Abstract
Background
Dl-3-n-butylphthalide (NBP) is an important medial therapy for acute ischemic stroke in China. Recent studied have revealed that NBP not only rescued the loss of dopaminergic neurons in cellular and animal models of Parkinson's disease (PD), but also could improve motor symptoms in PD patients. However, the protective mechanism is not fully understood. P53 is a multifunctional protein implicated in numerous cellular processes, including apoptosis, DNA repair, mitochondrial functions, redox homeostasis, autophagy and protein aggregations. In PD, p53 integrated with various neurodegeneration-related signals inducing neuronal loss, indicating the suppression of P53 might be a promising target for PD treatment. Therefore, the purpose of the current study was to systemically screen new therapeutic targets of NBP in PD.
Method
In our study, we constructed mpp + induced N2A cells to investigate the benefit effect of NBP in PD. MTT assay was performed to evaluate the cell viability; TMT-based LC–MS/MS was applied to determine the different expressed proteins (DEPs) of NBP pretreatment; online bioinformatics databases such as DAVID, STRING, and KEGG was used to construe the proteomic data. After further analyzed and visualized the protein–protein interactions (PPI) by Cytoscape, DEPs were verified by western blot.
Result
A total of 5828 proteins were quantified in the comparative proteomics experiments and 417 proteins were considered as DEPs (fold change > 1.5 and p < 0.05). Among the 417 DEPs, 140 were upregulated and 277 were downregulated in mpp + -induced N2A cells with NBP pretreatment. KEGG pathway analysis indicated that lysosome, phagosome, apoptosis, endocytosis and ferroptosis are the mainly enriched pathways. By using MCL clustering in PPI analysis, 48 clusters were generated and the subsequent KEGG analysis of the top 3 clusters revealed that P53 signaling pathway was recognized as the dominant pathway for NBP treatment.
Conclusion
NBP significantly relived mpp + -induced cell toxicity. The neuroprotective role of NBP was implicated with P53 signaling pathway in some extent. These findings will reinforce the understanding of the mechanism of NBP in PD and identify novel therapeutic targets.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is a multi-factorial age-related disorder which is characterized by irreversible impairment of normal movement coordination. Its cardinal motor impairment features are tremor, gait rigidity, bradykinesia and hypokinesia [1, 2]. The hallmark of PD pathology is the formation of Lewy bodies (LBs), which is consisted of misfolded and fibrillary forms of α-synuclein (α-syn) in surviving neurons [3]. At present, PD affect almost 1–2% of the world population, whereas the estimated prevalence rate would be double in 2040 [4,5,6]. Based on previous studies, the complicated mechanisms for PD development were ascribed to mitochondrial dysfunction, oxidative stress, apoptosis and neuroinflammation [3, 7, 8]. To date, many efforts have been made to explore the potential mechanism to counteract PD, however, it is still lack efficient therapy.
DL-3-n-butylphthalide (NBP), the first class I novel drug, which has been approved for the treatment of acute ischemia stroke in China since 2002 [9,10,11]. On account of the neuroprotective properties of NBP by eliminating free radicals, restoring mitochondrial function, reducing neuroinflammation and alleviating neuronal apoptosis, its therapeutic spectrum has expanded to various neurodegenerative diseases, such as Alzheimer disease (AD), Amyotrophic Laternal Sclerosis (ALS) and PD [12,13,14,15,16]. To date, many studies have clarified the benefit role of NBP in PD both in vitro and in vivo. Wang et al., demonstrated that NBP could rescue dopaminergic neurons by restoring mitochondrial function and alleviating NLRP3-mediated neuroinflammation [17]. It revealed NBP protected dopamine neurons by preventing the generation of ROS as well [18]. A preclinical study suggested the favorable effect of NBP in improving bradykinesia plus rigidity through UPDRS III motor evaluation [19]. However, there is still lack a systemic analysis to provide an insight into the protein profiling and functional pathways after NBP treatment in PD models.
Proteins are the crucial agent to execute various cell functions encoded by different genome, whereas the generation of high-quality protein expression profiling is much lagged behind RNA/DNA expression profiling [20, 21]. Traditionally, proteomic studies are engaged to investigate a large-scale of protein expressions, in order to provide new insight of the protein interactions, cellular functions and biological framework [22, 23]. Mass spectrometry (MS)-based quantitative proteomic studies using isobaric tags (eg., tandem mass tags, TMT; isobaric tags for absolute and relative quantification, ITRAQ) are well-established to identify differential expressed peptides with few missing values and precise quantification [2, 24, 25]. In addition, the quantified peptides are matched or identified using automated database searching (Uniprot is selected in our research) [26]. Biological function analysis is conducted through an online tool (Database for Annotation, Visualization, and Integrated Discovery, DAVID) and protein interactions are assayed through The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) [27]. Therefore, the reliable and deep-investigated protein data is well for discovering new therapeutic targets and the implicated biological pathways.
In the present study, we employed TMT-labeled global quantitative proteomic analysis to explore the DEPs in mpp + -induced N2A cells with or without NBP pretreatment. The biological functions and entirely canonical pathways of DEPs were conducted by GO, KEGG and protein–protein interaction (PPI) networks. As a result, we leverage the data to improve the understand of the neuroprotective role of NBP and its potential therapeutic mechanisms.
Materials and methods
Cell culture
The N2A cells which donated by Ji Jianguo lab from Peking University, were cultured in DMEM (Hyclone, USA)) containing 5% FBS (Hyclone, USA). The cells were settled in a 37 °C incubator with a humidified 95% air and 5% CO2. The cells were seeded in the 12-well microplates at a density of 1 × 104 cells/ml. We changed the culture medium every 1–2 days. The experimental groups were arranged as follows, control group (without NBP and MPP + treatment); MPP + -treated group (500 μM MPP + treated for 24 h); (3) NBP + MPP + treated group (5 μM NBP treated for 27 h and 500 μM MPP + for 24 h).
MTT assays to evaluate cell viability
The N2A cells were seeded in 96-well plates at a density of 1000 cells/well. Until the cells reached 50–60% confluence, different treatments were performed to the cells. Firstly, cells were maintained with 0, 40, 100, 250, 500 or 1000 μM MPP + for 24 h to detect the optimal concentration of MPP. Next, we selected different concentration of NBP (0, 0.5uM, 1uM, 2uM, 5uM and 10uM) for N2A cells treatment alone for 24 h to evaluate the cell toxicity of NBP. Last but not the least, cells were pretreated with varies concentration of NBP (0, 0.5uM, 1uM, 2uM, 5uM and 10uM) for 3 h and incubated with 500 μM MPP + for anther 24 h. MTT assay was carried out according to the manufacturer’s introduction (Beyotime Biotechnology, Shanghai, China). Briefly, MTT (5 mg/ml) solvent was added into each well (10 µl) and incubated at 37◦C for 4 h. After discarded the supernatant, 150 μL of DMSO was added into each well to dissolve the resulting MTT formazan. The absorbance at 570 nm was read by a microtiter plate reader (Multiskan FC, Thermo, USA).
Protein extraction and preparation
The N2A cells with different treatments were harvested and recovered by centrifugation (1000xg for 5 min at 4 °C). Four volumes of pre-chilled acetone were added into protein extractions overnight at − 20◦C to obtain precipitation. The protein samples were stored at − 80◦C for further analysis.
Trypsin/LysC Protein Digestion
60 μL 8 M urea was added to the precipitated proteins. The constructed protein suspensions were further broken using a Bioruptor Sonication Device. After determining the concentration of each protein sample using BCA assay, 60 ug protein sample was alkylated by incubation with 3 μL of 100 mM dithiothreitol (DTT; Sigma Aldrich) and 3 μL of 200 mM iodoacetamide (IAA; Sigma Aldrich) for 30 min at room temperature. Next, samples were digested into peptides by using lys-C (1:100 dilution) and trypsin (1:50 dilution) at 37◦C overnight.
Peptides cleanup
The digested peptides were acidified with Trifluoroacetic Acid (TFA) to a final concentration of 0.5% (pH was around 2–3). After that, samples were desalted sequentially for 1 min in small 1 ml C18 Sep-Pak columns (3 M EmporeTM, CA, USA) with acetonitrile (ACN), 0.1%TFA/70%ACN and 0.1% TFA followed by conditioning with methanol for 1 min. Finally, the desalted peptide mixture was dried by vacuum centrifugation via speedvac.
TMT-labeling
The peptides were resuspended with 100 mM tetraethylammonium bicarbonate (TEAB) and the concentration of peptides was determined using the Quantitative Colorimetric Peptide Assay (Cat. No. 23275, Thermo, USA). The TMT labeling reagent (0.8 mg) was dissolved in 41 μL of ACN. 12 μg peptides of each sample was labeled with 9 μL of the different TMT regent at room temperature for 60 min followed by quenching with eight microliter of 5% hydroxylamine for 20 min. In our study, we conducted three independent experiments, samples were labeled with TMT with reporter ions at m/z = 126, 128 (experiment 1); 127, 129 (experiment 2) and 127, 128 (experiment 3). In each experiment, the labeled channels were combined and dried down via speedvac.
Peptide fractionation
The combined TMT-labeled samples were dissolved in 1% formic acid and desalted in C18 Sep-Pak columns. Gradient elution was performed with 0.1% ammonium hydroxide (pH 10) (reagent A) and ACN (reagent B). The gradient conditions for the fractionation were 90% A/10% B, 87.5% A/12.5% B, 85%A/15% B, 82.5% A/17.5% B, 80% A/20% B, 77.5% A/22.5% B, 75% A/25% B, and 50% A/50% B. Therefore, peptides were eluted with the above solvents into seven fractions and dried via speedvac.
LC–MS/MS and data process
Samples were run on a Orbitrap Fusion Lumos mass spectrometer (Thermo, USA). a full MS survey scan (300–1500 m/z) was acquired at a resolution of 120,000 (at 200 m/z). The settings of the automatic gain control (AGC) target for MS1, maximum injection time, and a radio frequency (RF) lens were set as 1 × 106, 100 ms, and 30% respectively. The abundant ions with a charge state ≥ 2 were isolated in a 3 s cycle time. High-energy collision dissociation (HCD) MS/MS scans were set as follows: 37% collision energy, a mass resolution of 50,000, normalized AGC target at 1 × 105, isolation width of 1.2 m/z, dynamic exclusion at 30 s, and 10 parts per million (ppm) mass window.
The raw data was analyzed by using the SEQUEST algorithm implemented in the search engines: Proteome Discoverer (Version 2.2, Thermo Fisher Scientific) and searched in UniProt Database against the mus musculus FASTA files (August, 2013). The Sequest search parameters were include: trypsin digestion with two missed cleavages allowed; fixed modification, carbamidomethyl of cysteine; variable modification, oxidation of methionine, 10 ppm and 0.5 Da for MS tolerance; and a false discovery rate (FDR) < 1%. Protein identification required at least one unique peptide per protein group. Protein quantification was accomplished by using the quantification of TMT reporter ions.
GO and KEGG analysis
Firstly, the ratio of NBP/MPP + proteins was normalized to rectify the unequal protein content. DEPs were analyzed using a two-tailed t-test after log2 transformation. The p-values were adjusted using “BH method” (PD patients versus health donors). The cutoff of p < 0.05 from three replicates and Fold Change > 1.5 was used to define DEPs. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses for DEPs were constituted by DAVID (https://david.ncifcrf.gov, version 6.8).
Protein–Protein Interaction (PPI) Analysis
The PPI networks were explored using STRING v.11.5 (https://cn.string-db.org). Cytoscape v3.9.1 software (Cytoscape Consortium, San Diego, USA) was used to visualize the network and CytoHubba (one of the add-on APP of Cytoscape) was selected to explore the hub proteins by MCC method. In addition, MCL clusting with at least 3 inflation parameters was applied to find out the top 3 clusters. These protein clusters were further uploaded to cytoscape to visualize the complex networks.
Western blot
The total proteins from N2A cells were lysing with 1% SDS. Subsequently, the extracts were separated using SDS-PAGE on 10% gel and electro-transferred onto PVDF membranes (Bio-Rad, USA), then blocked with 5% nonfat milk at room temperature. The blots were incubated with the primary antibodies P53 (1:1000, abcam, ab246550) and Bax (1:1000, abcam, ab216494) overnight at 4 ℃. On the second day, the blots were incubated with secondary antibodies (4050–05 or 1031–05, Southern Biotech) for 1 h at room temperature, and visualized on amersham Imager 600. Data was normalized to β-actin and quantified using Image J software (NIH, Bethesda).
Statistical analysis
The proteomics statistical analyses were performed using R (v3.6.3) in the RStudio environment (v1.0.143). The WB statistical analysis were performed using SPSS 26.0 statistical software. Protein expression levels were clustered according to the protein Z-score before visualization using a heatmap. All data were presented as mean ± SEM, and analyzed by one-way analysis of variance (ANOVA) followed by Tukey-HSD test for intergroup differences. A value of P < 0.05 was considered statistical significance.
Result
The protective role of NBP in reducing mpp + -induced cytotoxicity
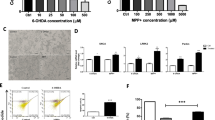
The viability of N2A cells in different treatment groups was first determined using MTT assays (Fig. 1). As indicated in Fig. 1A, the different concentration of NBP treatment alone had no significant effect on N2A cells (P > 0.05). In contrast, 24 h mpp + treatment induced irreversible cytotoxicity in N2A cells in a dose-dependent manner (Fig. 1B). Our result showed 500 uM mpp + resulted in almost 50% cell death compared with control group (58.1% ± 0.04 vs 98.6% ± 0.02, P = 0.03). Though 1000 uM mpp + treatment also decreased cell survival rate to 56.1% ± 0.03, the difference compared with 500 uM mpp + treatment group was not significant. The protective effect of NBP pretreatment on 500 uM mpp + -induced N2A cells was also detected and the result displayed in Fig. 1C showed that 0.1, 0.5, 1, 5, 10 uM NBP significantly increased survival rate after mpp + treatment, especially at 5 uM (77.6% ± 0.06, P = 0.01).
Effect of NBP on MPP + -induced cytotoxicity of N2A cells. (A) Cells were stimulated with NBP at a concentration of 0, 0.1, 0.5, 1, 5 or 10 μM for 24 h, and the cell viability was determined by MTT assay. (B) Different concentrations of MPP+ (0, 40, 100, 250, 500 and 1000 μM) was added to co-cultured with N2A cells for 24 h, and the cell viability was assayed with MTT. (C) Cells were pretreated with different concentrations of NBP (0, 0.1, 0.5, 1, 5 or 10 μM) for 24 h, and then these cells were subjected to MPP+ (500 μM) insults
The protein profile of mpp + -induced N2A Cells after NBP treatment
The workflow of the present quantitive proteomic analysis was demonstrated in Fig. 2A. We labeled mpp + -treated samples and NBP pretreated samples with TMT m/z = 126 N, 126C in three independent experiments. In experiment 1, 7273 proteins were identified, of which 6970 (95.83%) were quantified. In experiment 2 and 3, 6955 and 7483 proteins were identified, while 6657 (95.72%) and 7167 (95.79%) proteins were quantified, respectively. In conclusion, a total of 5828 proteins were quantified in the three replicates (Fig. 2B). Using log|fold-change|> 0.58 and P < 0.05 as the cutoff, 417 proteins were determined as DEPs. Among the 417 DEPs, 140 were upregulated and 277 were downregulated in NBP pretreatment group compared to control group (Fig. 2B). The complete list of DEPs is revealed in Table 1. The distribution of the log2 expression ratios (NBP + mpp + /mpp +) of the quantified proteins was roughly normal (Fig. 2C). Volcano plot showed DEPs between mpp + -treated group and NBP pretreatment group (Fig. 2D). In addition, the DEPs were also visualized by a heatmap in Fig. 2E.
The quantitative proteomics analysis of the protective effect of Dl-3-n-Butylphthalide (NBP). (A) The proteomics workflow for the current study. (B) In N2a cells which were stimulated with mpp+ (500 μM) in the absence or presence of NBP (40 μM) for 24 h, a total of 5829 proteins were identified in three replicates. Using the cut off of |fold-change|> 1.5 and p value < 0.05 to determine DEPs, we qualified 417 proteins and determined 277 increased and 140 decreased. (C) Overall distribution of the ratios of 5829 proteins in the quantitative proteomics with three replicates. (D) Volcano plot of mpp + -treated group and NBP group. The volcanic map was drawn using two factors, the fold change (Log2) between the two groups of samples and the p value (− Log10) obtained by the t-test, to show the significant difference in the data of the two groups of samples. The red dots in the figure are proteins that are significantly differently up-regulated, and the green dots are proteins that down-regulated. Gray dots indicate non-significantly DEPs. (E) Heat map of DEPs ratio, each row represents a protein, each column represents the ratio of a sample to a reference sample, and the ratio takes the value of log2
Bioinformatics analysis of the identified DEPs
GO and KEGG analysis of DEPs
To better understand the cellular location, functions and the involved biological pathways of the DEPs, Gene Oncology (GO) analysis were conducted (Fig. 3). According to our analysis, the DEPs were found to be mainly localized to the cytoplasm, cytosol, mitochondrion, actin cytoskeleton and actin filamentactin. The GO-biological processes (BP) analysis revealed that DEPs were related to actin cytoskeleton organization, actin filament organization, actin filament bundle assembly, positive regulation of tumor necrosis factor production, regulation of cell shape and so forth. Regarding to GO-molecular functions (MF), DEPs were biased towards actin binding, protein binding, actin filament binding, identical protein binding, SH3 domain binding and integrin binding. KEGG pathway enrichment analysis was also performed to understand the associated pathways of the DEPs (Fig. 4). The result indicated that lysosome, phagosome, apoptosis, endocytosis, cholesterol metabolism and ferroptosis were the mainly enriched pathways.
Ontological analysis of the DEPs. Classification of the DEPs based on cellular component (CC), biological process (BP) and molecular function (MF). We listed top 10 enriched GO pathways. The ordinate represents number of differential proteins contained in each classification and the abscissa represents the significantly enriched functional classification and pathway
Mapping STRING Protein–Protein Interaction (PPI) Network
The DEPs were uploaded to the STRING online tool to identify the protein–protein interactions and the biological significance of the identified proteins. Cytoscape was used to visualized the network. The result demonstrated that the DEPs constructed a complicated interaction network with 412 nodes and 1999 edges. The clustering coefficient was 0.388. As the expected edges number was 902, our result displayed a much higher value than the expected edges. The confident of PPI enrichment was p < 1 × 10−16. Likewise, the tight PPI network demonstrated in Fig. 5A also suggested the strong interaction between DEPs. Furthermore, screening the DEPs by CytoHubba App using the Maximum Clique Centrality (MCC) method in Cytoscape software, the top 10 hub proteins were recognized, namely, Integrin β2 (ITGB2), Ras-related C3 botulinum toxin substrate 2 (Rac2), Receptor-type tyrosine-protein phosphatase C (Ptprc), Nck-associated protein 1-like (Nckap 11), Hematopoietic lineage cell-specific protein (Hcls1), Isoform 2 of Tyrosine-protein phosphatase non-receptor type 6 (Ptpn6), Coronin-1A (Coro1a), Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 (Inpp5d), Neutrophil cytosol factor 4 (Ncf4) and Fermitin family homolog 3 (Fermt3)(Fig. 5B).
(A) Protein–protein interaction network consisting of 417 DEPs (B). The color saturation of the edges represents the confidence score of the association between modulated proteins. The top 10 proteins with the highest degree of PPI network connectivity were identified by the MMC method using CytoHubba. (C) The differentially regulated cluster I protein network is visualized using Cytoscape 3.8.0. The blue nodes indicate the significantly downregulated proteins and red nodes represent the significantly upregulated proteins (p < 0.05)
Cluster Analysis of DEPs
By using MCL clustering, 48 clusters were generated which contained more than 3 genes. We analyzed the top 3 generated clusters to recognize the therapeutic target and dominant pathway for NBP treatment. In Fig. 5C, there were 25 Proteins in cluster I namely, CD166 antigen (Alcam), C3a anaphylatoxin chemotactic receptor (C3ar1), Macrosialin (Cd68), SLAM family member 5 (Cd84), Macrophage colony-stimulating factor 1 receptor (Csf1r), Cathepsin S (Ctss), Vascular endothelial growth factor receptor 1 (Flt1), FYN-binding protein 1 (Fyb), H-2 class I histocompatibility antigen, D-B alpha chain(H2-D1), H-2 class I histocompatibility antigen, K-K alpha chain (H2-K1), Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 (Inpp5d), Lymphocyte antigen 86 (Ly86), T-lymphocyte surface antigen Ly-9 (Ly9), Lysozyme C-2 (Lyz2), COUP transcription factor 2 (Nr2f2), Isoform 2 of Tyrosine-protein phosphatase non-receptor type 6 (Ptpn6), Receptor-type tyrosine-protein phosphatase C (Ptprc), Leukocyte elastase inhibitor A (Serpinb1a), Serine peptidase inhibitor (Serpinb6a), NK13 (Serpinb6b), Signal-regulatory protein alpha (Sirpa), Src kinase-associated phosphoprotein 2 (Skap2), Signal-transducing adaptor protein 1 (Stap1), Toll-like receptor 2 (Tlr2), Toll-like receptor 7 (Tlr7). The enrichment KEGG pathways of cluster I proteins were involved Phagosome (FDR value = 0.0097), Cell adhesion molecules (FDR value = 0.0097), Antigen processing and presentation (FDR value = 0.0098) and Natural killer cell mediated cytotoxicity (FDR value = 0.0201).
The proteins In cluster II included Rho GTPase-activating protein 25 (Arhgap25), Rho GDP-dissociation inhibitor 2 (Arhgdib), Rho guanine nucleotide exchange factor 6 (Arhgef6), Coronin-1A (Coro1a), Cytochrome b-245 light chain (Cyba), Dedicator of cytokinesis protein 2 (Dock2), Docking protein 3 (Dok3), Engulfment and cell motility protein 1 (Elmo1), Fermitin family homolog 3 (Fermt3), G-protein-signaling modulator 3 (Gpsm3), Hematopoietic lineage cell-specific protein (Hcls1), Leupaxin (Lpxn), Neutrophil cytosol factor 1 (Ncf1), Neutrophil cytosol factor 2 (Ncf2),Neutrophil cytosol factor 4 (Ncf4), Pleckstrin (Plek), Tyrosine-protein phosphatase non-receptor type 18 (Ptpn18), Ras-related C3 botulinum toxin substrate 2 (Rac2), SAM and SH3 domain-containing protein 3 (Sash3), SH3 domain-binding glutamic acid-rich-like protein 3 (Sh3bgrl3), Proto-oncogene vav (Vav1)(Fig. 6A). The KEGG enrichment analysis was also conducted and the result showed that leukocyte transendothelial migration, chemokine signaling pathway and phagosome were the top 3 enriched terms of KEGG pathway. We deliberately explained the proteins involved in chemokine signaling pathway in Fig. 6B.
KEGG pathway enrichment analysis of cluster II proteins (A). The differentially regulated cluster II protein network in mpp+-stimulated N2A cells in response to NBP is visualized using Cytoscape 3.8.0. The blue nodes indicate the significantly downregulated cluster II proteins (p < 0.05). (B) The position of cluster III proteins in the KEGG “Chemokine signaling pathway” during the NBP treatment are shown in red color
In Fig. 7A, there were 20 Proteins in clusters III including Apoptosis regulatory protein Siva (Siva1), Protein mono-ADP-ribosyltransferase PARP3 (Parp3), Histone H1.5 (Hist1h1b), NAD-dependent protein deacylase sirtuin-5 (Sirt5), Histone H1.1 (Hist1h1a), Protein max (Max), Protein NDRG1 (Ndrg1), BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (Bnip3), BCL-6 corepressor (Bcor), Bcl2-associated agonist of cell death (Bad), Syntaxin-binding protein 2 (Stxbp2), Cellular tumor antigen p53 (Trp53), Lactadherin (Mfge8), Apoptosis regulator BAX (Bax), Tumor suppressor ARF (Cdkn2a), Caveolae-associated protein 2 (Cavin2/Sdpr), BH3-interacting domain death agonist (Bid), Cyclin-dependent kinase inhibitor 1B (Cdkn1b), Annexin A5 (Anxa5). The KEGG enrichment analysis demonstrated that the top 5 enriched pathway were Apoptosis, Measles, Platinum drug resistance, p53 signaling pathway and Chronic myeloid leukemia. We pointed out the DEPs in p53 signaling pathway in Fig. 7B.
KEGG pathway enrichment analysis of cluster III proteins. (A) The differentially regulated cluster III protein network. The blue nodes indicate the significantly downregulated cluster III proteins (p < 0.05). (B) The position of cluster III proteins in the KEGG “p53 signaling pathway” during the NBP treatment are shown in red color
The verification by Western blot
Based on the result of cluster analysis, P53 and Bax were associated with “P53 signaling pathway”. Therefore, P53 and Bax were selected for Western blotting validation (Fig. 8). The result revealed that compared with control group (n = 5), P53 and Bax were significantly upregulated in mpp + treated group (P53:1.00 ± 0.10 vs 2.38 ± 0.33, P = 0.003; Bax:1.00 ± 0.11 vs 3.39 ± 0.33, P < 0.001), whereas the pretreatment of NBP reversed this effect (P53:1.45 ± 0.19 vs 2.38 ± 0.33, P = 0.035; Bax:1.91 ± 0.33 vs 3.39 ± 0.33, P = 0.007). The WB validation of P53 and Bax is consistent with proteomic result.
Discussion
Parkinson’s disease (PD) is a motor related neurodegenerative disorder with cardinal clinical characters involving bradykinesias, tremor, postural instability and rigidityb [6]. The hallmarks of PD pathology are the loss of neurons in substantia nigra and the formation of Lewy bodies (LBs) in surviving neuron [5]. The aggregation of the misfolded and fibrillary α-synuclein (α-syn) are identified as the core mechanism of the formation of LBs [3]. As nigrostriatal dopaminergic pathway is primarily impaired in PD pathological process, dopamine replacement therapy based on the oral administration of L-DOPA is the first-line pharmacotherapy for PD to date [28]. In initial treatment, L-DOPA reverses these motor disturbances efficiently, whereas the long-term application may induce heterogenetic complications, particularly dyskinesia and motor fluctuations [28,29,30]. Therefore, the development of novel therapeutic strategies, especially those that target non-dopaminergic pathways are the urgent clinical quest.
Dl-3-n-butylphthalide (NBP; C12H14O2), extracted from the seeds of Apium graveolens, was first administrated as the therapy for patients suffered with acute ischemic stroke in 2002 in China [9, 11]. Albeit researches have documented that NBP displayed extensive pharmacological activities and exerted potentially beneficial effects in PD models both in vivo and in vitro, its precise mechanisms are still uncovered [17, 19, 31]. Therefore, we employed TMT-based LC–MS/MS to draw the differentially expressed proteins profiling of NBP pretreatment in mpp + -induced N2A cells. A total of 5828 proteins were quantified in the three replicates. Using fold-change > 1.5 and P < 0.05 as the cutoff to identified DEPs, compared to mpp + group, 417 proteins were determined as DEPs, among which 140 were upregulated and 277 were downregulated in mpp + -induced N2A cells with NBP pretreatment.
Subsequently, GO and KEGG analysis were performed to explore the cellular function and biological pathways enrichment of the DEPs. The result of GO-CC revealed that most DEPs were localized in cytosol and mitochondrion. As NBP held mito-protective effects on cerebral ischemia/reperfusion and cardiac ischemia models by reducing oxidative injury, alleviating mitochondrial apoptosis, and regulating mitochondrial biogenesis, we hypothesis that anti- mitochondrial injury might be one of the therapeutic approaches of NBP in PD as well. However, the exact mechanisms are required more work to illuminated. Based on the KEGG pathway enrichment analysis, “Lysosome”, “Phagosome”, “Apoptosis”, “Nucleotide metabolism”, “Ferroptosis” were all participated in the potential protective action of NBP in PD. In addition, “positive regulation of tumor necrosis factor production” and “apoptotic process” were also involved in GO-BP enrichment analysis. In this regard, the suppression of apoptotic process was supposed to be one of the salient pathways that related to the neuroprotective effect of NBP.
Next, the MCC method identified 10 hub proteins with the highest degree of connectivity, including Itgb2, Coro1a, Fermt3, Ptprc, Hcls1, Inpp5d, Ptpn6, Nckap11, Rac2 and Ncf4. Some of the proteins had been reported to be implicated in PD progress, even had been reported to be differentially expressed in PD patients. For instant, PTPRC (also named CD45) specifically dephopshorylated tyrosine residues. In AD, the deficiency of PTPRC promoted microglial activation and increased oligomeric Aβ accumulation [32, 33]. Recent study further revealed that PTPRC downregulated significantly in patients with PD [34]. ITGB2, which encoded the β2 integrin subunit, is implicated in defective adhesion and the subsequent inflammatory reaction [35]. Bioinformatic analysis revealed that ITGB2 was robust in microglia and played a role in the pathological mechanism of AD [36]. In addition, several network analyses reported ITGB2 altered in PD as well [37, 38]. Inpp5d is widely reported to be an AD risk gene [39]. Recent data processing of the available datasets further elucidated that Inpp5d was one of the hub genes involved in the overlapping molecular pathogenesis of AD and PD [37]. As Inpp5d was encoded for SHIP1, it acted as a negative regulator for PI3K/AKT signaling. Furthermore, previous study demonstrated that the depletion of SHIP1 could enhance the resistance to apoptosis [40]. Therefore, these findings indicated novel molecular mechanism of the neuroprotective effect of NBP in PD.
Cluster analysis suggested that p53 signaling pathway was one of the enriched pathways of the DEPs. In addition, p53 was also significantly differential expressed after NBP pretreatment. P53 was recognized as a tumor suppressor which encoded by the TP53 gene placed at 17p13.1 locus [41]. It was well known that p53 was essential for inducing apoptosis and was responsible for diverse cellular stresses. Pathogenic p53 integrated the cellular stresses comprising the generation ROS, inflammation, abnormal protein accumulation and Ca2+ overloading, to trigger cell death [42,43,44]. Recent study demonstrated that the specific deletion of p53 gene could eliminate dopaminergic neuronal cell death and further decrease motor deficits in MPTP-treated mice [45]. On one hand, the activated p53 interacted with antiapoptotic Bcl-2 family proteins and released Bax and Bad to open mPTP, which induced the intrinsic apoptotic pathway [46,47,48]. On the other hand, the accumulation of activated p53 might lead to significant impairment of autophagic clearance and promote α-syn aggregation [49]. In addition, p53 was also contributed to the regulation of DJ-1 as well as parkin both in mRNA and protein levels, which brought a feasible link between genetic and sporadic Parkinsonism in some extent [45]. Mitochondrial dysfunction was another hallmark of PD pathology. It had reported that p53 directly interacted with Parkin to inhibit its translocation to the damaged mitochondria, which aggravated the impairment of mitophagy and the consequent PD manifestations [50, 51]. In accordance with our result, NBP could break the 3-D structure of NQO1 and restrict p53 degradation in ischemia neurons [52]. Therefore, based on our result, the expression of P53 and its downstream factors including Bad and Bax were all reduced significantly after NBP pretreatment, indicating the suppression of p53 signaling pathway may be a potential therapeutic pathway for NBP. However, there is a dearth of studies to detect the association of NBP treatment and p53-medicated apoptosis in PD. Our research might shed light of novel therapeutic target of NBP in clinical PD treatment.
Conclusion
Taken together, using TMT-based quantitative proteomics, we revealed a signature of DEPs in N2A cells with NBP pretreatment when compared to untreated N2A cells. Among DEPs, essential proteins such as Itgb2, Coro1a, Fermt3, Ptprc, Hcls1, Inpp5d, Ptpn6, Nckap11, Rac2 and Ncf4 were identified as hub genes. The KEGG pathway and PPI network analysis provided preliminary information for discovering novel therapeutic targets and the implicated pathways for NBP treatment in PD. Therefore, our study warrants further studies to comprehensively and conclusively elucidate the role of these proteins or pathways in the potential clinical applications.
Availability of data and materials
The raw data has been submitted to the ProteomeX-change Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD037468. The datasets supporting the conclusions of this article are included within the article and its additional flies.
References
Jagadeesan AJ, Murugesan R, Vimala Devi S, Meera M, Madhumala G, Vishwanathan Padmaja M, Ramesh A, Banerjee A, Sushmitha S, Khokhlov AN, et al. Current trends in etiology, prognosis and therapeutic aspects of Parkinson’s disease: a review. Acta Biomed. 2017;88:249–62.
Zhao Y, Zhang Y, Zhang J, Yang G. Plasma proteome profiling using tandem mass tag labeling technology reveals potential biomarkers for Parkinson’s disease: a preliminary study. Proteomics Clin Appl. 2022;16: e2100010.
Zhao Y, Yang G. Potential of extracellular vesicles in the Parkinson’s disease - Pathological mediators and biomarkers. Neurochem Int. 2021;144: 104974.
Politi C, Ciccacci C, Novelli G, Borgiani P. Genetics and Treatment Response in Parkinson’s Disease: An Update on Pharmacogenetic Studies. Neuromolecular Med. 2018;20:1–17.
Zhao Y, Zhang Y, Zhang J, Yang G. Salvianolic acid B protects against MPP+-induced neuronal injury via repressing oxidative stress and restoring mitochondrial function. NeuroReport. 2021;32:815–23.
Ascherio A, Schwarzschild MA. The epidemiology of Parkinson’s disease: risk factors and prevention. Lancet Neurol. 2016;15:1257–72.
Gao XY, Yang T, Gu Y, Sun XH. Mitochondrial Dysfunction in Parkinson’s Disease: From Mechanistic Insights to Therapy. Front Aging Neurosci. 2022;14: 885500.
Thapa K, Khan H, Kanojia N, Singh TG, Kaur A, Kaur G. Therapeutic Insights on Ferroptosis in Parkinson’s disease. Eur J Pharmacol. 2022;930:175133.
Wang S, Ma F, Huang L, Zhang Y, Peng Y, Xing C, Feng Y, Wang X, Peng Y. Dl-3-n-Butylphthalide (NBP): A Promising Therapeutic Agent for Ischemic Stroke. CNS Neurol Disord Drug Targets. 2018;17:338–47.
Huang L, Wang S, Ma F, Zhang Y, Peng Y, Xing C, Feng Y, Wang X, Peng Y. From stroke to neurodegenerative diseases: The multi-target neuroprotective effects of 3-n-butylphthalide and its derivatives. Pharmacol Res. 2018;135:201–11.
Xu ZQ, Zhou Y, Shao BZ, Zhang JJ, Liu C. A Systematic Review of Neuroprotective Efficacy and Safety of DL-3-N-Butylphthalide in Ischemic Stroke. Am J Chin Med. 2019;47:507–25.
Wang CY, Xu Y, Wang X, Guo C, Wang T, Wang ZY. Dl-3-n-Butylphthalide Inhibits NLRP3 Inflammasome and Mitigates Alzheimer’s-Like Pathology via Nrf2-TXNIP-TrX Axis. Antioxid Redox Signal. 2019;30:1411–31.
Chen N, Zhou Z, Li J, Li B, Feng J, He D, Luo Y, Zheng X, Luo J, Zhang J. 3-n-butylphthalide exerts neuroprotective effects by enhancing anti-oxidation and attenuating mitochondrial dysfunction in an in vitro model of ischemic stroke. Drug Des Devel Ther. 2018;12:4261–71.
Abdoulaye IA, Guo YJ. A Review of Recent Advances in Neuroprotective Potential of 3-N-Butylphthalide and Its Derivatives. Biomed Res Int. 2016;2016:5012341.
Feng X, Peng Y, Liu M, Cui L. DL-3-n-butylphthalide extends survival by attenuating glial activation in a mouse model of amyotrophic lateral sclerosis. Neuropharmacology. 2012;62:1004–10.
Zhang D, Zheng N, Fu X, Shi J, Zhang J. Dl-3-n-butylphthalide attenuates myocardial ischemia reperfusion injury by suppressing oxidative stress and regulating cardiac mitophagy via the PINK1/Parkin pathway in rats. J Thorac Dis. 2022;14:1651–62.
Que R, Zheng J, Chang Z, Zhang W, Li H, Xie Z, Huang Z, Wang HT, Xu J, Jin D, et al. Dl-3-n-Butylphthalide Rescues Dopaminergic Neurons in Parkinson’s Disease Models by Inhibiting the NLRP3 Inflammasome and Ameliorating Mitochondrial Impairment. Front Immunol. 2021;12: 794770.
Xiong N, Huang J, Chen C, Zhao Y, Zhang Z, Jia M, Zhang Z, Hou L, Yang H, Cao X, et al. Dl-3-n-butylphthalide, a natural antioxidant, protects dopamine neurons in rotenone models for Parkinson’s disease. Neurobiol Aging. 2012;33:1777–91.
Zhou H, Ye M, Xu W, Yu M, Liu X, Chen Y. DL-3-n-butylphthalide therapy for Parkinson’s disease: A randomized controlled trial. Exp Ther Med. 2019;17:3800–6.
Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–30.
Liu Q, Zhang B. Integrative Omics Analysis Reveals Post-Transcriptionally Enhanced Protective Host Response in Colorectal Cancers with Microsatellite Instability. J Proteome Res. 2016;15:766–76.
Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, Jane-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell. 2020;180(387–402): e316.
Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13:1632–61.
Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–69.
Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–904.
Singh LK, Pandey M, Baithalu RK, Fernandes A, Ali SA, Jaiswal L, Pannu S, Neeraj, Mohanty TK, Kumaresan A, et al. Comparative Proteome Profiling of Saliva Between Estrus and Non-Estrus Stages by Employing Label-Free Quantitation (LFQ) and Tandem Mass Tag (TMT)-LC-MS/MS Analysis: An Approach for Estrus Biomarker Identification in Bubalus bubalis. Front Genet. 2022;13:867909.
Rodrigues BM, Mathias LS, Depra IC, Cury SS, de Oliveira M, Olimpio RMC, De Sibio MT, Goncalves BM, Nogueira CR. Effects of Triiodothyronine on Human Osteoblast-Like Cells: Novel Insights From a Global Transcriptome Analysis. Front Cell Dev Biol. 2022;10: 886136.
Dhall R, Kreitzman DL. Advances in levodopa therapy for Parkinson disease: Review of RYTARY (carbidopa and levodopa) clinical efficacy and safety. Neurology. 2016;86:S13-24.
Calabresi P, Di Filippo M, Ghiglieri V, Tambasco N, Picconi B. Levodopa-induced dyskinesias in patients with Parkinson’s disease: filling the bench-to-bedside gap. Lancet Neurol. 2010;9:1106–17.
Fisone G, Bezard E. Molecular mechanisms of l-DOPA-induced dyskinesia. Int Rev Neurobiol. 2011;98:95–122.
Li H, Wang H, Zhang L, Wang M, Li Y. Dl-3-n-Butylphthalide Alleviates Behavioral and Cognitive Symptoms Via Modulating Mitochondrial Dynamics in the A53T-alpha-Synuclein Mouse Model of Parkinson’s Disease. Front Neurosci. 2021;15: 647266.
Zhu Y, Hou H, Rezai-Zadeh K, Giunta B, Ruscin A, Gemma C, Jin J, Dragicevic N, Bradshaw P, Rasool S, et al. CD45 deficiency drives amyloid-beta peptide oligomers and neuronal loss in Alzheimer’s disease mice. J Neurosci. 2011;31:1355–65.
Tan J, Town T, Mori T, Wu Y, Saxe M, Crawford F, Mullan M. CD45 opposes beta-amyloid peptide-induced microglial activation via inhibition of p44/42 mitogen-activated protein kinase. J Neurosci. 2000;20:7587–94.
Bottero V, Santiago JA, Potashkin JA. PTPRC Expression in Blood is Downregulated in Parkinson’s and Progressive Supranuclear Palsy Disorders. J Parkinsons Dis. 2018;8:529–37.
Wen L, Marki A, Wang Z, Orecchioni M, Makings J, Billitti M, Wang E, Suthahar SSA, Kim K, Kiosses WB, et al. A humanized beta2 integrin knockin mouse reveals localized intra- and extravascular neutrophil integrin activation in vivo. Cell Rep. 2022;39: 110876.
Shu J, Li N, Wei W, Zhang L. Detection of molecular signatures and pathways shared by Alzheimer’s disease and type 2 diabetes. Gene. 2022;810: 146070.
Gupta R, Kumar P. CREB1(K292) and HINFP(K330) as Putative Common Therapeutic Targets in Alzheimer’s and Parkinson’s Disease. ACS Omega. 2021;6:35780–98.
Henderson AR, Wang Q, Meechoovet B, Siniard AL, Naymik M, De Both M, Huentelman MJ, Caselli RJ, Driver-Dunckley E, Dunckley T. DNA Methylation and Expression Profiles of Whole Blood in Parkinson’s Disease. Front Genet. 2021;12: 640266.
Xue F, Gao L, Chen T, Chen H, Zhang H, Wang T, Han Z, Gao S, Wang L, Hu Y, et al. Parkinson’s Disease rs117896735 Variant Regulates INPP5F Expression in Brain Tissues and Increases Risk of Alzheimer’s Disease. J Alzheimers Dis. 2022;89(1):67–77.
Liu Q, Sasaki T, Kozieradzki I, Wakeham A, Itie A, Dumont DJ, Penninger JM. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev. 1999;13:786–91.
Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83.
Wawryk-Gawda E, Chylinska-Wrzos P, Lis-Sochocka M, Chlapek K, Bulak K, Jedrych M, Jodlowska-Jedrych B. P53 protein in proliferation, repair and apoptosis of cells. Protoplasma. 2014;251:525–33.
Liu X, Fan L, Lu C, Yin S, Hu H. Functional Role of p53 in the Regulation of Chemical-Induced Oxidative Stress. Oxid Med Cell Longev. 2020;2020:6039769.
Luo Q, Sun W, Wang YF, Li J, Li DW. Association of p53 with Neurodegeneration in Parkinson’s Disease. Parkinsons Dis. 2022;2022:6600944.
Qi X, Davis B, Chiang YH, Filichia E, Barnett A, Greig NH, Hoffer B, Luo Y. Dopaminergic neuron-specific deletion of p53 gene is neuroprotective in an experimental Parkinson’s disease model. J Neurochem. 2016;138:746–57.
Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–8.
Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A, Morishita Y, Akira S, Taniguchi T, Tanaka N. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 2003;17:2233–8.
Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–9.
Karim MR, Liao EE, Kim J, Meints J, Martinez HM, Pletnikova O, Troncoso JC, Lee MK. alpha-Synucleinopathy associated c-Abl activation causes p53-dependent autophagy impairment. Mol Neurodegener. 2020;15:27.
da Costa CA, Sunyach C, Giaime E, West A, Corti O, Brice A, Safe S, Abou-Sleiman PM, Wood NW, Takahashi H, et al. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson’s disease. Nat Cell Biol. 2009;11:1370–5.
Talebi M, Talebi M, Kakouri E, Farkhondeh T, Pourbagher-Shahri AM, Tarantilis PA, Samarghandian S. Tantalizing role of p53 molecular pathways and its coherent medications in neurodegenerative diseases. Int J Biol Macromol. 2021;172:93–103.
Wang Y, Qi W, Zhang L, Ying Z, Sha O, Li C, Lu L, Chen X, Li Z, Niu F, et al. The novel targets of DL-3-n-butylphthalide predicted by similarity ensemble approach in combination with molecular docking study. Quant Imaging Med Surg. 2017;7:532–6.
Funding
This work was supported by Department of Health of Hebei Province (Grant No. 20200876).
Author information
Authors and Affiliations
Contributions
Yuan Zhao: Writing – original draft, Investigation, Writing – review & editing. Jian Zhang: Investigation. Yidan Zhang: Validation. Ya Gao: Data curation. Shuyue Li: Data curation. Cui Chang: Supervision. Lei Xu: Supervision. Guofeng Yang: Conceptualization, Methodology, Supervision. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhao, Y., Zhang, J., Zhang, Y. et al. Proteomic Analysis of Protective Effects of Dl-3-n-Butylphthalide against mpp + -Induced Toxicity via downregulating P53 pathway in N2A Cells. Proteome Sci 21, 1 (2023). https://doi.org/10.1186/s12953-022-00199-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12953-022-00199-x