and India (Table 5).
Relationship between miRNA-499 rs3746444 and cervical cancer
The pooled analysis showed that among the six genetic models, there was no correlation between miRNA-499 rs3746444 and the risk of cervical cancer (Table 4). Subgroup analysis indicated that this correlation remained absent in different genotyping assays and countries (Table 5).
Relationship between miRNA-196a2 rs11614913 and cervical cancer
The combined analysis showed that mutant TT was significantly associated with a lower risk of cervical cancer compared to the wild homozygous CC (OR = 0.641, 95% CI = 0.447-0.919, P = 0.016; TT vs. CC). The mutant gene T significantly reduced the risk of cervical cancer in both dominant (OR = 0.795, 95% CI = 0.636-0.994, P = 0.045; CT+TT vs. CC) and recessive (OR = 0.698, 95% CI = 0.532-0.917, P = 0.01; TT vs. CC+CT) genetic models. In the allele model, T significantly reduced the risk of cervical cancer as compared to C (OR = 0.783, 95% CI = 0.643-0.954, P = 0.015; T vs. C) (Table 4 and Fig. 3).
Subgroup analysis by genotyping method and country in the study of genotype analysis using TaqMan showed that the polymorphism of miRNA-196a2 rs11614913 significantly reduced the risk of cervical cancer in homozygous (OR = 0.762, 95% CI = 0.625-0.929, P = 0.007; TT vs. CC), recessive (OR = 0.807, 95% CI = 0.707-0.921, P = 0.001; TT vs. CT+CC), and allele (OR = 0.871, 95% CI = 0.795-0.955, P = 0.003; T vs. C) genetic models (Table 5). Studies in China showed that the polymorphism of miRNA-196a2 rs11614913 significantly reduces the risk of cervical cancer in homozygous (OR = 0.787, 95% CI = 0.672-0.921, P = 0.003; TT vs. CC), recessive (OR = 0.817, 95% CI = 0.724-0.923, P = 0.001; TT vs. CT+CC), and allele (OR = 0.883, 95% CI = 0.817-0.954, P = 0.002; T vs. C) genetic models. Furthermore, studies in India have shown that the risk of cervical cancer is significantly reduced in homozygous (OR = 0.167; 95% CI = 0.086-0.323; P < 0.001; TT vs. CC), dominant (OR = 0.389; 95% CI = 0.241-0.628; P < 0.001; CT+TT vs.CC), recessive (OR = 0.209; 95% CI = 0.114-0.381; P < 0.001; TT vs. CT+CC), and allele (OR = 0.362; 95% CI = 0.259-0.506; P < 0.001; T vs. C) genetic models.
Heterogeneity
MiRNA-146a rs2910164 showed no heterogeneity in the super-dominant genetic model, and heterogeneity in the homozygote and the dominant genetic models. The results of subgroup analysis showed that the heterogeneity decreased in the studies using the PCR-RFLP analysis. No heterogeneity was observed in the homozygous and recessive genetic models for miRNA-499 rs3746444, but considerable heterogeneity was observed in the remaining models. The heterogeneity persisted after subgroup analysis. MiRNA-196a2 rs11614913 showed no heterogeneity in heterozygote and hyper-dominant genetic models, but it showed heterogeneity in the homozygote, recessive, and allele models. Subgroup analysis showed that heterogeneity decreased in studies using TaqMan analysis (Table 4).
Sensitivity analysis
In order to explore whether the meta-analysis results were stable, sensitivity analysis was conducted. The meta-analysis results appeared to be stable.
Publication bias
Potential publication bias of this meta-analysis was tested by Begg’s test and Egger’s test (Table 4). The results shown in the table indicate that no publication bias existed among the polymorphisms of miRNA-146a rs2910164, miRNA-499 rs3746444, and miRNA-196a2 rs11614913 in all genetic models.
Discussion
Research on the impact of single nucleotide polymorphisms (SNPs) on the occurrence and development of diseases has increased over the years. With the development of molecular biology, discussions on genetic problems have become necessary. In the face of multiple pathogenic factors such as HPV infection, risky living environment, and host factors, which together lead to the occurrence of cervical cancer [2], while studying this process in depth, we found that the microenvironment formed by the regulation of certain genes in the human body seems to guide the occurrence of cervical cancer [26,27,28].
MiRNAs regulate gene expression at the post-transcriptional level, and participate in the regulation of various cellular functions. They affect cell proliferation, apoptosis, and carcinogenesis [29]. Variation of a certain miRNA, such as the conversion, insertion, deletion, or mismatch of a single base, can cause the polymorphism of the whole DNA sequence. This may in turn affect the function of cells and tissues and the whole host microenvironment [30]. Therefore, miRNAs may be used to predict the occurrence and development of cervical cancer [27]. We selected three kinds of miRNA polymorphisms (miRNA-146a rs2910164, miRNA-499 rs3746444, and miRNA-196a2 rs11614913) that have been studied more frequently in order to analyze their relationship with susceptibility to cervical cancer [16,17,18,19,20,21,22,23]. The results showed that the polymorphisms of miRNA-146a rs2910164 (mutant C allele) and miRNA-196a2 rs11614913 (mutant T allele) significantly reduced the risk of cervical cancer. There was no significant correlation between the polymorphisms of miRNA-499 rs3746444 and cervical cancer.
In the study using PCR-RFLP analysis, the polymorphism of miRNA-146a rs2910164 significantly reduced the risk of cervical cancer in homozygote, heterozygote, dominant, recessive, and allele models. With regard to the TaqMan analysis of the genotypes, the polymorphism of miRNA-196a2 rs11614913 significantly reduced the risk of cervical cancer in homozygous, recessive, and allelic models. According to the Chinese studies, miRNA-196a2 rs11614913 reduced cervical cancer risk in homozygous, recessive, and allelic models [16,17,18,19,20,21,22], whereas in Indian studies, it significantly reduced cervical cancer risk in homozygous, dominant, recessive, and allelic models [23]. Thus, using different genotype analysis methods and in different countries, the correlation between the polymorphisms of miRNA-146a rs2910164 and miRNA-196a2 rs11614913 and cervical cancer is somehow different. However, it cannot be denied that they can both reduce the risk of cervical cancer.
Meanwhile, a study by Yan et al. showed that the mutant T allele in the miRNA-126 rs4636297 polymorphism was associated with cervical cancer susceptibility, and the mutant T gene increased cervical cancer susceptibility compared to the wild C gene [21]. Wang et al. reported that the mutant C allele in the polymorphism of miRNA-149 rs2292832 was associated with cervical cancer susceptibility, and the mutant C gene increased cervical cancer susceptibility relative to the original T gene [22]. Furthermore, Chuntao noted that mutation of A allele to G allele in the polymorphism of miRNA-30c rs928508 could reduce the susceptibility to cervical cancer [20]. Fu K showed that miRNA-125 can negatively regulate the expression of VEGF in cervical cancer tissues and inhibit the PI3K/AKT signaling pathway to inhibit cell proliferation, invasion, and metastasis in cervical cancer, thus preventing the progression of cervical cancer [31]. The study of Lu X showed that miRNA-186-3p could inhibit the proliferation and migration of cervical cancer cell lines by inhibiting the expression of IGF1, and induce the apoptosis rate of cervical cancer cells [32]. Zhou H’s study showed that Mir-183-5p could mediate cirC_0018289 in vitro to regulate the development and angiogenesis of cervical cancer. Meanwhile, TMED5 plays a tumor-promoting role in the malignant development of cervical cancer by activating the Wnt7b/β-catenin signaling pathway, while Mir-183-5p can directly target TMED5 and inhibit its expression, so as to inhibit the occurrence of cervical cancer [33]. The above studies all indicate that miRNA expression is closely related to the occurrence and development of cervical cancer. However, due to few studies on the related miRNA polymorphisms, they were not included in the meta-analysis. These loci may present more in-depth studies in the future.
Persistent infection with high-risk HPV is a major factor in the development of cervical cancer [34, 35]. However, the occurrence of cervical cancer is a multigene, multistage abnormal regulation process [36, 37]. Studies have also shown that the abnormal expression of miRNAs affects the cervical epithelial cell carcinogenesis [38, 39]. Different miRNAs regulate different biological activities; some target to regulate the activity of tumor suppressor genes to inhibit development of the cancer cells. Others accelerate the growth and metastasis of cancer cells by stimulating proto-oncogenes and metastasis genes [39]. Studies have shown that the polymorphism of miRNA-146a rs2910164 G to C leads to increased expression of miR-146a, and the mutant CC genotype is associated with reduced risk of cervical cancer [40]. As a mediator of the pro-apoptotic transcription nuclear factor kappaB, miRNA-146a affects apoptosis and hence influences the development of cancer [41]. Each cancer tissue has its specific miRNA target [42], so the miRNA-146a polymorphism may affect the occurrence and development of cervical cancer through certain pathways. MiRNA-196a2 rs11614913 is a polymorphic site in the mature miR-196a2 sequence, and its mutation expression in situ gene C, the T allele, was found to be associated with reduced risk of cervical cancer [43]. Studies have shown that C gene increases the expression of mature miR-196a2 in cervical cancer patients, and this overexpression changes the state of binding of miR-196a2 to its target genes, thereby affecting the development of cervical cancer [19]. Tracking the changes of the two genetic polymorphism loci in human may help to understand how to reduce the incidence of cervical cancer, by changing the internal human environment. Therefore, these two sites may become the targets for the prevention and treatment of cervical cancer in the future. However, the biomarkers for targeted therapy of cervical cancer are complex and diverse, which need to be fully explored. Li S et al. also obtained 6 prognostic pivotal genes by WGCNA algorithm: SLC25A5, ENO1, ANLN, RIBC2, PTTG1, and MCM5. It also provides a broader direction for the research and treatment of cervical cancer [44].
However, this meta-analysis had several limitations: (1) Due to the fewer factors controlled by relevant studies, the relationship between age, number of abortions (Abortion refers to those who terminate before 28 weeks of pregnancy and whose fetal weight is less than 1000 g. This can be discussed in two categories: induced abortion and spontaneous abortion.), and the expression of the three miRNAs in cervical cancer patients was not analyzed. (2) The heterogeneity of some polymorphisms may be due to the patient population included (i.e., clinical heterogeneity), such as different types of cervical cancer, large age span, and the presence of other genetic diseases. At the same time, heterogeneity may also come from the research itself (i.e., methodological heterogeneity), such as different testing time and methods, and different experimental instruments. However, due to the lack of available data, all factors cannot be analyzed. It also reminds us that future studies should be designed more rigorously and record more comprehensive data to analyze results. (3) This meta-analysis did not establish whether the mixed effects of genetic polymorphisms were associated with susceptibility to cervical cancer.
Conclusion
In conclusion, this meta-analysis showed that the polymorphisms of miRNA-146a rs2910164 (mutant C allele) and miRNA-196a2 rs11614913 (mutant T allele) could reduce the susceptibility to cervical cancer.
Availability of data and materials
The data included in this study are all from the retrieved literature. For details, please refer to articles 16-23, in which the data are included in this study.
Abbreviations
- SNP:
-
Single nucleotide polymorphisms
- miRNA:
-
micro-RNA
- HPV:
-
Human papillomavirus
- NCTV:
-
non_coding_transcript_variant
- CIN:
-
Cervical intraepithelial neoplasia
- HWE:
-
Hardy-Weinberg equilibrium
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- HB:
-
Hospital-based
- PCR-RFLP:
-
Restriction fragment length polymorphism polymerase chain reaction
- PCR-LDR:
-
Polymerase chain reaction-ligase detection reaction
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA-Cancer J Clin. 2015;65:87–108.
Feng S. Analysis of risk factors and preventive health care of cervical cancer. Elect J Pract Gynecol Endocrinol. 2018;5:11–4.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN. Int J Cancer. 2012;136:E359–86.
Olusola P, Banerjee HN, Philley JV, Dasgupta S. Human papilloma virus-associated cervical cancer and health disparities. Cells. 2019;8(6):622. https://doi.org/10.3390/cells8060622 PMID: 31234354.
Shi Y, Li L, Hu Z, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45:918–22.
Hu Z, Ma D. The precision prevention and therapy of HPV-related cervical cancer: new concepts and clinical implications. Cancer Med. 2018;7(10):5217–36. https://doi.org/10.1002/cam4.1501 Epub 2018 Sep 14. PMID: 30589505; PMCID: PMC6198240.
Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol. 2014;35(6):5083–93. https://doi.org/10.1007/s13277-014-1737-z Epub 2014 Feb 20. PMID: 24554414.
Piccoli MT, Gupta SK, Thum T. Noncoding RNAs as regulators of cardiomyocyte proliferation and death. J Molcell Cardiol. 2015;89:59–67.
Chen Y, Fu LL, Wen X, Liu B, Huang J, Wang JH, et al. Oncogenic and tumor suppressive roles of microRNAs in apoptosis and autophagy. Apoptosis. 2014;19:1177–89.
Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019;20(24):6249. https://doi.org/10.3390/ijms20246249 PMID: 31835747; PMCID: PMC6941098.
Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G. Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 2019;20(5):1836–52. https://doi.org/10.1093/bib/bby054 PMID: 29982332; PMCID: PMC7414524.
Shen S, Zhang S, Liu P, Wang J, Du H. Potential role of microRNAs in the treatment and diagnosis of cervical cancer. Cancer Gene Ther. 2020;248-249:25–30. https://doi.org/10.1016/j.cancergen.2020.09.003 Epub 2020 Sep 22. PMID: 32987255.
Srivastava SK, Ahmad A, Zubair H, Miree O, Singh S, Rocconi RP, et al. microRNAs in gynecological cancers: small molecules with big implications. Cancer Lett. 2017;407:123–38.
Qi P, Wang L, Zhou B, Yao WJ, Xu S, Zhou Y, et al. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res. 2015;14:6289–96.
Kroliczewski J, Sobolewska A, Lejnowski D, Collawn JF, Bartoszewski R. microRNA single polynucleotide polymorphism influences on microRNA biogenesis and mRNA target specificity. Gene. 2018;640:66–72.
Zhou B, Wang K, Wang Y, Xi M, Zhang Z, Song Y, et al. Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol Carcinog. 2011;50:499–505.
Yue C, Wang M, Ding B, Wang W, Fu S, Zhou D, et al. Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecol Oncol. 2011;122:33–7.
Ma L. Relationship between the polymorphism of Mir-146a (RS2910164) gene and the differential expression of Mir-146A \ Mir-133b and the occurrence and development of cervical cancer. J Xinjiang Med Univ. 2015.
Ding B, Wang ML, Zhang ZD, Han SP, et al. Relationship between mir-196a2 T > C polymorphisms and susceptibility to cervical cancer. J Nanjing Med Univ. 2016;36:85–9.
Wang CT, Xu M, Zhang YQ, Shen X, Sui J, Liang YY, et al. A case-control study on the relationship between micRORNA-30C and 196A2 gene polymorphism and cervical cancer. J Environ Occup Med. 2016;33:1043–8.
Yan ZL, Zhou Z, et al. Polymorphisms in miRNA genes play roles in the initiation and development of cervical cancer. J Cancer. 2019;10(20):4747–53.
Wang SZ, Zhu H, Ding B, Feng X, Zhao W, Cui M, et al. Genetic variants in microRNAs are associated with cervical cancer risk. Pubmed. 2019;34(2):1–7.
Thakur N, Singhal P, mehrotra R, bharadwaj M. Impacts of single nucleotide polymorphisms in three microRNAs (miR-146a, miR-196a2 and miR-499) on the susceptibility to cervical cancer among Indian women. Pubmed. 2019;39(4):1–28.
Yao X, Florez ID, Zhang P, Zhang C, Zhang Y, Wang C, et al. Clinical research methods for treatment, diagnosis, prognosis, etiology, screening, and prevention: a narrative review. J Evid Based Med. 2020;13(2):130–6. https://doi.org/10.1111/jebm.12384 Epub 2020 May 22. PMID: 32445266.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5. https://doi.org/10.1007/s10654-010-9491-z.
Rao HC, Wei SD. Advances in research on susceptibility genes related to cervical cancer. Henan Medical Research. 2018;27:4087–9.
Yang XY, Wang J, Zhou H, Wan FS. Research progress on the relationship between abnormal miRNAs expression and development of cervical cancer. Chin J Cell Biol. 2020;42:166–72.
Zhu L, Tu H, Liang Y, et al. MiR-218 produces anti-tumor effects on cervical cancer cells in vitro. World J Surg Onc. 2018;16:204. https://doi.org/10.1186/s12957-018-1506-3.
Geng S, Guo HJ, Shi N, Tang Q, Chang LR. Advances in the study of the regulation mechanism of microRNA on tumor. Chin J New Drugs Clin Remed. 2020;39:193–7.
Duan X, Wang L, Sun G, Yan W, Yang Y. Understanding the cross-talk between host and virus in poultry from the perspectives of microRNA: Elsevier; 2020.
Fu K, Zhang L, Liu R, et al. MiR-125 inhibited cervical cancer progression by regulating VEGF and PI3K/AKT signaling pathway. World J Surg Onc. 2020;18:115. https://doi.org/10.1186/s12957-020-01881-0.
Lu X, Song X, Hao X, et al. MiR-186-3p attenuates tumorigenesis of cervical cancer by targeting IGF1. World J Surg Onc. 2021;19:207. https://doi.org/10.1186/s12957-021-02317-z.
Zou H, Chen H, Liu S, et al. Identification of a novel circ_0018289/miR-183-5p/TMED5 regulatory network in cervical cancer development. World J Surg Onc. 2021;19:246. https://doi.org/10.1186/s12957-021-02350-y.
Sun J, Wang XY. Value of human papillomavirus in cervical cancer screening. Chin Remed Clin. 2019;19:2625–6.
Zhou Z, Yang H, Yang L, et al. Human papillomavirus type 16 E6 and E7 gene variations associated with cervical cancer in a Han Chinese population. Elsevier. 2019;73:13–20. https://doi.org/10.1016/j.meegid.2019.04.008.
Wang HP, Gao ZW, Li RC, Liu C, Dong K, Zhang HZ, et al. Screening and functional analysis of differentially expressed genes in cervical cancer. Progr Mod Biomed. 2020;20:379–84.
Lee H, Kim KR, Cho NH, et al. MicroRNA expression profiling and Notch1 and Notch2 expression in minimal deviation adenocarcinoma of uterine cervix. World J Surg Onc. 2014;12:334. https://doi.org/10.1186/1477-7819-12-334.
Brandão BFC, Salviano-Silva A, Cristina BH, de Brajão OK, Adelman CG, Danielle M. From squamous intraepithelial lesions to cervical cancer: circulating microRNAs as potential biomarkers in cervical carcinogenesis. Pubmed. 2019;1872(2):188306. https://doi.org/10.1016/j.bbcan.2019.08.001.
Allegra A, Musolino C, Tonacci A, Pioggia G, Gangemi S. Interactions between the microRNAs and microbiota in cancer development: roles and therapeutic opportunities mDPI. 2020;12(4):805. https://doi.org/10.3390/cancers12040805.
Ren XM, Jia P, Xu PF, Fu L. The expression level of Mir-146a in cervical cancer and its relationship with clinicopathological parameters and prognosis. Mat Child Health Care Chin. 2019;34:1873–6.
Venuti A, Musarra-Pizzo M, Pennisi R, Tankov S, Medici MA, Mastino A, et al. HSV-1\EGFP stimulates miR-146a expression in a NF-κB-dependent manner in monocytic THP-1 cells. Sci Rep. 2019;9(1):51–7. https://doi.org/10.1038/s41598-019-41530-5 PMID: 30914680.
Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. https://doi.org/10.1016/j.critrevonc.2015.10.003 Epub 2015 Oct 8. PMID: 26481951.
Shruti S, Shalini S, Naseem F, Balraj M, Narain SA. Pre-microRNA gene polymorphisms and risk of cervical squamous cell carcinoma. Pubmed. 2017;11(9):GC01-GC04. https://doi.org/10.7860/JCDR/2017/25361.10543.
Li S, Han F, Qi N, et al. Determination of a six-gene prognostic model for cervical cancer based on WGCNA combined with LASSO and cox-PH analysis. World J Surg Onc. 2021;19:277. https://doi.org/10.1186/s12957-021-02384-2.
Acknowledgements
The authors would like to thank freescience (www.home-for-researchers.com) for the help with the English language.
Funding
This study was financially supported by the National Natural Science Foundation of China (No. 81774351).
Author information
Authors and Affiliations
Contributions
Conception and design: Qiang Zhang; Acquisition of data: Yihua Fan; Data extraction: Jingyu Xu, Junze Geng; Drafting of the manuscript: Jingyu Xu; Critical revision of the manuscript for important intellectual content: Yihua Fan, Zijun Qi, Tian Xia. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval and informed consent were not involved in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
: The PRISMA checklist.
Additional file 2: Table S2
: Search strategy table (PubMed).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, J., Geng, J., Zhang, Q. et al. Association of three micro-RNA gene polymorphisms with the risk of cervical cancer: a meta-analysis and systematic review. World J Surg Onc 19, 346 (2021). https://doi.org/10.1186/s12957-021-02463-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-021-02463-4