Abstract
Background
Several studies have reported the Cyclooxygenase 2 (COX-2) rs689466 polymorphism as a susceptibility locus of colorectal cancer (CRC), but their findings are inconsistent. Thus, this meta-analysis was performed to more accurately identify the effects of this polymorphism on CRC risk.
Methods
Potential case-control studies on EMBASE, Google Scholar, Web of Science, Cochrane Library, and PubMed were searched. The strength of association was quantified by pooled odds ratio and 95% confidence interval. Totally 16 articles involving 8998 cases and 11,917 controls were included.
Results
None of the five tested genetic models revealed an association between rs689466 polymorphism and CRC risk. Stratified analysis by ethnicity uncovered a positive association between this polymorphism and higher CRC risk in Caucasians, but not in Asians. In addition, we found that high expression of COX-2 was associated with better overall survival for all CRC patients.
Conclusion
To sum up, the COX-2 rs689466 polymorphism may be related with susceptibility to CRC in Caucasians. This finding should be verified by larger-size studies with different ethnic groups.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC), the second largest cause of cancer-induced death in the world [1], was estimated to cause 135,430 new cases and 50,260 deaths in the USA in 2017 [2]. However, the cause of CRC is unknown yet. Risk of CRC is significantly associated with diet, cigarette smoking, drinking, and other factors [3, 4]. The development of CRC also involves genetic factors [5].
Cyclooxygenases (COXs), including two isoforms of COX-1 and COX-2 identified so far, are critical in transforming arachidonic acid to the precursor of prostaglandins—prostaglandin H2 [6]. COX-2 overexpression in CRC tissues was associated with worse overall survival of CRC [7]. Paeonol, a COX-2 inhibitor, constrains prostaglandin E2 (PGE2) generation and COX-2 expression, thereby preventing human CRC cells from tumors [8]. COX-2 downregulation considerably eliminates the development, motion, and invasion of colon cancer [9]. These observations suggest COX-2 is pivotal in CRC development.
COX-2 containing 10 exon counts is located in the chromosome 1q31.1. Rs689466 polymorphism is in the promoter zone of COX-2 gene. The association between rs689466 polymorphism and CRC risk has been explored extensively [10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25]. However, the findings of these studies were inconclusive and inconsistent, which may be attributed to the small sample sizes, clinical heterogeneity, and ethnic differences. To solve the inconsistence among these studies, we designed this meta-analysis to clarify the potential association between COX-2 gene rs689466 polymorphism and CRC risk.
Materials and methods
Literature search and inclusion criteria
Two reviewers systematically and independently searched EMBASE, Google Scholar, Web of Science, Cochrane Library, and PubMed to find potential studies without any restriction. The key words included “polymorphism”, “single nucleotide polymorphism” or “SNP”, “colorectal cancer”, “colorectal tumor” or “CRC”, and “Cyclooxygenase-2” or “COX-2”. References of identified studies were manually screened to search any omitted article.
The inclusion criteria were (1) case-control studies, (2) enough data for computation of pooled odds ratio (OR) with 95% confidence interval (CI), (3) evaluation of association between COX-2 rs689466 polymorphism and CRC risk, and (4) target at humans.
Prognosis analysis
OncoLnc (http://www.oncolnc.org/) website was used to evaluate prognostic value of the mRNA expression (high vs. low expression, separated by 50%) of COX-2 gene. We analyzed the overall survival (OS) of CRC patients by calculating Log rank p value and hazard ratio (HR) with 95% confidence intervals.
Expression analysis
We also performed the expression quantitative trait loci (eQTL) analysis using GTEx portal web site (http://www.gtexportal.org/home/) to predict potential associations between the SNPs and gene expression levels.
Data isolation and quality assessment
Based on the inclusion criteria, two reviewers independently extracted the data of interest, including ethnicity, sample sizes (cases, controls), cancer type, name of first author, publication year, and country of origin. If data were unavailable in an article, we contacted the authors for relevant data. If more than one ethnicity were involved in one article, we collected genotype data separately.
The quality of each included study was assessed using the Newcastle-Ottawa Scale (NOS) [26]. Generally, a score from 5 to 9 stars indicates high methodological quality while a score from 0 to 4 means slow quality. Disagreements between the two reviewers were solved by discussion or consultancy with a third reviewer.
Statistical analysis
Statistical analyses were carried out using Stata 11.0 (StataCorp, College Station, USA). Stratified analyses of ethnicity, source of control (SOC), Hardy–Weinberg equilibrium (HWE), and genotyping methods were also conducted. Regarding potential heterogeneity among studies, we defined significant heterogeneity at the levels p < 0.10 and I2 > 50%. A random-effect model was used in case of significant heterogeneity; otherwise, a fixed-effect model was used [27]. The effect on heterogeneity test and the stability of results were evaluated via sensitivity analysis by eliminating one study each time. HWE in the controls was examined via Pearson’s χ2 test. The positive findings were evaluated by calculating false-positive report probability (FPRP). An FPRP threshold of 0.2 and a prior probability of 0.1 were set to detect an OR for a correlation with the tested genotype. FPRP < 0.2 implied a possible relationship. Publication bias was tested by visually inspecting the symmetry of Begg’s funnel plot and assessing Egger’s test [28]. The Power and Sample Size Program software was used to calculate power and sample size. The following parameters are used: α, the type I error probability for a two-sided test; P0, the probability of exposure in controls; N is the number of case patients; m, the ratio of control to experimental subjects; Ψ, odd ratio of exposure in cases relative to controls. Statistical significance was set at p < 0.05.
Results
Characteristics of included articles
The initial search returned 161 articles. Then, 43 duplicated articles were excluded, and 75 articles were omitted after title and abstract examination. Of the remaining 43 articles, full-text review denied 27 articles. Finally, 16 studies with 8998 cases and 11917 controls were included [10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25]. The process of article selection is illustrated in Fig. 1. The characteristics of the included studies are listed in Table 1. Two ethnicities were involved, including Caucasians (12 studies) [12,13,14, 16,17,18,19,20,21, 23,24,25] and Asians (4 studies) [10, 11, 15, 22]. Two studies failed to conform to HWE [15, 22]. The NOS scores range from 5 to 7 stars, suggesting that the included studies are all of high quality.
Quantitative analysis
We evaluated the association between COX-2 gene expression and CRC prognosis using the OncoLnc website. Our data showed that high expression of COX-2 was associated with better OS for all CRC patients (HR, 0.66; 95% CI, 0.45–0.98; p = 0.0357, Fig. 2). We speculated that COX-2 may be a tumor suppressor gene.
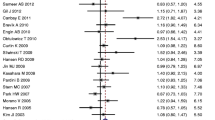
We also conducted a meta-analysis between an important single nucleotide polymorphism (SNP) of COX-2 gene and CRC risk, and found that the COX-2 rs689466 polymorphism is not associated with CRC risk (G vs. A: OR = 1.06 (95% CI 0.94–1.19), p = 0.363, Fig. 3; GG + AG vs. AA 1.08 (0.95–1.24), p = 0.237; GG vs. AG + AA 1.06 (0.84–1.32), p = 0.627; GG vs. AA 1.10 (0.84–1.44), p = 0.478; GA vs. AA 1.07 (0.95–1.21), p = 0.453; Table 2). Nevertheless, an association between CRC risk and COX-2 rs689466 polymorphism was obtained in Caucasians (G vs. A OR = 1.15 (95% CI 1.02–1.29), p < 0.05, Fig. 4) but not in Asians. Stratified analysis by HWE and genotyping methods revealed no association either in HWE-positive studies (GG + AG vs. AA OR, 1.12 (95% CI 0.99–1.27), p = 0.068, Table 3) or HWE-negative studies. Stratified analysis of SOC showed that rs689466 polymorphism was associated with increased risk for hospital-based population. In addition, we found that rs689466 polymorphism was associated with the expression of COX-2 gene according to the result on GTEx portal data (Supplementary Figure 1).
Sensitivity analysis and publication bias
Sensitivity analysis shows no single study largely influenced the pooled data, indicating that our results are statistically robust. Neither Begg’s funnel plot (GG + AG vs. AA, Fig. 5) nor Egger’s test finds any publication bias.
Power analysis and FPRP analyses
The power analysis revealed that this study had a power of 89.8% to detect the effect of rs689466 polymorphism on CRC susceptibility among Caucasians, assuming an OR of 1.15. Table 4 presented the FPRP values at different p level. The FPRPs for positive associations were much larger, indicating some possible bias due to limited sample size. Larger-sized studies are needed to confirm these findings.
Discussion
This meta-analysis showed no relationship between COX-2 rs689466 polymorphism and CRC risk in the whole populations. However, stratified analyses of ethnicity and SOC indicated that rs689466 polymorphism was associated with higher CRC risk among Caucasians and hospital-based populations.
CRC is the third leading cancer, but its occurrence and death rates vary largely among different areas in the world [30]. The lifetime risk of CRC development is ~ 5% in many regions [1]. About 45% of diagnosed CRC patients die, regardless of therapy [1]. The COX-2 mRNA levels are over-expressed in almost 80% of CRC patients [31]. COX-2 inhibitors are promising candidates for chemoprevention of CRC in clinic [32, 33]. The use of COX-2 inhibitor may help to improve the outcomes of stage III CRC patients [34]. Abovementioned data suggested that COX-2 may participate in the development of CRC. Rs689466 polymorphism is a pivotal SNP of COX-2 gene. The G allele rs689466 polymorphism was reported to transcriptionally activate COX-2 in colon cancer cells [29]. Thus, we assumed this SNP may be associated with the risk of CRC. Recently, a host of studies investigated the relationship between COX-2 rs689466 polymorphism and CRC risk [10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25]. A case-control study from the Netherlands observed no association between this SNP and CRC risk [16], which was consistent with the findings of some Caucasian studies. However, positive association was also obtained among other Caucasians [12, 18, 19]. Of the four studies from China, two studies found no association between COX-2 rs689466 polymorphism and CRC risk [11, 22], while the other two studies demonstrated a correlation between this polymorphism and lower CRC risk [10, 15]. To solve these inconsistencies, Wang et al. conducted a meta-analysis involving 5 studies (1854 cases and 2950 controls) and concluded COX-2 rs689466 polymorphism was not associated with CRC susceptibility [35]. Similarly, another meta-analysis also suggested that COX-2 rs689466 polymorphism was not associated with CRC risk in the overall population or in the stratified analyses of ethnicity, cancer location, SOC, or HWE [36]. We think the previous two meta-analyses have some limitations. Firstly, Wang et al. [35] omitted three studies meeting the inclusion criteria [10, 15, 16] and did not conduct stratified analyses of SOC or HWE. Secondly, Peng et al. omitted a study [36] and did not analyze the origin of heterogeneity. Therefore, their findings should be interpreted with caution. To date, several emerging studies have been reported since these meta-analyses. Consequently, it is necessary to conduct a comprehensive meta-analysis that included these new studies to determine whether the COX-2 rs689466 polymorphism was associated with CRC risk.
Herein, we included 16 studies with larger sample sizes (8998 cases and 11,917 controls) in this meta-analysis. Although our results suggested that COX-2 rs689466 polymorphism was not associated with a higher CRC risk in the overall population, subgroup analysis of ethnicity showed that COX-2 rs689466 polymorphism was associated with increased CRC risk in Caucasians, but not in Asians, suggesting different racial inheritance for Caucasians and Asians. The ethnic difference may be explained by the different allele frequency of this polymorphism. Asians have higher A allele frequency than Caucasians (European) (0.494 vs. 0.194). Another reason may be the differences among ethnic groups in sample sizes. In this meta-analysis, the sample sizes of Caucasians and Asians were different. Furthermore, varied living environments and diets may be important factors. In addition, clinical heterogeneity may also contribute to contradictory findings. As reported, G allele of rs689466 polymorphism could transcriptionally activate COX-2 [29]. We supposed this SNP may regulate COX-2 gene transcription and protein translation, thereby involving in the development of CRC. Additionally, we found that high expression of COX-2 was associated with better OS for CRC patients. To be frank, the development of CRC is attributed to multiple genes, genetic background, and environmental factors. Further studies that considered environmental and genetic factors were urgently needed.
This meta-analysis has several limitations. Firstly, subgroup analyses of age, sex, smoking, drinking status, or tumor size were not conducted due to data shortage. Secondly, estimates of confounding factors were unadjusted, which might affect the final results. Thirdly, possible gene-gene and gene-environment interactions were ignored because of data insufficiency. Fourthly, only Asians and Caucasians were included, and the findings may be inapplicable to other racial groups. Fifthly, we did not explore the association between rs689466 polymorphism and COX-2 protein. Lastly, only four Asian studies were included in this meta-analysis.
In conclusion, this meta-analysis confirms an association between COX-2 gene rs689466 polymorphism and increased CRC risk among Caucasians. Nevertheless, this finding should be validated by further studies in other ethnicities.
Availability of data and materials
The relevant data could be available when the corresponding author was contacted.
Abbreviations
- COX-2:
-
Cyclooxygenase-2
- CRC:
-
Colorectal cancer
- COXs:
-
Cyclooxygenases
- PGE2:
-
Prostaglandin E2
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- OS:
-
Overall survival
- NOS:
-
Newcastle-Ottawa
- SOC:
-
Scale source of control
- HWE:
-
Hardy–Weinberg equilibrium
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
Sandhu MS, Dunger DB, Giovannucci EL. Insulin, insulin-like growth factor-I (IGF-I), IGF binding proteins, their biologic interactions, and colorectal cancer. J Natl Cancer Inst. 2002;94(13):972–80.
Giovannucci E. Insulin, insulin-like growth factors and colon cancer: a review of the evidence. J Nutr. 2001;131(11 Suppl):3109S–20S.
Valle L. Genetic predisposition to colorectal cancer: where we stand and future perspectives. World J Gastroenterol. 2014;20(29):9828–49.
DeWitt DL. Prostaglandin endoperoxide synthase: regulation of enzyme expression. Biochim Biophys Acta. 1991;1083(2):121–34.
Peng L, Zhou Y, Wang Y, Mou H, Zhao Q. Prognostic significance of COX-2 immunohistochemical expression in colorectal cancer: a meta-analysis of the literature. PLoS One. 2013;8(3):e58891.
Li M, Tan SY, Wang XF. Paeonol exerts an anticancer effect on human colorectal cancer cells through inhibition of PGE(2) synthesis and COX-2 expression. Oncol Rep. 2014;32(6):2845–53.
Che XH, Chen CL, Ye XL, Weng GB, Guo XZ, Yu WY, et al. Dual inhibition of COX-2/5-LOX blocks colon cancer proliferation, migration and invasion in vitro. Oncol Rep. 2016;35(3):1680–8.
Zhang Y, Liu C, Peng H, Zhang J, Cai X, Zhang Q. Promoter polymorphisms of cyclooxygenase-2 gene and the susceptibility to colorectal cancer. World Chin J Digestol. 2012;12(17):1579–84.
Ruan Y, Sun J, Wu F, Jiang S. Relationship between COX-2 gene polymorphism and the risk of colorectal cancer. Int J Digest Dis. 2013;11(04):260–3 72.
Vogel LK, Saebo M, Hoyer H, Kopp TI, Vogel U, Godiksen S, et al. Intestinal PTGS2 mRNA levels, PTGS2 gene polymorphisms, and colorectal carcinogenesis. PLoS One. 2014;9(8):e105254.
Tomitao MTP, Nahas SC, Kubrusly MS, Furuya TK, Diniz MA, Marie SKN, et al. Cyclooxygenase-2 gene polymorphisms and susceptibility to colorectal cancer in a Brazilian population. J Gastrointest Oncol. 2017;8(4):629–35.
Thompson CL, Plummer SJ, Merkulova A, Cheng I, Tucker TC, Casey G, et al. No association between cyclooxygenase-2 and uridine diphosphate glucuronosyltransferase 1A6 genetic polymorphisms and colon cancer risk. World J Gastroenterol. 2009;15(18):2240–4.
Tan W, Wu J, Zhang X, Guo Y, Liu J, Sun T, et al. Associations of functional polymorphisms in cyclooxygenase-2 and platelet 12-lipoxygenase with risk of occurrence and advanced disease status of colorectal cancer. Carcinogenesis. 2007;28(6):1197–201.
Siezen CL, Tijhuis MJ, Kram NR, van Soest EM, de Jong DJ, Fodde R, et al. Protective effect of nonsteroidal anti-inflammatory drugs on colorectal adenomas is modified by a polymorphism in peroxisome proliferator-activated receptor delta. Pharmacogenet Genomics. 2006;16(1):43–50.
Siezen CL, Bueno-de-Mesquita HB, Peeters PH, Kram NR, van Doeselaar M, van Kranen HJ. Polymorphisms in the genes involved in the arachidonic acid-pathway, fish consumption and the risk of colorectal cancer. Int J Cancer. 2006;119(2):297–303.
Shomaf M, Yousef AL, Ababna N, Bobali Y. Cyclooxygenase-2 (COX2) gene polymorphisms and the risk of sporadic colorectal cancer and polyps among Jordanian population. Turk J Gastroenterol. 2015;26(2):154–8.
Pereira C, Queiros S, Galaghar A, Sousa H, Pimentel-Nunes P, Brandao C, et al. Genetic variability in key genes in prostaglandin E2 pathway (COX-2, HPGD, ABCC4 and SLCO2A1) and their involvement in colorectal cancer development. PLoS One. 2014;9(4):e92000.
Pereira C, Pimentel-Nunes P, Brandao C, Moreira-Dias L, Medeiros R, Dinis-Ribeiro M. COX-2 polymorphisms and colorectal cancer risk: a strategy for chemoprevention. Eur J Gastroenterol Hepatol. 2010;22(5):607–13.
Makar KW, Poole EM, Resler AJ, Seufert B, Curtin K, Kleinstein SE, et al. COX-1 (PTGS1) and COX-2 (PTGS2) polymorphisms, NSAID interactions, and risk of colon and rectal cancers in two independent populations. Cancer Causes Control. 2013;24(12):2059–75.
Li S, Zhao X, Wu Z, Li Y, Zhu L, Cui B, et al. Polymorphisms in arachidonic acid metabolism-related genes and the risk and prognosis of colorectal cancer. Familial Cancer. 2013;12(4):755–65.
Hoff JH, te Morsche RH, Roelofs HM, van der Logt EM, Nagengast FM, Peters WH. COX-2 polymorphisms -765G-->C and -1195A-->G and colorectal cancer risk. World J Gastroenterol. 2009;15(36):4561–5.
Andersen V, Ostergaard M, Christensen J, Overvad K, Tjonneland A, Vogel U. Polymorphisms in the xenobiotic transporter multidrug resistance 1 (MDR1) and interaction with meat intake in relation to risk of colorectal cancer in a Danish prospective case-cohort study. BMC Cancer. 2009;9:407.
Andersen V, Holst R, Kopp TI, Tjonneland A, Vogel U. Interactions between diet, lifestyle and IL10, IL1B, and PTGS2/COX-2 gene polymorphisms in relation to risk of colorectal cancer in a prospective Danish case-cohort study. PLoS One. 2013;8(10):e78366.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Wassen L, Jertborn M. Influence of exogenous reproductive hormones on specific antibody production in genital secretions after vaginal vaccination with recombinant cholera toxin B subunit in humans. Clin Vaccine Immunol. 2006;13(2):202–7.
Pereira C, Sousa H, Silva J, Brandao C, Elgueta-Karstegl C, Farrell PJ, et al. The -1195G allele increases the transcriptional activity of cyclooxygenase-2 gene (COX-2) in colon cancer cell lines. Mol Carcinog. 2014;53(Suppl 1):E92–5.
Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, Young GP, et al. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64(10):1637–49.
Roelofs HM, Te Morsche RH, van Heumen BW, Nagengast FM, Peters WH. Over-expression of COX-2 mRNA in colorectal cancer. BMC Gastroenterol. 2014;14:1.
Kraus S, Naumov I, Arber N. COX-2 active agents in the chemoprevention of colorectal cancer. Recent Results Cancer Res. 2013;191:95–103.
Kim SH, Margalit O, Katoh H, Wang D, Wu H, Xia D, et al. CG100649, a novel COX-2 inhibitor, inhibits colorectal adenoma and carcinoma growth in mouse models. Investig New Drugs. 2014;32(6):1105–12.
Ng K, Meyerhardt JA, Chan AT, Sato K, Chan JA, Niedzwiecki D, et al. Aspirin and COX-2 inhibitor use in patients with stage III colon cancer. J Natl Cancer Inst. 2015;107(1):345.
Wang J, Guo X, Zhang J, Song J, Ji M, Yu S, et al. Cyclooxygenase-2 polymorphisms and susceptibility to colorectal cancer: a meta-analysis. Yonsei Med J. 2013;54(6):1353–61.
Peng Q, Yang S, Lao X, Tang W, Chen Z, Lai H, et al. Meta-analysis of the association between COX-2 polymorphisms and risk of colorectal cancer based on case-control studies. PLoS One. 2014;9(4):e94790.
Acknowledgements
None.
Funding
None
Author information
Authors and Affiliations
Contributions
Conceptualization, Chen Chen and Mohammad Amzad Ali; Methodology, Yong-Chen Zhang; Validation, Yong-Chen Zhang and Hui Zhao; Formal analysis, Yong-Chen Zhang and Hui Zhao; Investigation, Yong-Chen Zhang and Hui Zhao; Resources, Chen Chen and Yong-Chen Zhang; Data curation, Hui Zhao and Mohammad Amzad Ali; Writing-original draft preparation, Chen Chen and Mohammad Amzad Ali; Writing-review and editing, Yong-Chen Zhang, Chen Chen, and Mohammad Amzad Ali. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not required since previously published data were analyzed.
Consent for publication
No patient was recruited to the present study.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
The association between rs689466 polymorphism and the expression of COX-2 gene.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, YC., Zhao, H., Chen, C. et al. COX-2 gene rs689466 polymorphism is associated with increased risk of colorectal cancer among Caucasians: a meta-analysis. World J Surg Onc 18, 192 (2020). https://doi.org/10.1186/s12957-020-01957-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12957-020-01957-x