Abstract
Background
Scavenger receptor BI (SR-BI) is a classic high-density lipoprotein (HDL) receptor, which mediates selective lipid uptake from HDL cholesterol esters (HDL-C). Apolipoprotein M (ApoM), as a component of HDL particles, could influence preβ-HDL formation and cholesterol efflux. The aim of this study was to determine whether SR-BI deficiency influenced the expression of ApoM.
Methods
Blood samples and liver tissues were collected from SR-BI gene knockout mice, and serum lipid parameters, including total cholesterol (TC), triglyceride (TG), high and low-density lipoprotein cholesterol (HDL-C and LDL-C) and ApoM were measured. Hepatic ApoM and ApoAI mRNA levels were also determined. In addition, BLT-1, an inhibitor of SR-BI, was added to HepG2 cells cultured with cholesterol and HDL, under serum or serum-free conditions. The mRNA and protein expression levels of ApoM were detected by RT-PCR and western blot.
Results
We found that increased serum ApoM protein levels corresponded with high hepatic ApoM mRNA levels in both male and female SR-BI−/− mice. Besides, serum TC and HDL-C were also significantly increased. Treatment of HepG2 hepatoma cells with SR-BI specific inhibitor, BLT-1, could up-regulate ApoM expression in serum-containing medium but not in serum-free medium, even in the presence of HDL-C and cholesterol.
Conclusions
Results suggested that SR-BI deficiency promoted ApoM expression, but the increased ApoM might be independent from HDL-mediated cholesterol uptake in hepatocytes.
Similar content being viewed by others
Background
Atherosclerosis is now one of the most common causes of death around the world, especially in advanced countries. High-density lipoprotein (HDL), which is always considered as a protective factor against atherosclerosis, serves as a vehicle for delivering excess cholesterol esters from peripheral tissues or cells to the liver and steroidgenic tissues. The process is called reverse cholesterol transport (RCT). Scavenger receptor class B type I (SR-BI) is a physiological high-affinity HDL receptor [1, 2]. It is most abundant in the liver, where it mediates selective cholesterol uptake from HDL-cholesterol (HDL-C) to hepatocytes in the later stage of RCT [3, 4]. Studies with mice showed that over-expression of SR-BI results in decreased HDL and total cholesterol (TC) concentrations, suppressing atherosclerosis by increasing RCT [5,6,7,8,9]. In contrast, down-regulation of hepatic SR-BI results in elevated HDL concentration, exaggerating atherosclerosis [10,11,12]. While in human, previous studies have identified that humans who carry mutations in a SCARB1 gene (a gene encoding human SR-BI), exhibited relatively higher plasma HDL-C levels and increased risk of cardiovascular disease [13, 14]. The above observations verified that a key role for hepatic SR-BI in both rodent and human cholesterol metabolism.
Apolipoprotein M (ApoM), an approximately 26 kDa protein, which is a constituent of plasma HDL [15], is mainly expressed in hepatocytes of the liver and tubular epithelial cells in kidney [16, 17]. Recently, many investigations have demonstrated that ApoM plays a critical role in lipid and lipoprotein metabolism. ApoM could delay oxidation of low-density lipoprotein cholesterol (LDL-C), influence preβ-HDL formation, hence mediating the antioxidant effect of HDL and acting as an acceptor of sphingosine-1-phosphate (S1P) [18,19,20,21]. It has been also suggested that ApoM is one of the important regulators of cholesterol metabolism and RCT, because ApoM-enriched HDL increases the ability of HDL to mobilize cellular cholesterol, and accelerates cholesterol efflux from macrophages in vivo [22]. Obviously, ApoM does promote the process of RCT. Since SR-BI is a key factor in RCT, this study was designed to investigate whether deficiency of SR-BI could affect the expression of ApoM, as well as its potential mechanisms.
Methods
Animals
SR-BI−/− mice on a C57BL/6 background were kindly provided by Nanjing Medical University. The genotypes of the offspring of these mice were analyzed using the base-quenched probe method and gel electrophoresis, and all of them were housed in mesh stainless steel cages, maintained on a 12 h light-dark cycle with ad libitum access to a regular chow diet and tap water. The animals used in the experiments were 9–10 weeks old, and were picked out randomly. After 12 h fasting, blood samples for serum lipid and ApoM lipoprotein analysis were drawn into tubes with Na2EDTA and centrifuged for 10 min at 800×g. Serum samples were stored at − 80 °C until analysis. Livers were removed and immediately frozen after animals were sacrificed.
Cell culture
HepG2 cells (American Type Culture Collection, Manassas, VA, USA) were maintained in DMEM containing 10% fetal calf serum. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2. The HepG2 cells were seeded at a density of 6 × 105/well on 6-well plates, and were grown to 70–80% confluence for 24 h before adding BLT-1 (Merck-Millipore, Darmstadt, Germany). Prior to the experiment, cells were washed twice with phosphate-buffered saline (PBS). Cells were cultured in the above medium or serum-free medium with 1% bovine serum albumin (BSA), containing one or more additives, i.e., BLT-1, HDL, and cholesterol, at different concentrations as described in the figure legends. HDL was purchased from Prospect Biosystems (Newark, NJ, USA), and cholesterol was obtained from Sigma-Aldrich (St. Louis, MO, USA).
Genotypic analysis
Genomic DNA was isolated from mouse tail biopsies. Genotyping was performed by the base-quenched probe method [23], followed by melting curve analysis. The results were confirmed by gel electrophoresis [11]. Primers and probes specific for the targeted and wild-type SR-BI alleles are presented in Table 1.
Total RNA extraction and real time RT-PCR
Total RNA from mouse liver tissues or HepG2 cells was isolated according to the manufacturer’s instructions using a total RNA purification kit (Shanghai Shenergy Biotech Co. Ltd., Shanghai, China). First-strand cDNA was synthesized from 2 μg of total RNA using the RevertAid™ first strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). Primer Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA) was used to design the mouse and human ApoM primers and probe used in the TaqMan assay, shown in Table 2. Relative standard curves for ApoM, ApoAI, and GAPDH were generated to compensate for the efficiency of PCR. ApoM and ApoAI mRNA levels were quantified and expressed relative to GAPDH mRNA levels, and were performed on a LightCycler480®II in a final volume of 25 μL. Optimum reaction conditions were obtained with 0.1 μL of 100 μM of each primer and probe, 2.5 μL of 10× PCR buffer, 1.5 μL of 25 mM MgCl2, 0.5 μL of 10 mM 4× dNTPs, 0.25 μL of 5 U/μL Taq DNA polymerase, and 2 μL template cDNA. Finally 17.95 μL ddH2O was added to the reaction mixture. The mixture was preheated at 95 °C for 1 min to activate Taq polymerase, followed by 40 cycles at 95 °C for 5 s and 60 °C for 15 s. Samples were amplified simultaneously in duplicate in one assay run. The threshold cycle (CT) was defined as the fractional cycle number at which the reporter fluorescence reached a certain level.
Western blot analysis
ApoM concentrations in serum and HepG2 cells were determined by western blot analyses [24]. In brief, serum was diluted in PBS (1:50), then 20 μL diluted samples or 20 μg protein extracted from HepG2 cells were separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). After transferring onto polyvinylidene difluoride (PVDF) membranes, the blots were incubated at 37 °C with anti-ApoM or anti-GAPDH antibodies for 2 h, then with horseradish peroxidase-conjugated goat anti-rabbit IgG for 1 h. Bands were visualized by an ECL + Plus western blotting detection system (CW Biotech, Beijing, China) and quantified using a scanner with Quantity One software (Version 4.2.1, Bio-Rad Laboratories, Hercules, CA, USA).
Biochemical analysis
Serum TC, triglyceride (TG), HDL-C and LDL-C were detected by standard clinical chemical methods.
Statistical analysis
Data are expressed as the mean ± SD. Statistical analyses were conducted using GraphPad Prism (version 5.0). Comparisons among groups were analyzed by one-way ANOVA followed by unpaired Student’s t-test. A p value < 0.05 was considered statistically significant.
Results
Genotyping results of SR-BI gene knockout mice
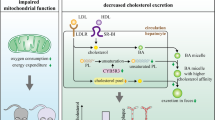
The SR-BI+/− and SR-BI−/− mice looked normal (weight, general appearance, and behavior). Therefore, we designed primers and probes to amplify the targeted segment by two means: the base-quenched probe method and gel electrophoresis. According to the base-quenched probe technique, the mutant allele achieved a perfect match resulting in a high melting temperature of 62.25 ± 0.31 °C, compared with the WT allele (53.25 ± 0.81 °C), while the SR-BI+/− mice showed two melting valleys (Fig. 1a and b). For gel electrophoresis, the amplification of a 330 bp fragment represented the deletion of SR-BI allele, whereas a fragment of 427 bp represented the WT allele, and the presence of both fragments suggested heterozygous mice (Fig. 1c). The results of over 200 mice showed consistency between the two methods.
PCR-based genotyping analysis, confirmed by gel electrophoresis. a Curves of fluorescence (F) versus temperature (T) for sequence-specific base-quenched probe complementary to the SR-BI knockout gene sequence. b Derivative melting curves (-dF/dT vs. T) that depict the same data shown in panel A, wild-type (SR-BI+/+), heterozygous (SR-BI+/−), and homozygous (SR-BI−/−) mutant mice. c Two sets of primer pairs specific for the wild-type (primers 2 and 3) or targeted mutant (primers 2 and 4) alleles were used to screen genomic DNA by PCR as described. Representative results from wild-type (SR-BI+/+, lanes 3 and 4), heterozygous (SR-BI+/−, lanes 5 and 6), and homozygous mutant (SR-BI−/−, lanes 1 and 2) animals are shown
Serum lipid concentrations in mice
As shown in Table 3, serum TC levels in the SR-BI+/− group were significantly increased relative to WT controls, (both males and females, p < 0.01) and extremely higher in the SR-BI−/− group (both males and females, P < 0.001); HDL-C concentrations were also raised in the SR-BI−/− group (males, P < 0.05 and females, P < 0.001); but there was no significant difference in the serum TG levels or in LDL-C concentrations among the different genotypes. However, the SR-BI−/− group showed an obviously abnormal lipid metabolism.
Serum ApoM protein and hepatic ApoM mRNA levels in the different groups
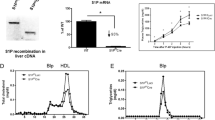
To investigate whether knockout of SR-BI affected the expression of ApoM in vivo, we detected the protein ApoM in the serum of the different groups. Serum ApoM protein levels were significantly higher in the SR-BI−/− group compared to WT controls (P < 0.01) and the SR-BI+/− group (P < 0.05), but there was no significant difference between the SR-BI+/− group and WT controls (Fig. 2a). However, serum ApoAI protein concentrations showed no differences among them (data not shown). Consistent with this, hepatic ApoM mRNA levels were much higher in the SR-BI−/− group than the others. In the livers of SR-BI−/− mice, ApoM levels increased by 186% and 170% in males and females relative to WT controls, respectively (P < 0.001) (Fig. 2b). Similarly to serum, there were no obvious differences in hepatic ApoAI mRNA levels among the groups (data not shown).
Serum ApoM and hepatic ApoM mRNA levels in mice. a Western blot analysis of serum ApoM levels in different groups of SR-BI mice. The SR-BI+/+ males or SR-BI+/+ females were set at 100% as respective controls. b Hepatic ApoM mRNA levels in different groups of SR-BI mice. The SR-BI+/+ males or SR-BI+/+ females were set at 100% as respective controls. n = 5–6 per group, mean ± SD, **P < 0.01 vs. SR-BI+/+ group; *P < 0.05 vs. SR-BI+/+ group; aP < 0.001 vs. SR-BI+/+ group; bP < 0.01 vs. SR-BI+/− group; cP < 0.001 vs. SR-BI+/+ group; dP < 0.001 vs. SR-BI+/+ group
Effects of BLT-1 on the expression of ApoM in HepG2 cells
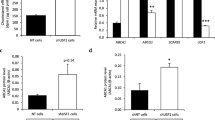
To further validate the effects of SR-BI deficiency on ApoM expression, we treated HepG2 cells with BLT-1, a specific inhibitor of SR-BI. When cells were incubated with 10% fetal calf serum, the mRNA levels of ApoM increased in HepG2 cells treated with 10 μM BLT-1 (P < 0.05) compared to controls (Fig. 3a). Consistent with this, ApoM protein levels were significantly elevated (P < 0.05) (Fig. 3b). When cells were cultured in serum-free medium with 1% BSA, there was no significant change in the expression of ApoM, either in mRNA or in protein levels (Fig. 3c and d).
Effects of BLT-1 treatment on ApoM expression in serum-containing medium or serum-free medium. HepG2 cells were treated with 0, 0.1, 1, 10, or 50 μM BLT-1 in serum-containing medium (a and b) or serum-free medium with 1% BSA (c and d) for 12 h. ApoM mRNA levels were determined by RT-PCR (a and c). ApoM protein levels were measured by western blotting (b and d). Each group contained six replicates, and data are presented as the mean ± SD. The 0 μM group was set at 100% as control. *P < 0.05 vs. 0 μM group; **P < 0.01 vs 0 μM group; ***P < 0.001 vs 0 μM group
Effects of BLT-1, HDL and cholesterol on the expression of ApoM in HepG2 cells
As shown in Fig. 4, there was no difference in mRNA levels of ApoM among groups including cells treated with BLT-1 alone, with HDL-C and cholesterol, and with HDL-C, cholesterol together with BLT-1.
Effects of HDL-C, cholesterol, and BLT-1 on ApoM expression in HepG2 cells. ApoM mRNA levels in cells cultured with HDL-C, cholesterol and BLT-1 for 12 h were measured by RT-PCR. The control group without HDL-C, cholesterol or BLT-1 was set as 100%. n = 6 for each group, data are presented as the mean ± SD
Discussion
The aim of the present study was to explore whether lack of SR-BI could affect the expression of ApoM. Our results indicated that the absence of SR-BI did up-regulate ApoM expression, both in mRNA and protein levels in SR-BI gene knockout mice (Fig. 2), which suggested SR-BI could regulate expression of ApoM. To investigate the potential mechanism, we used BLT-1, a specific inhibitor of SR-BI, to treat HepG2 hepatoma cells. The results showed the mRNA levels of ApoM increased significantly in HepG2 cells treated with 10 μM BLT-1 in presence of 10% fetal calf serum, but not in absence of serum (Fig. 3a). These results strongly suggested that BLT-1 may prevent some serum factors not yet identified transferring into cells, which could influence the expression of ApoM. It should be point out that the high concentrations of BLT-1 induced cell death, for cells treated with 50 μM BLT-1 were mostly detached (Fig. 3). To further validate whether the regulation of ApoM is due to RCT induced by SR-BI in hepatocytes, we treated HepG2 cells with HDL-C, cholesterol and BLT-1 in serum-free medium with 1% BSA, respectively. The results showed that ApoM expression was not affected by HDL-C and cholesterol, which indicated that cholesterol metabolism might not regulate ApoM expression. Even though BLT-1 was used to inhibit the function of SR-BI, ApoM expression was not changed significantly (Fig. 4). It suggested SR-BI-mediated selective cholesterol uptake did not involve in regulation of ApoM.
In the present study, we found lack of SR-BI caused TC and HDL-C accumulation, which was consistent with previous studies [11]. As s constituent of HDL, serum ApoM level also increased in SR-BI deficient mice. We consider that this phenomenon may due to the absence of SR-BI, which causes negative feedback and leads to up-regulation of HDL-C, including ApoM. Further investigation demonstrated that inhibition of SR-BI by BLT-1 also elevated ApoM expression in HepG2 cells treated with medium containing fetal calf serum. Interestingly, the phenomenon was not observed in medium without serum. These results implied there may be some feedback regulators existing in serum. In conclusion, deficiency of SR-BI could up-regulate ApoM, which may be due to negative feedback rather than HDL-mediated cholesterol uptake in hepatocytes. The detailed mechanism remains to be elucidated.
Abbreviations
- ApoM:
-
Apolipoprotein M
- DMEM:
-
Dulbecco’s Modified Eagle’s Medium
- FBS:
-
Fetal bovine serum
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- HDL-C:
-
High density lipoprotein-cholesterol
- LDL-C:
-
Low density lipoprotein-cholesterol
- PBS:
-
Phosphate buffered saline
- PCR:
-
Polymerase chain reaction
- TC:
-
Total cholesterol
- TG:
-
Triglyceride
References
Acton SL, Scherer PE, Lodish HF, Krieger M. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J Biol Chem. 1994;269:21003–9.
Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–20.
Cho S, Park EM, Febbraio M, Anrather J, Park L, Racchumi G, Silverstein RL, Iadecola C. The class B scavenger receptor CD36 mediates free radical production and tissue injury in cerebral ischemia. J Neurosci. 2005;25:2504–12.
Out R, Hoekstra M, Spijkers JA, Kruijt JK, van Eck M, Bos IS, Twisk J, Van Berkel TJ. Scavenger receptor class B type I is solely responsible for the selective uptake of cholesteryl esters from HDL by the liver and the adrenals in mice. J Lipid Res. 2004;45:2088–95.
Wang N, Arai T, Ji Y, Rinninger F, Tall AR. Liver-specific overexpression of scavenger receptor BI decreases levels of very low density lipoprotein ApoB, low density lipoprotein ApoB, and high density lipoprotein in transgenic mice. J Biol Chem. 1998;273:32920–6.
Ueda Y, Royer L, Gong E, Zhang J, Cooper PN, Francone O, Rubin EM. Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J Biol Chem. 1999;274:7165–71.
Ueda Y, Gong E, Royer L, Cooper PN, Francone OL, Rubin EM. Relationship between expression levels and atherogenesis in scavenger receptor class B, type I transgenics. J Biol Chem. 2000;275:20368–73.
Kozarsky KF, Donahee MH, Glick JM, Krieger M, Rader DJ. Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arterioscler Thromb Vasc Biol. 2000;20:721–7.
Arai T, Wang N, Bezouevski M, Welch C, Tall AR. Decreased atherosclerosis in heterozygous low density lipoprotein receptor-deficient mice expressing the scavenger receptor BI transgene. J Biol Chem. 1999;274:2366–71.
Varban ML, Rinninger F, Wang N, Fairchild-Huntress V, Dunmore JH, Fang Q, Gosselin ML, Dixon KL, Deeds JD, Acton SL, et al. Targeted mutation reveals a central role for SR-BI in hepatic selective uptake of high density lipoprotein cholesterol. Proc Natl Acad Sci U S A. 1998;95:4619–24.
Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci U S A. 1997;94:12610–5.
Mardones P, Quinones V, Amigo L, Moreno M, Miquel JF, Schwarz M, Miettinen HE, Trigatti B, Krieger M, VanPatten S, et al. Hepatic cholesterol and bile acid metabolism and intestinal cholesterol absorption in scavenger receptor class B type I-deficient mice. J Lipid Res. 2001;42:170–80.
Brunham LR, Tietjen I, Bochem AE, Singaraja RR, Franchini PL, Radomski C, Mattice M, Legendre A, Hovingh GK, Kastelein JJ, Hayden MR. Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans. Clin Genet. 2011;79:575–81.
Khovidhunkit W. A genetic variant of the scavenger receptor BI in humans. N Engl J Med. 2011;364:1375–6. author reply 1376
Christoffersen C, Nielsen LB, Axler O, Andersson A, Johnsen AH, Dahlback B. Isolation and characterization of human apolipoprotein M-containing lipoproteins. J Lipid Res. 2006;47:1833–43.
Zhang XY, Dong X, Zheng L, Luo GH, Liu YH, Ekstrom U, Nilsson-Ehle P, Ye Q, Xu N. Specific tissue expression and cellular localization of human apolipoprotein M as determined by in situ hybridization. Acta Histochem. 2003;105:67–72.
Zhang XY, Jiao GQ, Hurtig M, Dong X, Zheng L, Luo GH, Nilsson-Ehle P, Ye Q, Xu N. Expression pattern of apolipoprotein M during mouse and human embryogenesis. Acta Histochem. 2004;106:123–8.
Wolfrum C, Poy MN, Stoffel M. Apolipoprotein M is required for prebeta-HDL formation and cholesterol efflux to HDL and protects against atherosclerosis. Nat Med. 2005;11:418–22.
Christoffersen C, Jauhiainen M, Moser M, Porse B, Ehnholm C, Boesl M, Dahlback B, Nielsen LB. Effect of apolipoprotein M on high density lipoprotein metabolism and atherosclerosis in low density lipoprotein receptor knock-out mice. J Biol Chem. 2008;283:1839–47.
Christoffersen C, Obinata H, Kumaraswamy SB, Galvani S, Ahnstrom J, Sevvana M, Egerer-Sieber C, Muller YA, Hla T, Nielsen LB, Dahlback B. Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M. Proc Natl Acad Sci U S A. 2011;108:9613–8.
Elsoe S, Ahnstrom J, Christoffersen C, Hoofnagle AN, Plomgaard P, Heinecke JW, Binder CJ, Bjorkbacka H, Dahlback B, Nielsen LB. Apolipoprotein M binds oxidized phospholipids and increases the antioxidant effect of HDL. Atherosclerosis. 2012;221:91–7.
Elsoe S, Christoffersen C, Luchoomun J, Turner S, Nielsen LB. Apolipoprotein M promotes mobilization of cellular cholesterol in vivo. Biochim Biophys Acta. 2013;1831:1287–92.
Luo G, Zheng L, Zhang X, Zhang J, Nilsson-Ehle P, Xu N. Genotyping of single nucleotide polymorphisms using base-quenched probe: a method does not invariably depend on the deoxyguanosine nucleotide. Anal Biochem. 2009;386:161–6.
Xu N, Nilsson-Ehle P, Hurtig M, Ahren B. Both leptin and leptin-receptor are essential for apolipoprotein M expression in vivo. Biochem Biophys Res Commun. 2004;321:916–21.
Acknowledgements
We thank International Science Editing for language editing.
Ethical approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (81201352), Jiangsu Provincial Natural Science Foundation of China (BK2012154, BK20151179), Changzhou High-Level Medical Talents Training Project (No: 2016ZCLJ002 and 2016ZCLJ019), and Changzhou International Scientific and Technological Cooperation Project (CZ20150012).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
YHF performed the experiments, analyzed the results and wrote the manuscript. LZ supervised the study and provided technical support. JW, MMY, JZ helped in performing the study. GHL designed the study, and did proof read the paper. NX provided guidance on the whole study. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Feng, YH., Zheng, L., Wei, J. et al. Increased apolipoprotein M induced by lack of scavenger receptor BI is not activated via HDL-mediated cholesterol uptake in hepatocytes. Lipids Health Dis 17, 200 (2018). https://doi.org/10.1186/s12944-018-0849-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12944-018-0849-7