Abstract
Background
Traditional preclinical echocardiography (ECHO) modalities, including 1-dimensional motion-mode (M-Mode) and 2-dimensional long axis (2D-US), rely on geometric and temporal assumptions about the heart for volumetric measurements. Surgical animal models, such as the mouse coronary artery ligation (CAL) model of myocardial infarction, result in morphologic changes that do not fit these geometric assumptions. New ECHO technology, including 4-dimensional ultrasound (4D-US), improves on these traditional models. This paper aims to compare commercially available 4D-US to M-mode and 2D-US in a mouse model of CAL.
Methods
37 mice underwent CAL surgery, of which 32 survived to a 4 week post-operative time point. ECHO was completed at baseline, 1 week, and 4 weeks after CAL. M-mode, 2D-US, and 4D-US were taken at each time point and evaluated by two separate echocardiographers. At 4 weeks, a subset (n = 12) of mice underwent cardiac magnetic resonance (CMR) imaging to serve as a reference standard. End systolic volume (ESV), end diastolic volume (EDV), and ejection fraction (EF) were compared among imaging modalities. Hearts were also collected for histologic evaluation of scar size (n = 16) and compared to ECHO-derived wall motion severity index (WMSI) and global longitudinal strain as well as gadolinium-enhanced CMR to compare scar assessment modalities.
Results
4D-US provides close agreement of ESV (Bias: -2.55%, LOA: − 61.55 to 66.66) and EF (US Bias: 11.23%, LOA − 43.10 to 102.8) 4 weeks after CAL when compared to CMR, outperforming 2D-US and M-mode estimations. 4D-US has lower inter-user variability as measured by intraclass correlation (ICC) in the evaluation of EDV (0.91) and ESV (0.93) when compared to other modalities. 4D-US also allows for rapid assessment of WMSI, which correlates strongly with infarct size by histology (r = 0.77).
Conclusion
4D-US outperforms M-Mode and 2D-US for volumetric analysis 4 weeks after CAL and has higher inter-user reliability. 4D-US allows for rapid calculation of WMSI, which correlates well with histologic scar size.
Similar content being viewed by others
Background
Echocardiography (ECHO) is a highly efficient and reliable tool for clinical and pre-clinical evaluation of cardiac function [1,2,3]. Advances in high-frequency and high-resolution image acquisition continue to increase temporal and spatial resolution in small rodent models [1, 4, 5]. However, traditional ECHO modalities for evaluating ventricular size and function, including 1-dimensional motion-mode (M-mode) and 2-dimensional (2D-US) long-axis analysis, make geometric assumptions about the heart that limit their accuracy, particularly in the setting of heart disease, where regional ventricular shape abnormalities may exist [6,7,8]. For example, mouse models of cardiac pathology, particularly myocardial infarction (MI), frequently result in abnormal ventricular remodeling that deviates from the geometric assumption that the left ventricle has an ellipsoid shape, limiting the utility of conventional echocardiography in such settings and requiring careful consideration of modality choice [9, 10].
Three-dimensional (3D) ECHO has gained favor recently in clinical and pre-clinical models, allowing full volume visualization of the heart by stacking concentric short-axis images to form a 3D representation at static time points [4, 11, 12]. 3D ECHO has been further improved by combining respiratory- and ECG-gated image acquisition to create 3D images throughout the cardiac cycle, referred to as 4-dimensional ultrasound (4D-US) [13, 14]. 4D-US has recently became commercially available for pre-clinical models and has already been validated against cardiac magnetic resonance (CMR) imaging in wild type mice and a mouse model of hypertrophy [15]. While CMR imaging remains the gold standard for assessment of cardiac function in mice [16,17,18], recent studies in 3D ECHO and 4D-US have shown considerable advances in scanning time and reliability [13,14,15].
In this study, we aimed to validate commercially available 4D-US in a mouse model of myocardial infarction. We performed coronary artery ligation (CAL) surgery on mice to evaluate traditional M-mode, 2D-US, and 4D-US using the Vevo 3100 preclinical imaging system against CMR imaging to compare the assessment of volumetric parameters at 4 weeks after CAL.
Methods
Coronary artery ligation and mouse model
All animal use was performed at the University of Pittsburgh in compliance with the National Institutes of Health Guide for Care and Use of Experimental Animals and was approved by the University of Pittsburgh Animal Care and Use Committee. Previously characterized mice harboring a floxed TFAM gene were crossed with a-myosin heavy chain Cre transgene (MHC-Cre) to generate mice that were MHC-Cre (+) x Flox-TFAM (n = 22) and MHC-Cre (−) x Flox-TFAM (n = 15). Both males and females were used in this study. Mice were aged to twelve weeks, at which point they underwent baseline echocardiography followed by coronary artery ligation surgery. Mice were anesthetized and ventilated prior to open thoracotomy through the 4th rib followed by opening of the pericardium and coronary artery ligation (CAL) by suture placement around the proximal coronary artery [19]. All surviving mice underwent follow-up echocardiography at 1 and 4 weeks.
Echocardiography
Mice were anesthetized using isoflurane delivered by nose cone at the following timepoints: baseline (n = 37), 1 week following CAL (n = 32), and 4 weeks following CAL (n = 32). At each timepoint, depilatory cream was applied to the thorax to remove hair. Animals were maintained at 37 °C via heating pad and rectal probe and were monitored using surface ECG limb electrodes throughout imaging. Transthoracic echocardiography was performed using the Vevo 3100 imaging systems (FUJIFILM VisualSonics, Toronto, Canada) with a probe attached to a step motor. The Visualsonic MX400 (20–46 MHz, 50 μm axial resolution) linear array transducer was used for all image acquisition. Heart rate was maintained between 400 and 500 bpm during imaging by adjusting isoflurane concentration to a final concentration of 1–2%. M-mode and B-mode images of the heart were obtained for at least ten cardiac cycles in the parasternal long axis and mid-papillary muscle level short axis views. For 4D image acquisition, the step motor was positioned just below the apex and the motor aligned to take concentric short axis images in 0.2 mm steps. At each position, a complete cardiac cycle was recorded using automated ECG and respiratory gating. 4D images were constructed using Vevo 4D image software. Image analysis was performed independently by two blinded sonographers (CR and BM). End-diastolic volume (EDV), End-systolic volume (ESV), and Ejection Fraction (EF), were calculated using the following formulas:
M-Mode (short-axis):
2D-US analysis of the parasternal long-axis was completed using operator-defined LV trace function of Vevo LAB software (v3.2.0) and calculations of EDV, ESV, and EF made by Simpson’s method. 4D-US measurements were calculated directly from volumetric measurements based on operator-defined edge-tracing using Vevo 4D imaging software.
The Vevo Strain software was used to measure both longitudinal and radial strain by using long-axis images and semi-automated border tracking. The LV was visualized during end-diastole and the endocardial and epicardial borders were determined. A minimum of five cardiac cycles were then traced automatically by the speckle-tracking software and reviewed by the user. Respiratory variation was excluded from these cycles. Global peak longitudinal and radial strain were calculated using Vevo Strain software on images taken from the long-axis. Global peak circumferential and radial strain were calculated from images from the short axis. Further, the LV was automatically divided into six segments in the long-axis images by the software. The standard deviation of the strain among the individual segments was calculated as a measure of LV dyssynchrony.
WMSI
WMSI was calculated using a 16-segment model collected from 3 short axis views collected during 4D-US image acquisition (n = 32). Short axis views were obtained 1 mm, 3 mm, and 5 mm from the apex for standardization. The most distal image was divided into 4 sections and the remaining sections divided into 6 sections as previously described (Fig. 3) [20, 21]. Individual sections were graded as: 1- normal, 2- hypokinetic, 3- akinetic, 4- dyskinetic, 5- aneurysmal. WMSI was calculated as average of all 16-segment motion scores.
CMR
Mice (n = 12) were anesthetized with 4% isoflurane mixed with room air in an induction box for 1 to 3 min. The depth of anesthesia was monitored by toe reflex, extension of limbs, and spine positioning. Anesthesia was maintained by 1.5 to 2% isoflurane and 100% oxygen via a nose cone. Respiration waveforms were continuously monitored using a small pneumatic pillow under the animal’s diaphragm connected to a magnet-compatible pressure transducer (SA Instruments, Stony Brook, NY). CMR was performed on a Bruker Biospec 7 T/30 system (Bruker Biospin MRI, Billerica, MA) with a 35-mm quadrature coil for both transmission and reception. The Bruker Intragate module was used for image-gated cine MRI with retrospective navigation. Subcutaneous injection of Multi-Hance (Gadobenate dimeglumine, 529 mg/ml, Bracco Diagnostics, Inc., Monroe Twp, NJ 08831) was administered immediately before the CMR acquisition at 0.1 mmol Gd/kg bodyweight. T1-weighted images to highlight LGE were acquired 15–20 min after the subcutaneous administration of Multi-Hance. Eight T1-weighted short-axis imaging planes covering the whole ventricular volume with no gaps were acquired with the following parameters: Field of view (FOV) = 2.5 cm X 2.5 cm, slice thickness = 1 mm, in-plane resolution = 0.97 μm, flip angle (FA) = 10 degrees, echo time (TE) = 3.059 msec, repetition time (TR) = 5.653 msec. White-blood cine movies with 20 cardiac phases were acquired for each mouse with equivalent temporal resolution for the cine loops was about 16.5–21.5 ms per frame. Eight short-axis imaging planes covering the whole ventricular volume with no gaps and one long-axis plane were acquired with the following parameters: Field of view (FOV) = 2.5 cm X 2.5 cm, slice thickness = 1 mm, in-plane resolution = 0.97 μm, flip angle (FA) = 30 degrees, echo time (TE) = 1.872 msec, repetition time (TR) = 38.293 msec.
The extent of myocardial infarction was defined by the percentage of the myocardium displaying hyperintensity 15–20 min after Gd administration. To obtain the proportion of myocardial infarction, the area of hyperintensity was manually traced by a blinded operator on the Paravision 5.1 Xtip software (Bruker Biospin MRI, Billerica, MA). The extent of myocardial hemorrhage was defined by dark hypointensity on the cine images. To obtain the proportion of myocardial hemorrhage, the area of hypointensity was manually traced by a blinded operator on the software. The left ventricular endocardium and epicardium boundaries of each imaging slice at the end-systole (ES) and the end-diastole (ED) were manually traced by a blinded operator in the software to calculate the following functional parameters: left ventricular blood volume (LVV), left ventricular wall volume (LV wall), LV mass, stroke volume (SV), ejection fraction, heart rate (HR), cardiac output (CO), longitudinal shortening, and radial shortening. LVV is calculated by summation of all the short-axis slices. The EF was calculated using the following equation: \( EF=\frac{\sum \limits_i{A}_i^{es}{h}_i}{\sum \limits_i{A}_i^{ed}{h}_i}\times 100\% \), where \( {A}_i^{es} \) is the internal left ventricle area of slice i at end systole, \( {A}_i^{ed} \) the internal left ventricle area of slice i at end diastole, and hi is the thickness of each scanned slice.
Tissue histology
LV tissue (n = 16) was fixed overnight in 10% formaldehyde at 4 °C. Tissues were then washed with PBS and transferred to 70% EtOH and stored at room temperature. After fixation, tissues were brought to the Department of Pathology Histology Core at the University of Pittsburgh and sectioned into 10 μm slices at 1 mm intervals throughout the myocardium. Sections were stained with hematoxylin and eosin (H&E) or Masson’s Trichrome stains and images obtained on a TissueFAXS Histo (TissueGnostics, Vienna, Austria) upright brightfield microscope utilizing HistoQuest software. Image analysis was performed by automated red and blue channel separation using Image Measurement 9.0 (Bersoft Imaging, Cologne, Germany).
Statistical analysis
Differences between imaging modalities were evaluated by Bland-Altman analysis and are expressed as % bias and 95% level of agreement (LOA). Bland-Altman percentage bias was calculated as (100*(B-A)/Average vs Average) where “B” represents measurements from 4D-US, 2D-US, or M-mode, and “A” represents measurements from CMR. Intraclass correlation was used to evaluate reliability between single measurements of EDV, ESV, and EF between two users (CR and BM). Normality was assessed using D’Agostino-Pearson omnibus K2 testing for each data set. Regression analysis was performed using Spearman rank correlation to compare scar size among several methods (Fig. 4, Supplemental Table 2). Correlation was graded as poor (0.0–0.5), moderate (0.5–0.7), strong (0.7 to 0.9), or very strong (0.9–1.0). Supplemental Fig. 2 data are expressed and mean ± standard error. Power analysis was completed based on linear regression analysis to confirm group size (two-sided test, α = 0.05, based on values for EDV). For all statistical tests, p ≤ 0.05 was considered significant. All normality, regression and statistical tests were completed using Graphpad Prism 7 software (San Diego, CA) except for ICC, which was calculated using Microsoft Excel (Redmond, WA), and power analysis which was calculated using StataCorp Stata 16.0 (College Station, Texas).
Results
Animal model of myocardial infarction
32 of the initial 37 mice survived to 4 weeks post-CAL (86.4%, Table 1). The mouse background was MHC-CRE (+) x Flox-TFAM or MHC-CRE (−) x Flox-TFAM. Subgroup analysis was performed between MHC (+) and MHC (−) groups and there was no significant difference noted in survival, scar size, baseline EDV, ESV, or EF, as well as 4-week EDV, ESV, or EF between groups by any ECHO modality (Supplemental Fig. 2, Supplemental Table 1).
Comparison of ECHO modalities to CMR
To compare imaging modalities following infarction, mice were evaluated by ECHO using traditional M-mode, 2D-US, and 4D-US (representative images in Fig. 1 with mean values reported in Table 2) at baseline, 1 week following CAL, and 4 weeks following CAL. One-day after the 4-week ECHO was performed, a subset of randomly chosen mice underwent CMR to serve as reference standard for volumetric measurements (Fig. 1). Bland-Altman analysis of M-mode, 2D-US, and 4D-US was used to compare the percentage difference of ECHO modalities to CMR (Fig. 2). Of the three ECHO modalities, 4D-US demonstrated the lowest bias when comparing ESV (4D-US Bias: 2.55% LOA: − 61.55 to 66.66, 2D-US Bias: 29.84% LOA: − 43.10 to 102.8, M-Mode Bias: 43.57% LOA: − 21.87 to 109.0; Fig. 2) and EF (4D-US Bias: -11.23% LOA: − 56.26 to 33.80, 2D-US Bias: -24.42% LOA: − 87.99 to 39.16, M-Mode Bias: -12.90% LOA: − 53.80 to 28.00). 4D-US was outperformed by 2D-US when evaluating EDV (4D-US Bias: -13.21% LOA: − 47.71 to 21.28, 2D-US Bias: 5.84% LOA: − 24.47 to 36.15, M-Mode Bias: 35.17% LOA: 0.73 to 69.61). Linear regression was also performed between ECHO modalities and CMR, which demonstrates strong to very strong correlation between 4D-US and CMR (EDV: r = 0.887, ESV: r = 0.943, EF: r = 0.811; Supplemental Fig. 1). 2D-US has lower correlation values for every modality (EDV: r = 0.806, ESV: r = 0.793, EF: r = 0.406). M-Mode correlation is slightly higher than 4D-US for EDV, but lower for ESV and EF (EDV: r = 0.902, ESV: r = 0.882, EF: r = 0.747). All ECHO modalities had similar HRs at the time of image acquisition (4D-US: 459.5 ± 6.3, 2D-US: 459.7 ± 9.2, M-Mode: 451.7 ± 8.3; Table 1) though HR was lower during CMR (356.2 ± 8.2) due to imaging constraints. 4D-US demonstrated the highest ICC for estimation of EDV (4D-US: 0.91, 2D-US: 0.72, M-Mode: 0.80) and ESV (4D-US: 0.93, 2D-US: 0.72, M-Mode: 0.79). M-mode had a higher ICC for evaluation EF (4D-US: 0.64, 2D-US: 0.31, M-Mode: 0.69).
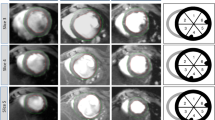
Representive Images Using 4D-US and CMR. a 4D-US cube reconstruction of concentric stacked images in an infarcted heart. b Representative wire tracing of 4D-US reconstruction in an infarcted heart 4 weeks after CAL. c Representative short-axis images taken during end-diastole (left column) and end-systole (right column) at 1 mm intervals from the apex (top) to base (bottom) of an infarcted heart 4 weeks after CAL. d Representative mid-ventricular CMR images of an infarcted heart during end-diastole (top) and end-systole (bottom)
Bland Altman Analysis and Inter-user Variability of ECHO Modalities. a Representative ECHO modalities for 4D-US, 2D-US, and M-Mode. b Bland-Altman analysis demonstrating the difference between selected measurement modality (4D-US, 2D-US, and M-Mode) and CMR. Data are presented as % bias and dotted lines represent 95% confidence interval. X-axes represent CMR volumetric measurements and Y-axes represent % Difference of ECHO values over CMR values. c Inter-user variability between two users for each ECHO modality as calculated by intraclass correlation
Wall motion severity index (WMSI), scar sizing, and strain analysis
To quantify scar size following CAL, we utilized gadolinium-enhanced CMR, step-wise short-axis images obtained via 4D-US, long-axis strain analysis, and trichrome staining of tissue sections, which was used as the gold standard (Fig. 3). Following sacrifice 4 weeks after CAL, a subset of all mice underwent histologic staining to evaluate scar size. Scar size was estimated histologically on trichrome-stained cross sections (Table 1, Fig. 3). The mean scar volume was 21.54 ± 4.07% (Range 4.02–57.00%, Table 1). Late gadolinium-enhanced CMR images yielded a mean volume of 13.53 ± 2.22%% (range 5.52–28.10%, Table 2). WMSI average scores are 1.32 ± 0.05 (range 1.07–2.07, Table 2, n = 32). Global longitudinal strain was calculated throughout the myocardium with average strain − 5.27 ± 1.10% (range − 13.29% – -1.072%, Table 2). Spearman rank correlation was used to assess the association of scar size between WMSI, CMR hyperintense regions/LV area, or global peak longitudinal strain to histologic stain assessments (Fig. 4). When compared to histologic scar size, WMSI correlates strongly (r = 0.77, p < 0.01), CMR hyperintense volume correlates very strongly (r = 0.90, p < 0.001), and longitudinal strain correlates strongly (r = 0.74, p = 0.01). Additional strain measurements, including global radial strain, LV dyssynchrony from long-axis imaging, and both circumferential and radial strain from short axis imaging were also calculated and a full comparison of modalities assessed via Spearman correlation (Supplemental Table 2).
Representative Concentric Images of a Single Heart by CMR, 4D-US, and Histology. CMR (top row), 4D-US (middle row), and trichrome-stained histologic slides taken from a single infarcted heart at ~ 1 mm steps from the apex (left) through basal-ventricle (right). 4D-US images (middle row) are divided into 16 sections for wall-motion score index, labeled as anterior (A), lateral (L), inferior (I), septal (S), anterior-lateral (AL), inferior-lateral (IL), inferior-septal (IS), and anterior-septal (AS)
Correlation between WMSI and CMR to Histology. Regression analysis with Spearman rank correlation comparing wall motion score index (WMSI) to histological infarct size (left), CMR hyperintense tissue to histologic infarct size (middle), and global peak longitudinal strain to histologic scar size (right)
Discussion
Over the past two decades, 3D- and 4D-US have been gaining clinical favor as the technology has advanced and now offers rapid and cost-effective high-resolution imaging [22]. Variations of 3D echocardiography have been shown to have quick and reliable volumetric assessment using semi-automated tracing [23], low inter-user variability [24], and have strong agreement with CMR in normal and abnormal LV’s [25]. Real-time 3D-ultrasound has been shown to outperform 2D-US when compared to single-photon emission computed tomography following myocardial infarction [26] and has allowed for increased detection of LV dyssynchrony [27]. Our work shows that commercially available 4D-US in similarly beneficial in a pre-clinical CAL model, demonstrating higher agreement with CMR and lower inter-user variability than traditional 2D-ECHO modalities.
This study evaluated 4D-US in a mouse model of myocardial infarction 4 weeks after CAL and compared 4D-US endpoints to 2D-US, M-mode, CMR, and histologic parameters. The mouse CAL model presents a challenge for evaluation using traditional echocardiography, given that wall motion abnormalities and dyssynchronous contraction can cause abnormal LV geometry. Traditional M-mode and 2D-US assume ellipsoid geometry for volumetric measurements [6]. Following CAL, mice typically have a hyperdynamic base, static apex, and increased sphericity that limit the effectiveness of the geometric assumptions [17, 28,29,30]. 4D-US allows for real-time image analysis at multiple planes encompassing the LV that is gated by ECG and respiration [13, 14] to minimize motion artifact during image reconstruction. The result is a data-driven volumetric model that eliminates spatial and temporal assumptions about LV morphology. Our data demonstrate that for calculation of EF and ESV, 4D-US provides overall closer agreement when compared to benchmark CMR than other ECHO modalities 4 weeks after CAL. Additionally, we show that 4D-US has lower inter-user variabilty when measuring ventricular volumes at 4 weeks. Finally, 4D-US allows for simplified evaluation of WMSI, which correlates well with scar size by histologic analysis.
In agreement analyses, we found that 4D-US had lower percentage bias by Bland-Altman analysis than 2D-US and M-mode compared to CMR for the evaluation of ESV and EF at 4 weeks (Fig. 2), suggesting more accurate assessment of volumetric parameters. 4D-US was outperformed by 2D-US in the evaluation of EDV at this time point. However, across the three endpoints evaluated (EDV, ESV, and EF), 4D-US was the most reliable modality in terms of bias and 95% level of agreement. 4D-US has much lower bias than M-mode, which demonstrated > 40% bias in the estimation of ESV. 2D-US bias was improved compared to M-mode, but was characterized by the highest inter-user variability as demonstrated by lowest ICC for the evaluation of EDV, ESV, and EF (Fig. 2c). Inter-user variabilty was lowest using 4D-US for evaluation of EDV and ESV, but was mildly outperformed by M-Mode when evaluating EF (Fig. 2c). In total, our data is consistent with other studies that found that 4D-US provides better accuracy and less inter-user variability when comparing to CMR than 2D-US or M-mode values, making it the prefered echo modality for surgical animal studies.
In considering the limitations of 4D-US, we noted that 4D-US uniformly underestimated EDV and ESV when compared to CMR, though the changes were more pronounced in EDV (Fig. 2b, Table 2). The 4D-US measurements may suffer limitations in surgical mouse models, as the base of the heart is difficult to clearly visualize given the overlying scar following thoracotomy required in the CAL surgery, and may result in lower total volumes. A similar underestimation of volumes was previously noted in evaluation of round “phantoms” using identical software [15], suggesting that a systemic underestimation result of volumes may be present in this modality. Regardless of this underestimation, the modality still represents an direct improvement over other traditional ECHO modes.
Recent work by Russo et al. has shown that automated step-wise short-axis ECHO imaging performed on CAL mice strongly correlates with CMR [17], providing reproducible volumetric evaluations nearly on par with CMR, but at a fraction of the time and cost required for CMR, as well as increased portability and accessibility. Our data support the increased value of this step-wise approach, while incorporating ECG- and respiratory-gating and the higher resolution of the Vevo-3100 for enhanced temporal and spatial resoluation. Soepriatna et al. previously demonstrated similar benefits of gated 4D-US in infarcted mice using manual 3-D reconstruction throughout the cardiac cycle [14]. Previous work by Grune et al. compared wild type mice using 4D-US obtained on Vevo 3100 hardware utilizing custom software analysis to volumetric data from CMR, and found excellent agreement between the modalities [15]. While these studies demonstrated the value of the hardware system and imaging throughout the cardiac cycle, here we use an easily-accessible commercial semi-automated system in a mouse CAL model, which utilizes edge-tracing software to simplify volumetric calculation for users, resulting in both rapid analysis and low inter-user variability.
The 4D-US imaging modality can to quickly and reproducibly quantify scar characteristics using clinical scales of wall-motion, increasing the translational relevance of pre-clinical assessments. Previous works have quantified scar size using ECHO based on wall-motion abnormalities from 2D and 3D-US reconstruction, demonstrating good comparison between ECHO estimations and histologic scar size [31, 32]. We evaluated WMSI using a 16-segment model across three short axis views used in the clinical settings as previously described [20, 21]. We found that WMSI correlates strongly with histologic analysis of scar size (r = 0.77, Fig. 4), whereas longitudinal peak strain also correlates strongly (r = 0.74) and gadolinium-enhanced CMR correlated very strongly (r = 0.90). It should be noted that this pre-clinical model utilizes permanent coronary occlusion. Models of transient occlusion may be complicated by myocardial stunning, which may impair the value of WMSI and its correlation to scar size, and requires further study.
The 4D-US method allows for easy standardization on the WMSI across animals, as the automated step function can be utilized to standardize the height of short-axis imaging. In this study, we used three short-axis images taken 1 mm, 3 mm, and 5 mm from the apex to quantify WMSI. Unfortunately, the 4D-US step-wise images are not compatable with Vevo Strain software, which would allow the user to rapidly obtain staged short-axis strain measurements, which has previously been validated as a predictor of scar size [33]. A full comparison table assessing the correlation between a number of previously validated markers of scar size, including histology, WMSI, CMR, global longitudinal strain, global radial strain, and LV dyssynchrony have been included in Supplemental Table 2.
Potential caveats
A potential confounder in the study is the use of multiple genotypes on a single background. Specifically, we utilized transgenic mice for the evaluation of volumetric measurements, pooling data from α-MHC-Cre (+) x Flox-TFAM and α-MHC-Cre (−) x Flox-TFAM mice. Because we found no difference between survival, scar size, baseline and 4-week EDV, ESV, or EF between groups (Supplemental Fig. 1, Supplemental Table 1), we included both models for this data set. We believe that the transgenic background does not limit the correlations drawn in this paper between volumetric studies and agrees with the NIH’s effort to reduce animal suffering by utilizing mice readily available in our lab. The CMR and ECHO measurements were completed only on mice following CAL and not on sham animals in this study, and only a limited subset of animals (n = 12) underwent all imaging modalities.
It should be noted that there was a HR discrepancy between the ECHO modalities and CMR imaging. While the HR remained consistent between 4D-US, 2D-US, and M-Mode, the HR was nearly 100 bpm slower when undergoing CMR (Table 1) as necessitated by the CMR system. Maintaining physiologic HR is a well known limitation of CMR imaging in the mouse [16]. Sub-physiologic HR’s are associated with elevated ESV, EDV, and decreased EF in healthy mice [34, 35]. However, previous work in a mouse CAL model has shown that EF does not correlate with HR in the CAL mouse in CMR, suggesting that the HR discrepancy may not be as important in the CAL model as it is in in healthy mice [36].
Conclusions
In this paper we demonstrate that commercially available semi-automated 4D-US provides quick and reliable volumetric measurements of the heart following CAL that compares favorably to CMR values. 4D-US correlates better with CMR than 2D-US and M-Mode for evaluation of volumetric parameters 4 weeks after CAL. 4D-US also allows simple evaluation of WMSI, which is a clinically relevant metric that correlates well with scar size by histologic analysis.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 2D-US:
-
2-Dimensional Ultrasound
- 3D:
-
3-Dimensional
- 4D-US:
-
4-Dimensional Ultrasound
- A:
-
Anterior
- AL:
-
Anterior Lateral
- AS:
-
Anterior Septal
- CAL:
-
Coronary Artery Ligation
- CMR:
-
Cardiac Magnetic Resonance
- CO:
-
Cardiac Output
- Echocardiography:
-
ECHO
- ED:
-
End Diastole
- EDV:
-
End Diastolic Volume
- EF:
-
Ejection Fraction
- ES:
-
End Systole
- ESV:
-
End Systolic Volume
- FA:
-
Flip Angle
- FOV:
-
Field of View
- H&E:
-
Hematoxylin and Eosin
- I:
-
Inferior
- ICC:
-
Intraclass Correlation
- IL:
-
Inferior Lateral
- IS:
-
Inferior Septal
- L:
-
Lateral
- Level of agreement:
-
LOA
- LV:
-
Left Ventricle
- LVV:
-
Left Ventricular blood volume
- MI:
-
Myocardial Infarction
- M-Mode:
-
Motion-mode
- S:
-
Septal
- SV:
-
Stroke Volume
- TE:
-
Echo Time
- TR:
-
Repetition Time
- WMSI:
-
Wall Motion Severity Index
References
Scherrer-Crosbie M, Thibault H. Echocardiography in translational research: of mice and men. J Am Soc Echocardiogr. 2008;21(10):1083–92.
Gardin JM, Siri FM, Kitsis RN, Edwards JG, Leinwand LA. Echocardiographic assessment of left ventricular mass and systolic function in mice. Circ Res. 1995;76:907–14.
Rottman JN, Ni MDG, Brown M. Imaging in Basic Science. Mouse-Echo : From Bedside to Bench: Part II Echocardiographic Evaluation of Ventricular Function in Mice. 2007;24(1).
Dawson D, Lygate CA, Saunders J, et al. Quantitative 3-Dimensional Echocardiography for Accurate and Rapid Cardiac Phenotype Characterization in Mice. 2004;108(suppl IV):1632–1637.
Benavides-Vallve C, Corbacho D, Iglesias-Garcia O, et al. New strategies for echocardiographic evaluation of left ventricular function in a mouse model of long-term myocardial infarction. PLoS One. 2012;7(7):1–9.
Parisi F, Moynihan PF, Feldman CL. Approaches to determination of left ventricular volume and ejection fraction by real-time two-dimensional echocardiography. Clin Cardiol. 1979;2:257–63.
Lang RM, Badano LP, Mor-avi V, et al. Recommendations for Cardiac Chamber Quantification by Echocardiography in Adults : An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr. 2015;28(1):1–39.e14.
Lindsey ML, Kassiri Z, Virag JAI, de Castro Brás LE, Scherrer-Crosbie M. Guidelines for measuring cardiac physiology in mice. Am J Physiol Circ Physiol. 2018;314(4):H733–52.
Gao X, Dart AM, Dewar E, Jennings G, Du X. Serial echocardiographic assessment of left ventricular dimensions and function after myocardial infarction in mice, vol. 45; 2000. p. 330–8.
Patten RD, Aronovitz MJ, Deras-mejia LUZ, et al. Ventricular remodeling in a mouse model of myocardial infarction. Am J Physiol Heart Circ Physiol. 1998;274(5):1812–20.
Lindsey ML, Kassiri Z, Virag JAI, de Castro Bras LE, Scherrer-Crosbie M. Guidelines for Measuring Cardiac Physiology in Mice. Am J Physiol Circ Physiol. 2018;ajpheart(00339):2017.
Dorosz JL, Lezotte DC, Weitzenkamp DA, Allen LA SE. Performance of 3-dimensional echocardiography in measuring left ventricular volumes and ejection fraction a systematic review and meta-analysis. JACC 2012;59(20):1799–1808.
Damen FW, Berman AG, Soepriatna AH, et al. High-frequency 4-dimensional ultrasound (4DUS): a reliable method for assessing murine cardiac function. Tomography. 2017;3(4):180–7.
Soepriatna AH, Damen FW, Vlachos PP, Goergen CJ. Cardiac and respiratory-gated volumetric murine ultrasound Arvin. Int J Cardiovasc Imaging. 2018;34(5):713–24.
Grune J, Blumrich A, Brix S, et al. Evaluation of a commercial multi-dimensional echocardiography technique for ventricular volumetry in small animals. Cardiovasc Ultrasound. 2018;16(1):1–13.
Price AN, Cheung KK, Cleary JO, Campbell AE, Riegler J, Lythgoe MF. Cardiovascular Magnetic Resonance Imaging in Experimental Models. Open Cardiovasc Med J. 2010;4:278–92.
Russo I, Micotti E, Fumagalli F, et al. A novel echocardiographic method closely agrees with cardiac magnetic resonance in the assessment of left ventricular function in infarcted mice. Sci Rep. 2019;9(1):1–10.
Akki A, Gupta A, Weiss RG. Magnetic resonance imaging and spectroscopy of the murine cardiovascular system. Am J Hear Ciculation Physiol. 2013;304(5):633–48.
Mcgaffin KR, Witham WG, Yester KA, et al. Cardiac-specific leptin receptor deletion exacerbates ischaemic heart failure in mice. Eur Soc Cardiol. 2011:60–71.
Takagawa J, Zhang Y, Wong ML, et al. Myocardial infarct size measurement in the mouse chronic infarction model: comparison of area- and length-based approaches. J Appl Physiol. 2007;102(6):2104–11.
Morgan EE, Faulx MD, McElfresh TA, et al. Validation of echocardiographic methods for assessing left ventricular dysfunction in rats with myocardial infarction. Am J Physiol Circ Physiol. 2004;287(5):H2049–53.
Lang RM, Addetia K, Narang A, Mor-avi V. 3-dimensional echocardiography. J Am Coll Cardiol Cardiovasc Imaging. 2018;11(12):1854–78.
Jacobs LD, Salgo IS, Goonewardena S, et al. Rapid online quantification of left ventricular volume from real-time three-dimensional echocardiographic data. Eur Heart J. 2006;27:460–8.
Jenkins C, Bricknell K, Hanekom L, Marwick TH. Reproducibility and accuracy of echocardiographic measurements of left ventricular parameters using real-time three-dimensional echocardiography. J Am Coll Cardiol. 2004;44(4):878–86.
Kuhl HP, Schreckenberg M, Rulands D, et al. High-resolution transthoracic quantitation of cardiac volumes and. J Am Coll Cardiol. 2004;43(11):2083–90.
Arai K, Hozumi T, Matsumura Y, et al. Accuracy of measurement of left ventricular volume and ejection fraction by new real-time three-dimensional echocardiography in patients with wall. Am J Cardiol. 2004;94:2–8.
Kapetanakis S, Kearney MT, Siva A, Gall N, Cooklin M, Monaghan MJ. Real-time three-dimensional echocardiography a novel technique to quantify global left ventricular mechanical Dyssynchrony. Circulation. 2005;112:992–1000.
Kanno S, Lerner DL, Schuessler RB, Betsuyaku T, Yamada KA, Saffitz JE. Echocardiographic evaluation of ventricular remodeling in a mouse model of myocardial infarction. J Am Soc Echocardiogr. 2002;27(1):601–9.
St MG, Sutton J, Sharpe N. Clinical cardiology : new Frontiers left ventricular remodeling after myocardial infarction pathophysiology and therapy. Circulation. 2000;101:2981–8.
Salto-Tellez M, Yung Lim S, El-Oakley RM, Tang TP, Almsherqi ZALS. Myocardial infarction in the C57BL/6J mouse: A quantifiable and highly reproducible experimental model. Cardiovasc Physiol. 2004;13(2).
Scherrer-Crosbie M, Steudel W, Hunziker PR, Liel-Cohen N, Ullrich R, Zapol WMPM. Three-dimensional echocardiographic assessment of left ventricular wall motion abnormalities in mouse myocardial infarction. J Am Soc Echocardiogr. 1999;12(10):834–40.
Rodrigues AC, Hataishi R, Ichinose F, Bloch K, Duremeaux G, Picard MS-CM. Relationship of systolic dysfunction to area at risk and infarction size after ischemia-reperfusion in mice. J Am Soc Echocardiogr. 2004;17(9):948–53.
Bhan A, Sirker A, Zhang J, et al. High-frequency speckle tracking echocardiography in the assessment of left ventricular function and remodeling after murine myocardial infarction. Am J Hear Ciculation Physiol. 2014;306(9):H1371–83.
Franco F, Dubois SK, Peshock RM, et al. Magnetic resonance imaging accurately estimates LV mass in a transgenic mouse model of cardiac hypertrophy. Am J Hear Ciculation Physiol. 1998;43:H679–83.
Yang X, Liu Y, Rhaleb N, et al. Echocardiographic assessment of cardiac function in conscious and anesthetized mice. Am J Physiol Heart Circ Physiol. 1999;46:H1967–74.
Berry CJ, Thedens DR, Light-mcgroary K, et al. Journal of cardiovascular magnetic effects of deep sedation or general anesthesia on cardiac function in mice undergoing cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2009;11(6):1–7.
Acknowledgements
We would like to thank Dr. Andrew Althouse for guidance of statistical analyses.
Funding
Research reported in this manuscript was supported by: American Heart Association Transformation Project Award 18TPA34230048 and NIH Instrument Grant for Advanced High-Resolution Rodent Ultrasound Imaging System 1S10OD023684-01A1.
Author information
Authors and Affiliations
Contributions
CR obtained, analyzed and interpreted ECHO data and was a major contributor to writing the manuscript. GC, BK, and FV performed manuscript review and helped write the discussion portion. BM contributed to ECHO analysis. LG performed all animal surgeries. YW performed and analyzed CMR. SMN contributed to statistical analysis. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal use was performed at the University of Pittsburgh in compliance with the National Institutes of Health Guide for Care and Use of Experimental Animals and was approved by the University of Pittsburgh Animal Care and Use Committee.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Linear Regression Analysis of 4-Week ECHO Modalities Compared to CMR. Linear regression with correlation coefficients evaluating volumetric measurements of 4D-US, 2D-US, and M-Mode at 4 weeks to CMR at 4 weeks.
Additional file 2: Figure S2
. Transgenic Mouse Lines have No Differences in EDV, ESV, or EF. α-MHC-Cre (+) x Flox-TFAM and α-MHC-Cre (−) x Flox-TFAM mice were compared at baseline and Week 4 following CAL by 4D-US, 2D-US, and M-mode and demonstrate no significant changes by any modality when comparing EDV, ESV, and EF.
Additional file 3: Table S1.
Comparison of ESV, EDV, EF, Scar Size, and survival between transgenic groups.
Additional file 4: Table S2.
Spearman Correlation Values and p-Values Between Modalities for Assessing Scar Size. Histologic Sections, WMSI from 4D-US, Hyperintense Tissue on CMR, Longitudinal Strain, Long Axis Radial Strain, Long-Axis LV Dyssynchrony, Short Axis Radial Strain, and Short Axis Circumferential Strain are compared to each other modality
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Rutledge, C., Cater, G., McMahon, B. et al. Commercial 4-dimensional echocardiography for murine heart volumetric evaluation after myocardial infarction. Cardiovasc Ultrasound 18, 9 (2020). https://doi.org/10.1186/s12947-020-00191-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12947-020-00191-5