Abstract
Objective
This meta-analysis was implemented to evaluate the association between hypoxia-inducible factor-1α (HIF-1α) C1772T/G1790A polymorphisms and susceptibility to head and neck cancer (HNC).
Material and methods
This meta-analysis has been registered on PROSPERO platform (CRD42021257309). The PubMed, Embase and Web of Science databases were searched to retrieve eligible published papers. STATA software was used to calculate the pooled odds ratios (ORs) and corresponding 95% confidence intervals (CIs) to assess the correlation strength.
Results
Our results demonstrated that the HIF-1α C1772T polymorphism was significantly related to an increased HNC risk (OR = 2.27, 95% CI = 1.17–4.42 for the homozygous model; OR = 11.53, 95% CI = 1.11–120.4 for the recessive model), especially in Caucasians (OR = 2.16, 95% CI = 1.09–4.27 for the homozygous model; OR = 2.28, 95% CI = 1.15–5.51 for the recessive model). Similarly, a remarkable correlation was discovered between the G1790A polymorphism and HNC risk (OR = 72.11, 95% CI = 2.08–2502.4 for the homozygous model; OR = 58.05, 95% CI = 1.70–1985.77 for the recessive model). Moreover, in the subgroup analysis by source of controls, a statistically significant correlation was discovered in the population-based (PB) subgroup (OR = 9.43, 95% CI = 1.20–73.9 for allelic model; OR = 72.11, 95% CI = 2.08–2502.4 for the homozygous model; OR = 3.22, 95% CI = 1.28–8.08 for the heterozygous model; OR = 7.83, 95% CI = 1.48–41.37 for the dominant model; OR = 58.05, 95% CI = 1.70–1985.8 for the recessive model) but not in the hospital-based (HB) subgroup.
Conclusion
Our study found that both HIF-1α C1772T and G1790A polymorphisms might be a higher risk of HNC, especially in the Caucasian group with the C1772T polymorphism.
Similar content being viewed by others
Introduction
Head and neck cancer (HNC), which includes oropharyngeal cancer, nasopharyngeal cancer, laryngeal cancer and tongue cancer, is the eighth most common cancer according to the latest data reported in the Global Cancer Statistics 2018 [1, 2]. Based on statistics, more than 550,000 new cases of HNC occur worldwide each year, with 300,000 deaths [3]. Although the treatment of HNC patients is progressing, the age of patients has gradually decreased in recent years [4], which may be due to the infection of human papillomavirus (HPV), environmental pollution and unhealthy living habits [5, 6]. Because tumours have low survival rates and high mortality, the quality of life of patients with cancer is greatly reduced [7]. HNC is caused by multiple factors, of which smoking and drinking alcohol are recognized as major risk factors [8,9,10,11,12]. For instance, the study has shown that tobacco increases mutations in cancer [13]. In addition, infection of HPV can cause a variety of cancers, such as cervical cancer and oropharyngeal cancer, and has recently attracted the attention of scientists as another important risk factor for HNC [14, 15]. However, not every individual exposed to the above conditions will have HNC, which indicates that individual genetic susceptibility is also an important factor in the occurrence of HNC [6, 16, 17].
Hypoxia initiates a series of cellular responses, such as angiogenesis, proliferation and glucose and energy metabolism, which might result in the occurrence and development of tumours [18]. Hypoxia-inducible factor-1 (HIF-1) can regulate cellular adaptations to hypoxia [19]. Moreover, it has been reported that HIF-1 can activate numerous genes that play a key role in the critical biological behaviour of tumours [20, 21]. HIF-1α has the ability to determine the activity of HIF-1 and can regulate the expression level of genes related to angiogenesis and metastasis. Many researchers have demonstrated that high HIF-1α expression is found in most human tumours, such as breast carcinoma, hepatocellular cancer, cervical cancer and colorectal tumours [22,23,24,25], and HIF-1α may also be a prognostic marker in patients with oral cancer [26]. Moreover, carbonic anhydrase IX, a hypoxia-induced enzyme, is related to HIF-1α activity, as its overexpression is associated with poor prognosis in a variety of tumours, especially neuroblastoma [27]. Under normal oxygenation conditions, HIF-1α is modified by the enzyme prolyl hydroxylase (PHD), bound by von Hippel-Lindau factor (VHL), ubiquitinated and degraded by the proteasome. Alterations of this system predispose patients to a higher susceptibility to the development of tumours caused by mutations inactivating VHL, with a false signal of hypoxia [28]. In addition, recent studies have discovered that HIF-1α is related to poor prognosis in most tumours [29,30,31].
HIF-1 gene polymorphisms mediate genetic predisposition to cancer, of which C1772T and G1790A are two common single nucleotide polymorphisms (SNPs) of the HIF-1 gene [32]. Both polymorphisms have been reported to result in increased HIF-1α transcription activity under hypoxic conditions [33, 34]. Additionally, it has been reported that both are related to increased cancer microvessel density, and they are crucial in the progression of different cancers [23, 24, 34, 35].
In recent years, several researchers have reported the potential relationship between HIF-1α polymorphisms and susceptibility to HNC, but the results have been conflicting [32, 34, 36,37,38,39,40]. Thus, it is essential to conduct a comprehensive meta-analysis with high statistical power to study the role of HIF-1α C1772T and G1790A polymorphisms in the progression of HNC.
Methods
Selection of relevant studies
The meta-analysis was guided in strict accordance with the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement [41] (Additional file 1). The meta-analysis has been registered on PROSPERO platform (https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42021257309) with the registration number CRD42021257309. We used the "PICOs" strategy to guide the development of the research question (P: HNC patients; I: T(C1772T) and A(G1790A); C: C(C1772T) and G(G1790A); O: the risk of HNC; S: case–control study). The computerized literature retrieval was conducted using the PubMed, Embase and Web of Science databases to identify qualified studies with the following terms: ‘hif-1α’, or ‘hypoxia-inducible factor-1α’, or ‘hif-1’, or ‘hypoxia-inducible factor-1’, or ‘rs11549465’, or ‘C1772T’, or ‘P582S’, or ‘rs11549467’, or ‘G1790A’, or ‘A588T’ And ‘mutation’, or ‘mutations’, or ‘variants’, or ‘variant’, or ‘polymorphism’, or ‘polymorphisms’ And ‘carcinoma’, or ‘neoplasm’, or ‘tumour’, or ‘cancer’, or ‘carcinogenesis’ And ‘head and neck’, or ‘HNC’, or ‘oral’, or ‘oral cavity’, or ‘pharyngeal’, or ‘laryngeal’, or ‘laryngopharyngeal’, or ‘hypopharyngeal’, or ‘nasopharyngeal’, or ‘oropharyngeal’. The retrieval time was from database establishment to 5 November, 2020. Finally, all the included studies were carefully reviewed by the researchers (WT and BBT) to determine eligible studies, and another researcher (LL) discussed any differences.
Inclusion and exclusion criteria
Literature that satisfied the following criteria was included: (1) case-controlled, (2) described the correlation between the polymorphisms of HIF-1α C1772T and G1790A and HNC risk, and (3) data provided by the study were available. Literature that conformed to the following criteria were not enrolled: (1) nonhuman studies, reviews, editorials, commentaries and case reports; (2) full text could not be found; and (3) no sufficient information was provided.
Data extraction and quality assessment
The following data were retrieved: first author, publication year, country, ethnicity, genotyping methods, sources of controls, counts of case group and control group, genotype and allele frequencies for cases and controls, cancer site and P value of Hardy-Weinberg equilibrium (HWE) in controls. The Newcastle-Ottawa Scale (NOS) was selected for literature quality assessment, including population selection, comparability between groups, and exposure factors [42]. The Office of Health Assessment and Translation (OHAT) risk of bias rating tool was applied to evaluate the bias risk of the included articles [43,44,45]. Data extraction and quality assessment were conducted independently by two researchers (WT and BBT), and disagreements were discussed with a third researcher (LL).
Statistical analysis
STATA 11.0 (College Station, TX 77845, USA) was used for our meta-analysis. The strength of the correlation between HNC risk and HIF-1α C1772T/G1790A polymorphisms was assessed by ORs along with the corresponding 95% CIs. In our meta-analysis, we examined the relationship using allelic, homozygous, heterozygous, dominant and recessive genetic models. The genotyping method, ethnicity, source of control group and tumour site were used to perform subgroup analysis to determine whether certain factors were correlated with the overall ORs. Cochran's Q test and I2 test were used for heterogeneity analysis. When P < 0.10 or I2 > 50%, we believed that there was heterogeneity among the studies, and the DerSimonian and Laird random effects model was used for analysis; otherwise, the Mantel-Haenszel fixed effect model was used for analysis. Publication bias was evaluated by Begg's test and Egger's test, and the stability of the results was evaluated by sensitivity analysis. A Z test was conducted to analyse the statistically significant results, and a P value less than 0.05 was regarded as statistically significant.
Results
Literature search and characteristics of studies
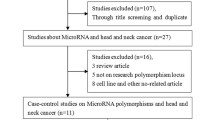
After excluding animal studies, reviews, repeated studies, conference studies and reading the full text, 7 original studies [32, 34, 36,37,38,39,40] were ultimately included. The article selection process is shown in Fig. 1.
Among the 7 enrolled studies, 7 studies were ultimately correlated with the C1772T polymorphism [32, 34, 36,37,38,39,40], and 6 studies were related to the G1790A polymorphism [30, 32, 37,38,39,40]. Overall, the meta-analysis included five articles conducted on oral cancer (OC), one article on glottic laryngeal cancer (GLC), and one on HNC. Of the 7 studies on the C1772T polymorphism, the genotype distributions in the controls in 4 articles complied with HWE, and 3 studies did not [32, 38, 40]. All studies related to the HIF-1α G1790A polymorphism showed that the genotype distribution of the control was in line with HWE. The main characteristics of the enrolled studies are shown in Table 1. The results of the researchers' scoring of the included studies according to the NOS scale are shown in Additional file 2, Table S1. The results of risk of bias assessment according to the OHAT risk of bias rating tool are shown in Additional file 2, Table S2.
Quantitative synthesis
The results of the meta-analysis, namely, the relationship between HIF-1α C1772T and G1790A polymorphisms and HNC, are shown in Table 2.
HIF-1α C1772T polymorphism analysis
For the HIF-1α C1772T polymorphism, a random-effects model was adopted due to the obvious heterogeneity among the studies. We evaluated the association of the C1772T polymorphism with HNC risk in all genetic models except allelic and recessive models. The overall results demonstrated that the C1772T polymorphism was significantly related to a higher HNC risk under the homozygous and recessive genetic models (OR = 2.27, 95% CI = 1.17–4.42 for the homozygous model; OR = 11.53, 95% CI = 1.11–120.4 for the recessive model, Fig. 2) but not under other genetic models (P > 0.05). In the subgroup analyses, we found that the C1772T polymorphism could significantly increase HNC risk in the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) genotyping method subgroup (OR = 2.27, 95% CI = 1.17–4.42 for the homozygous model; OR = 11.53, 95% CI = 1.11–120.4 for the recessive model). Moreover, a significant relationship was discovered between the C1772T polymorphism and an increased HNC risk for Caucasians (OR = 2.16, 95% CI = 1.09–4.27 for the homozygous model; OR = 2.28, 95% CI = 1.15–5.51 for the recessive model).
HIF-1α G1790A polymorphism analysis
For the HIF-1α G1790A polymorphism, we still applied a random effects model to count all the genetic models. We noticed a substantial relationship between the G1790A polymorphism and the increased risk of HNC for the homozygous and recessive genetic models (OR = 72.11, 95% CI = 2.08–2502.4 for the homozygous model; OR = 58.05, 95% CI = 1.70–1985.8 for the recessive model). In the stratified analyses, a substantial relationship was observed for the PCR-RFLP genotyping method subgroup (OR = 72.11, 95% CI = 2.08–2502.4 for the homozygous model; OR = 7.00, 95% CI = 1.18–41.68 for the dominant model; OR = 58.05, 95% CI = 1.70–1985.8 for the recessive model), population-based study subgroup (OR = 9.43, 95% CI = 1.20–73.9 for allelic model, Fig. 3; OR = 72.11, 95% CI = 2.08–2502.4 for the homozygous model; OR = 3.22, 95% CI = 1.28–8.08 for the heterozygous model; OR = 7.83, 95% CI = 1.48–41.37 for the dominant model; OR = 58.05, 95% CI = 1.70–1985.8 for the recessive model) and OC (P < 0.05 under all genetic models).
Sensitivity analysis and publication bias
After omitting one article at a time, no significant change was observed in the pooled ORs in the sensitivity analysis (Fig. 4, TT vs. CC of HIF-1α C1772T). Egger tests and Begg’s funnel plots were used to assess publication bias. The P value in the Egger test demonstrated statistical evidence for no substantial publication under all genetic models (P = 0.188 for T vs. C; P = 0.539 for TT vs. CC; P = 0.934 for TC vs. CC; P = 0.979 for TT + TC vs. CC; P = 0.329 for TT vs. TC + CC; P = 0.871 for A vs. G; P = 0.785 for AA vs. GG; P = 0.643 for AG vs. GG; P = 0.700 for AA + AG vs. GG; P = 0.606 for AA vs. AG + GG). In addition, the shape of Begg’s funnel plot appeared to be symmetric (Fig. 5, TT + TC vs. CC of HIF-1α C1772T), which indicated low risk of publication bias.
Discussion
HIF-1 acts as a vital component in the progression and metastasis of cancer by activating numerous genes associated with angiogenesis regulation, energy metabolism and cell survival [34, 35]. Moreover, high HIF-1α expression has been demonstrated in various tumours [22,23,24,25]. Certain polymorphisms in the HIF-1α gene have been linked to an individual's predisposition to cancers [46]. Among them, HIF-1α C1772T and G1790A polymorphisms were recently confirmed. The potential correlation between HIF-1α C1772T/G1790A polymorphisms and HNC susceptibility has been reported by some investigators, but the results were inconclusive [32, 34, 36,37,38,39,40]. This might be because of the limitations of these studies, such as ethnic differences, control source differences, small sample sizes and different methodologies. Meta-analysis, as a powerful tool, could bridge these difficulties and provide a more precise and reliable conclusion than a single article.
To the best of our knowledge, no studies have evaluated HIF-1αC1772T in the progression of HNC. In this meta-analysis, seven studies were ultimately enrolled for the C1772T polymorphism [32, 34, 36,37,38,39,40], and six studies were included for the G1790A polymorphism [32, 34, 37,38,39,40]. Overall, the results demonstrated that the HIF-1α C1772T polymorphism is an important factor in determining the increased risk of HNC (OR = 2.27, 95% CI = 1.17–4.42 for the homozygous model; OR = 11.53, 95% CI = 1.11–120.4 for the recessive model). In addition, a statistically significant correlation between the HIF-1α G1790A polymorphism and a higher risk of HNC was discovered for the homozygous and recessive genetic models (OR = 72.11, 95% CI = 2.08–2502.4 for the homozygous model; OR = 58.05, 95% CI = 1.70–1985.8 for the recessive model). These results indicated that these two polymorphisms play an important role in the progression and development of HNC.
In the stratification analysis of the C1772T polymorphism by the genotyping method, the relevance of the PCR-RFLP genotyping subgroup in the homozygous and recessive models was statistically significant. Regarding the stratification analysis by ethnicity, under homozygous and recessive genetic models, the association between the HIF-1α C1772T polymorphism and increased risk of HNC in the Caucasian population was very significant, but not in the Asian population, which demonstrated genetic diversity among different ethnic groups. This could be explained by the following reasons. First, different ethnic populations live a variety of lifestyles. Second, various environmental factors may be correlated with different ethnicities. Third, various ethnic populations carry different genetic traits.
In the subgroup analysis of the HIF-1α G1790A polymorphism by the genotyping method, we found that there was a statistically significant correlation between PCR-RFLP genotyping method subgroups in the homozygote, dominant and recessive genetic models. This might be because the relationship can be influenced by various genotyping methods, indicating that it is necessary to identify a genotyping method with high specificity and sensitivity to raise the reliability of results. In the subgroup analysis according to the source of controls, a statistically significant correlation was discovered in the PB subgroup but not in the HB subgroup. The reasons for the inconsistent results in HNC risk remain unknown. We supposed that certain selection bias could exist in the HB subgroup because patients without HNC were included, and they might be less representative of the general population than the populations in the PB subgroup. The location of HNC includes the oral cavity, pharynx, cheek and larynx, and different locations have different characteristics; thus, further subgroup analysis was carried out according to tumour types. Regarding the subgroup analysis by tumour type, the HIF-1α G1790A polymorphism was substantially related to a higher risk of OC in the five genetic models. This could be due to different tumour sites being exposed to different microenvironments. HIF-1α expression profiles could be regulated or influenced by the different microenvironments, and the same polymorphism might therefore play different roles in different sites [47].
However, there were some inevitable limitations in this meta-analysis. First, the size of the sample in some subgroups was small, and the results from certain subgroup analyses therefore did not have sufficient power to confirm the relationship. Second, there may have been publication bias because some qualified unpublished articles were not included in our study. Third, subgroup analyses by age, gender, alcohol, smoking or other variables were not performed because of information limitations. Therefore, it is necessary to study the role of HIF-1α C1772T and G1790A polymorphisms in HNC risk with more data and a larger sample size.
Despite these shortcomings, our study has several advantages. First, the latest data were contained in the analysis to evaluate the correlation between C1772T and G1790A polymorphisms in HIF-1α and HNC susceptibility. Second, no publication bias was observed, and evidence for the overall robustness of the results was provided by sensitivity analysis. Additionally, to our knowledge, this is the first meta-analysis to summarize the role of HIF-1α C1772T polymorphisms in HNC susceptibility. However, a previous meta-analysis [48] evaluated the relationship between the G1790A polymorphism and HNC. However, our research has advantages in the following aspects. Our study included five genetic models, and subgroup analyses by genotyping methods and cancer type were performed. Moreover, in this study, we conducted publication bias analysis and sensitivity analysis, which showed that our conclusion was reliable.
Conclusions
In conclusion, the HIF-1α C1772T and G1790A polymorphisms were significantly related to susceptibility to HNC. Moreover, we found for the first time that the C1772T polymorphism could statistically increase HNC risk among Caucasians. In addition, the HIF-1α G1790A polymorphism was strongly related to a higher risk of HNC, especially OC. However, further well-designed papers with larger sample sizes are needed to confirm our results.
Availability of data and materials
The current study was based on the results of relevant published studies.
Abbreviations
- HNC:
-
Head and neck cancer
- HIF-1:
-
Hypoxia-inducible factor-1
- PHD:
-
Prolyl hydroxylase
- VHL:
-
von Hippel-Lindau factor
- HWE:
-
Hardy-Weinberg equilibrium
- OC:
-
Oral cancer
- GLC:
-
Glottic laryngeal cancer
- SNPs:
-
Single nucleotide polymorphisms
- NOS:
-
Newcastle-Ottawa Scale
- OHAT:
-
The Office of Health Assessment and Translation
- HPV:
-
Human papillomavirus
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. https://doi.org/10.3322/caac.20107.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Saba NF, Goodman M, Ward K, Flowers C, Ramalingam S, Owonikoko T, et al. Gender and ethnic disparities in incidence and survival of squamous cell carcinoma of the oral tongue, base of tongue, and tonsils: a surveillance, epidemiology and end results program-based analysis. Oncology. 2011;81(1):12–20. https://doi.org/10.1159/000330807.
Bray F, Haugen M, Moger TA, Tretli S, Aalen OO, Grotmol T. Age-incidence curves of nasopharyngeal carcinoma worldwide: bimodality in low-risk populations and aetiologic implications. Cancer Epidemiol Biomark Prev. 2008;17(9):2356–65. https://doi.org/10.1158/1055-9965.EPI-08-0461.
Rettig EM, D'Souza G. Epidemiology of head and neck cancer. Surg Oncol Clin N Am. 2015;24(3):379–96. https://doi.org/10.1016/j.soc.2015.03.001.
Bover Manderski MT, Black K, Udasin IG, Giuliano AR, Steinberg MB, Ohman Strickland P, et al. Risk factors for head and neck cancer in the World Trade Center Health Program General Responder Cohort: results from a nested case-control study. Occup Environ Med. 2019;76(11):854–60. https://doi.org/10.1136/oemed-2019-105890.
Brunotto M, Zarate AM, Bono A, Barra JL, Berra S. Risk genes in head and neck cancer: a systematic review and meta-analysis of last 5 years. Oral Oncol. 2014;50(3):178–88. https://doi.org/10.1016/j.oraloncology.2013.12.007.
Marron M, Boffetta P, Zhang ZF, Zaridze D, Wunsch-Filho V, Winn DM, et al. Cessation of alcohol drinking, tobacco smoking and the reversal of head and neck cancer risk. Int J Epidemiol. 2010;39(1):182–96. https://doi.org/10.1093/ije/dyp291.
Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. https://doi.org/10.1038/nrc2982.
Kim L, King T, Agulnik M. Head and neck cancer: changing epidemiology and public health implications. Oncology (Williston Park). 2010;24(10):915–9 24.
Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99(10):777–89. https://doi.org/10.1093/jnci/djk179.
Sankaranarayanan R, Masuyer E, Swaminathan R, Ferlay J, Whelan S. Head and neck cancer: a global perspective on epidemiology and prognosis. Anticancer Res. 1998;18(6B):4779–86.
Batta N, Pandey M. Mutational spectrum of tobacco associated oral squamous carcinoma and its therapeutic significance. World J Surg Oncol. 2019;17(1):198. https://doi.org/10.1186/s12957-019-1741-2.
Berman TA, Schiller JT. Human papillomavirus in cervical cancer and oropharyngeal cancer: one cause, two diseases. Cancer. 2017;123(12):2219–29. https://doi.org/10.1002/cncr.30588.
Fakhry C, Blackford AL, Neuner G, Xiao W, Jiang B, Agrawal A, et al. Association of oral human papillomavirus DNA persistence with cancer progression after primary treatment for oral cavity and oropharyngeal squamous cell carcinoma. JAMA Oncol. 2019;5(7):985–92. https://doi.org/10.1001/jamaoncol.2019.0439.
Lacko M, Braakhuis BJ, Sturgis EM, Boedeker CC, Suarez C, Rinaldo A, et al. Genetic susceptibility to head and neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2014;89(1):38–48. https://doi.org/10.1016/j.ijrobp.2013.09.034.
Munshi T, Heckman CJ, Darlow S. Association between tobacco waterpipe smoking and head and neck conditions: a systematic review. J Am Dent Assoc. 2015;146(10):760–6. https://doi.org/10.1016/j.adaj.2015.04.014.
Hill RP, Marie-Egyptienne DT, Hedley DW. Cancer stem cells, hypoxia and metastasis. Semin Radiat Oncol. 2009;19(2):106–11. https://doi.org/10.1016/j.semradonc.2008.12.002.
Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–90. https://doi.org/10.1038/28867.
Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270(3):1230–7. https://doi.org/10.1074/jbc.270.3.1230.
Dery MA, Michaud MD, Richard DE. Hypoxia-inducible factor 1: regulation by hypoxic and non-hypoxic activators. Int J Biochem Cell Biol. 2005;37(3):535–40. https://doi.org/10.1016/j.biocel.2004.08.012.
Naidu R, Har YC, Taib NA. Associations between hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms and risk of developing breast cancer. Neoplasma. 2009;56(5):441–7. https://doi.org/10.4149/neo_2009_05_441.
Hsiao PC, Chen MK, Su SC, Ueng KC, Chen YC, Hsieh YH, et al. Hypoxia inducible factor-1alpha gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase susceptibility to hepatocellular carcinoma. J Surg Oncol. 2010;102(2):163–9. https://doi.org/10.1002/jso.21539.
Kim YH, Park IA, Park WY, Kim JW, Kim SC, Park NH, et al. Hypoxia-inducible factor 1alpha polymorphisms and early-stage cervical cancer. Int J Gynecol Cancer. 2011;21(1):2–7. https://doi.org/10.1097/IGC.0b013e318204f6e6.
Frank B, Hoffmeister M, Klopp N, Illig T, Chang-Claude J, Brenner H. Single nucleotide polymorphisms in Wnt signaling and cell death pathway genes and susceptibility to colorectal cancer. Carcinogenesis. 2010;31(8):1381–6. https://doi.org/10.1093/carcin/bgq082.
Zhou J, Huang S, Wang L, Yuan X, Dong Q, Zhang D, et al. Clinical and prognostic significance of HIF-1alpha overexpression in oral squamous cell carcinoma: a meta-analysis. World J Surg Oncol. 2017;15(1):104. https://doi.org/10.1186/s12957-017-1163-y.
Pacino GA, Salvatore C, Antonino M, Cristina DMM, Piero P, Giacomo S. Advanced olfactory neuroblastoma in a teenager: a clinical case and short review of literature. Childs Nerv Syst. 2020;36(3):485–9. https://doi.org/10.1007/s00381-020-04514-9.
Serra A, Caltabiano R, Giorlandino A, Musumeci A, Conti A, Zanghì G, et al. Nasal metastasis as the first manifestation of a metachronous bilateral renal cell carcinoma. Pathologica. 2017;109(4):421–5.
Baldewijns MM, van Vlodrop IJ, Vermeulen PB, Soetekouw PM, van Engeland M, de Bruine AP. VHL and HIF signalling in renal cell carcinogenesis. J Pathol. 2010;221(2):125–38. https://doi.org/10.1002/path.2689.
Min JH, Yang H, Ivan M, Gertler F, Kaelin WG Jr, Pavletich NP. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296(5574):1886–9. https://doi.org/10.1126/science.1073440.
Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–32. https://doi.org/10.1038/nrc1187.
Munoz-Guerra MF, Fernandez-Contreras ME, Moreno AL, Martin ID, Herraez B, Gamallo C. Polymorphisms in the hypoxia inducible factor 1-alpha and the impact on the prognosis of early stages of oral cancer. Ann Surg Oncol. 2009;16(8):2351–8. https://doi.org/10.1245/s10434-009-0503-8.
Fu XS, Choi E, Bubley GJ, Balk SP. Identification of hypoxia-inducible factor-1alpha (HIF-1alpha) polymorphism as a mutation in prostate cancer that prevents normoxia-induced degradation. Prostate. 2005;63(3):215–21. https://doi.org/10.1002/pros.20190.
Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T, et al. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis. 2003;24(11):1779–83. https://doi.org/10.1093/carcin/bgg132.
Smaldone MC, Maranchie JK. Clinical implications of hypoxia inducible factor in renal cell carcinoma. Urol Oncol. 2009;27(3):238–45. https://doi.org/10.1016/j.urolonc.2007.12.001.
Prasad J, Goswami B, Gowda SH, Gupta N, Kumar S, Agarwal K, et al. Does hypoxia-inducible factor -1 alpha (HIF-1alpha) C1772T polymorphism predict short-term prognosis in patients with oral squamous cell carcinoma (OSCC)? J Oral Pathol Med. 2018;47(7):660–4. https://doi.org/10.1111/jop.12718.
Shieh TM, Chang KW, Tu HF, Shih YH, Ko SY, Chen YC, et al. Association between the polymorphisms in exon 12 of hypoxia-inducible factor-1alpha and the clinicopathological features of oral squamous cell carcinoma. Oral Oncol. 2010;46(9):e47–53. https://doi.org/10.1016/j.oraloncology.2010.04.009.
Mera-Menendez F, Hinojar-Gutierrez A, Guijarro Rojas M, de Gregorio JG, Mera-Menendez E, Sanchez JJ, et al. Polymorphisms in HIF-1alpha affect presence of lymph node metastasis and can influence tumor size in squamous-cell carcinoma of the glottic larynx. Clin Transl Oncol. 2013;15(5):358–63. https://doi.org/10.1007/s12094-012-0930-z.
Chen MK, Chiou HL, Su SC, Chung TT, Tseng HC, Tsai HT, et al. The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol. 2009;45(12):e222–6. https://doi.org/10.1016/j.oraloncology.2009.07.015.
Alves LR, Fraga CAC, Oliveira MVM, Sousa AA, Jorge ASB, Marques-Silva L, et al. High HIF-1α expression genotypes increase odds ratio of oral cancer. Head neck oncology. 2012;4(5):87.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. https://doi.org/10.1136/bmj.b2700.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5. https://doi.org/10.1007/s10654-010-9491-z.
Rooney AA, Boyles AL, Wolfe MS, Bucher JR, Thayer KA. Systematic review and evidence integration for literature-based environmental health science assessments. Environ Health Perspect. 2014;122(7):711–8. https://doi.org/10.1289/ehp.1307972.
Handbook for conducting a literature-based health assessment using OHAT approach for systematic review and evidence integration. National Institute of Environmental Health Sciences; 2015. https://ntp.niehs.nih.gov/ntp/ohat/pubs/handbookjan2015_508.pdf.
OHAT Risk of Bias Rating Tool for Human and Animal Studies. NTP (National Toxicology Program); 2015. https://ntp.niehs.nih.gov/ntp/ohat/pubs/riskofbiastool_508.pdf.
Konac E, Onen HI, Metindir J, Alp E, Biri AA, Ekmekci A. An investigation of relationships between hypoxia-inducible factor-1 alpha gene polymorphisms and ovarian, cervical and endometrial cancers. Cancer Detect Prev. 2007;31(2):102–9. https://doi.org/10.1016/j.cdp.2007.01.001.
Saidak Z, Mentaverri R, Brown EM. The role of the calcium-sensing receptor in the development and progression of cancer. Endocr Rev. 2009;30(2):178–95. https://doi.org/10.1210/er.2008-0041.
Zhou Y, Lin L, Wang Y, Jin X, Zhao X, Liu D, et al. The association between hypoxia-inducible factor-1α gene G1790A polymorphism and cancer risk: a meta-analysis of 28 case-control studies. Cancer Cell Int. 2014;14(1):37. https://doi.org/10.1186/1475-2867-14-37.
Acknowledgements
We would like to acknowledge the Department of Surgical Oncology and General Surgery, The First Hospital of China Medical University and the Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumours (China Medical University), Ministry of Education, 155 North Nanjing Street, Heping District, Shenyang, 110001, China for providing technical support.
Funding
None
Author information
Authors and Affiliations
Contributions
WT is the first author of the manuscript and was mainly responsible for the literature search, data acquisition, data analysis, statistical analysis, manuscript preparation and manuscript editing. LL and BBT were involved in the literature search, data acquisition, data analysis and statistical analysis. ZZT proposed the research design and was responsible for manuscript editing and manuscript review. LRY was involved in the statistical analysis, manuscript preparation and manuscript editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
PRISMA 2009 Checklist
Additional file 2.
Table Supplementary 1. Quality assessment of included studies (Newcastle-Ottawa Scale). Table Supplementary 2. The risk of bias of included studies on the basis of the OHAT.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, T., Zhang, Zt., Li, L. et al. Correlation between hypoxia-inducible factor-1α C1772T/G1790A polymorphisms and head and neck cancer risk: a meta-analysis. World J Surg Onc 19, 210 (2021). https://doi.org/10.1186/s12957-021-02324-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-021-02324-0