Abstract
Background
Escherichia coli (E. coli) O25b/ST131 clone causes urinary tract infection (UTI) and is associated with a broad spectrum of other infections, such as intra-abdominal and soft tissue infections, that can be affecting bloodstream infections. Therefore, since O25b/ST131 has been reported in several studies from Iran, in the current study, we have investigated the molecular characteristics, typing, and biofilm formation of O25b/ST131 clone type E. coli collected from UTI specimens.
Methods
A total of 173 E. coli isolates from UTI were collected. The susceptibility to all fourth generations of cephalosporins (cefazolin, cefuroxime, ceftriaxone, cefotaxime, ceftazidime, cefepime) and ampicillin, ampicillin-sulbactam and aztreonam was determined. Class A ESBLs, class D ESBL and the presence of pabB gene screenings to detect of O25b/ST131 clone type were performed by using of PCR. Biofilm formation was compared between O25b/ST131 isolates and non-O25b/ST131 isolates. Finally, ERIC-PCR was used for typing of ESBL positive isolates.
Results
Ninety-four ESBL positive were detected of which 79 of them were O25b/ST131. Antimicrobial susceptibility test data showed that most antibiotics had a higher rate of resistance in isolates of the O25b/ST131 clonal type. Biofilm formation showed that there was a weak association between O25b/ST131 clone type isolates and the level of the biofilm formation. ERIC-PCR results showed that E. coli isolates were genetically diverse and classified into 14 groups.
Conclusion
Our results demonstrated the importance and high prevalence of E. coli O25b/ST131 among UTI isolates with the ability to spread fast and disseminate antibiotic resistance genes.
Similar content being viewed by others
Introduction
Uropathogenic Escherichia coli (UPEC) strains associated with a high incidence of community-acquired and hospital-acquired UTIs [1]. There are 150 million UTIs worldwide each year and drug-resistant infections typically require more complex treatment regimens and are more likely to occur if treatment is unsuccessful [2,3,4]. Furthermore, 70–95% of community-acquired UTIs are caused by UPEC which is the second most common infection in the community [4, 5]. The UTIs is the main cause of E.coli bloodstream infections leading, to 40,000 deaths from sepsis every year in the US [2, 6]. Moreover, E. coli, which especially causes extraintestinal infections, becomes resistant to every class of antibiotics used to treat such infections [7]. Unfortunately, the capacity of UPEC to obtain multiple drug resistance, particularly board-spectrum β-lactamases, may impede the therapeutic control of infections [1]. The clonal extension is an essential factor related to the diffusion of extended-spectrum beta-lactamases (ESBLs) producing E. coli isolates, mainly giving rise to the spread of multidrug-resistant (MDR) strains [8]. The CTX-M-type beta-lactamase enzyme, especially CTX-M-15, is the predominant ESBL, and is often found in E. coli sequence type 131 (ST131) [9]. E. coli ST131 was first discovered in 2008 based on the sequences inside seven E. coli housekeeping genes described as Multilocus Sequence Typing (MLST). According to MLST and molecular methods, such as PCR, studies have shown that the ST131 clone is an important human pathogen worldwide [10]. The E. coli ST131 clone causes UTI and is associated with a wide spectrum of other infections, such as bloodstream, soft tissue infections, and intra-abdominal, as well as epididymal-orchitis, meningitis, and septic shock [11]. In addition, E.coli forms a biofilm that is associated with the pathogenesis of diarrheagenic E. coli [12]. Biofilm formation by extraintestinal pathogenic E. coli (ExPEC) was largely observed in UPEC [13]. Therefore, as the few studies reported O25b/ST131 from Iran and the purpose of this study is to investigate the genetic characteristics, types, and biofilm formation methods of E. coli in O25b/ST131 clones to determine the extent of resistance and distribution of the most resistant clone.
Methods and materials
Bacterial strains
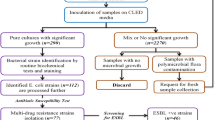
A total of 173 clinical isolates were collected from two selective hospitals in Tehran. The 94 ESBLs isolates were selected among them which, recovered from 33 male and 61 female. These isolates were collected from both outpatients and hospitalized patients over a period of 9 months from October 2018 to June 2019. In addition, all isolates recovered from urine and one strain per patient was investigated.
Isolation and identification of E. coli isolates
All strains were isolated on MacConkey’s agar (Conda lab, Spain) and genotypically confirmed by amplification of the 16S rDNA gene by using ECO primers (Table 1) [14]. The Metabion (Germany) primers were used in this study.
ESBL-confirmatory testing
The antibiotic disks BD (USA) and Mast (UK) were used to determining susceptibility profiles using the disk diffusion method. Furthermore, by using cefotaxime and ceftazidime with and without clavulanic acid disks determination of the ESBLs isolates was performed in accordance with Clinical and Laboratory Standards Institute (CLSI, 2020) guidelines. Increasing of ≥ 5-mm in zone diameter of the ceftazidime-clavulanic acid compared to the zone diameter of ceftazidime considered as ESBLs [15, 16].
Antimicrobial susceptibility testing
We also performed additional susceptibility testing on positive ESBL isolates using the following disks: cefazolin (30 μg), cefuroxime (30 μg), ceftriaxone (30 μg), cefepime (30 μg), aztreonam (30 μg), ampicillin (10 μg), and ampicillin-sulbactam (10/10 μg).
Detection of antibiotic resistance genes
DNA was extracted by the boiling method using TE buffer as previously described [17]. All 94 ESBLs isolates were screened for class A ESBLs (blaGES, blaSHV, blaCTX-M, blaVEB, blaPER, and blaTEM) and class D ESBL (blaOXA-10) using PCR. In addition, the type of clone O25b/ST131 was detected by confirming the presence of the pabB gene [18, 19]. All the primers for the mentioned β-lactamase genes are listed in Table 1.
Biofilm formation assay and quantification
The biofilm formation was performed in a 96-well polystyrene plate containing 10 O25b/ST131 positive strains, 10 randomly selected negative O25b/ST131 strains and the E.coli ATCC 25922 as control. Isolates were incubated overnight in Tryptic Soy Broth (TSB) media (Conda lab, Spain) and then the optical density (OD) of each isolate was adjusted between 0.4 and 0.6 at 600 nm. Furthermore, 190 µl of TSB broth containing of 10 µl of bacterial suspension was added to each well. Incubation was performed overnight at 37 ℃ with continuous shaking at 30 rpm. Biofilm assay for each isolates was performed in triplicate using TSB broth as the negative control. Moreover, after incubation the wells were washed with distilled water, stained with 0.1% crystal violet, and left at room temperature 10 min. After incubation, wells were washed 3 times with distilled water. Eventually, 200 µl of 95% ethanol was added to wells, and the OD was measured at 492 nm using an ELISA reader had measured. The OD values were considered as an index of biofilm formation. Quantitative analysis to evaluate the biofilm formation was performed by calculating the average absorbance of the control wells (Ac) that subtracted from the A492 nm of all test wells. Mean values and standard deviations were calculated for all experiments. Isolates characterized as (4 × Ac) < A = strong biofilm producer, (2 × Ac) < A ≤ (4 × Ac) = moderate biofilm producer, Ac < A ≤ (2 × Ac) = weak biofilm producer and A ≤ Ac = no biofilm producer [20, 21].
ERIC-PCR typing
ERIC-PCR was performed to evaluate the genetic relationship between ESBLs isolates. Each PCR reaction mixture in a total volume of 20 µL contained: 1 μl of each primer, 10 μl of the master mix (Ampliqon, Denmark), 3.5 μl of template DNA, and 4.5 μl of deionized water. The reaction was as follows: initial denaturation at 94 °C for 1 min, with the 30 cycles, denaturation step at 94 °C for 30 s, annealing at 52 °C for 35 s, extension at 72 °C for 4 min, and final extension for 5 min at 72 °C. The amplicon was electrophoresed on a 1.2–1.5% (w/v) agarose gel containing a safe stain (Yekta Tajhiz Azma, Iran) at 90 V for 90 min. The [DM2100] ExcelBand 100 bp DNA Ladder (Smobio, Taiwan) were used as marker [22, 23].
To calculate the placement and visibility of the gels were assessed by ERIC-PCR according to their molecular weights and molecular markers. Electrophoretic patterns were calculated using BioNumerics gel analysis software (Applied Maths, Belgium). Gel-to-gel banding pattern comparison was performed, to ensure adequateness; the analysis contains a normalization step, that makes each lane an equal length. The “band scoring” process identifies bands of each lane that combined to generate the fingerprint-based on the threshold of stringency and optimization settings, set at 1.0%. By using the Bionumerics, the design of a phylogenetic tree for isolated strains was performed via the presence of a broad range of genetic heterogeneities among their populations. The cut-off for cluster definition was 50%.
Statistical analysis
All of E. coli isolates data were collected and entered into SPSS software, v. 22.0 (SPSS inc., USA) for analysis. Interpretation of demographic information was based on frequency. The association between different genes, antibiotic resistance, and O25b/ST131 clone type were evaluated by using the chi-square (χ2) test. The eta (η) correlation ratio was determined to investigate the association between the O25b/ST131 clone type and the level of biofilm formation. The level of statistical significance was set at p ≤ 0.05.
Results
Antibiotic susceptibility and resistance determinants
In this study, among 173 E. coli isolated from UTI, ESBL-producer isolates were included in the study for further testing. Of the 173 isolates causing UTI, 94 (54.3%) isolates were resistant to one of four-generation of cephalosporins. Of 94 isolates, 35 (37.2%) were isolated from men and 59 (62.8%) were isolated from women. The highest resistance in all isolates was observed to ampicillin (97.9%). Most of the tested antibiotics had susceptibility rate between 1 and 10.6% also, highest susceptibility observed in ceftazidime, cefepime and aztreonam with rates of 72.3%, 52.1% and 47.9%, respectively. Resistance to cefazolin as the first generation of cephalosporin cefuroxime as the second generation was 93.6% and 90.4%, respectively. Although 87.2% and 90.4% of the isolates were resistant to ceftriaxone and cefotaxime, respectively, as the third generation of cephalosporins, only 25.5% of the isolates were resistant to ceftazidime. Resistance to cefepime, a fourth-generation cephalosporin, was also confirmed in 47.9% of isolates. Details of the antibiotic susceptibility test can be found in Table 2.
Prevalence of O25b/ST131 clonal group and ESBLs-encoding genes
Based on the fact that 94 isolates were resistant to one of the cephalosporins, they underwent ESBL production identification test, which revealed that all of these isolates were phenotypically ESBL-producing. Molecular technique for determining the presence of the ESBL genes showed that 52 (55.3%), 79 (84%), 89 (94.6%), 20 (21.3%), 19 (20.2%), 83 (88.3%), and 4 (4.3%) isolates harbored of blaTEM, blaSHV, bla-CTX-M-1, bla-CTX-M-2, bla-CTX-M-9, bla-CTX-M-15, and blaPER, genes, respectively. On the other hand, bla-CTX-M-8, bla-CTX-M-25, and blaVEB genes were not found in any of the isolates. Also, the study of blaGES and blaOXA genes, revealed that none of the isolates carrying these two carbapenemase genes. Using a special primer (pabB) 79 (84%) isolates were determined to belong to the O25b/ST131 clone type.
Biofilm formation assay
Six out of 10 E. coli O25b/ST131 isolates, had ability to form a biofilm, of which 3, 2, and 1 isolates could form strong, moderate, and weak biofilms, respectively. In addition, the weak relationship between O25b/ST131 clone type isolates and the level of the biofilm formation was observed (Eta = 0.243).
Statistical analysis
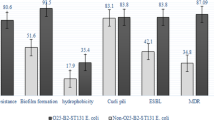
Analysis of antibiotic susceptibility test data showed that, with the exception of ampicillin and ampicillin-sulbactam, the O25b/ST131 clone type isolates had a higher rate of resistance to other antibiotics than the ESBLs samples (Table 2). In addition, it was found that the percentage of resistance genes in O25b/ST131 clone type isolates was higher than in non-O25b/ST131 isolates, and statistical analysis of the data showed a significant association between the presence of bla-CTX-M-1, bla-CTX-M-2, bla-CTX-M-9, bla-CTX-M-15, blaPER, genes and O25b/ST131 clone type isolates (P < 0.05). Figure 1 details of the percentage of resistance genes present in O25b/ST131 clone type isolates compared to non-O25b/ST131 isolates.
ERIC-PCR profiles of ESBLs isolates
Bands were calculated for each sample and a high genetic diversity of E.coli was found. The genotyping profiles of 84 ESBL E. coli strains in accordance with ERIC-PCR fingerprinting is shown in Fig. 2, and fourteen groups were formed using ERIC-PCR fingerprinting, but 10 strains were not typable. In the studied strains, 22, belonged to the E10 cluster, and the minimum, 1, belonged to the E3 and E4 cluster. Two of the strains were in the E1, E2, and E7. Fourteen were in the E14, 6 strains in the E5, 8 strains in the E12, 3 strains in the E8, 5 strains in the E9, 7 strains in the E11 and the E13 cluster. The predominant DNA fingerprints fragments was identified with the size 750 bp, which was found in 70 strains, and the least frequent, a size of 170 bp, was observed in 2 strains. ERIC-PCR demonstrated that the isolates investigated in the this study had a wide range of genetic diversities and this method showed a good sensitivity in detecting slight differences between isolates. Studies and comparisons of dendrograms and antibiotic susceptibility test results have provided valuable results; i.e., samples in the E10 cluster were sensitive to ceftazidime, cefepime, and aztreonam. The ERIC-PCR banding patterns have shown 0 to 30 bands encompassing 150 bp to 3000 bp.
Discussion
The number of ESBL-producing bacteria has increased over the past decade, and to control the infections and selection of the most suitable antibiotics showed the importance of detection of these isolates. Furthermore, new policies are required to restrict these isolates spread, especially in a hospital environment [30, 33]. Until the 1990s, the ESBL genes were mostly detected in Klebsiella pneumoniae rather than E. coli but, in recent years, it has been mainly found in E. coli isolates [34]. Previous studies also a study Iran report well-establish relation between ST131 and ESBL production [35, 36] therefore, this study designed to investigate the prevalence of ESBL-producing bacteria belonged to O25b/ST131 clone type among clinical isolates collected from two selective hospitals in Tehran. Of 173 isolates, 94 (54.3%) carried the ESBL genes with the most detected gene (94.6%) being bla-CTX-M-1. In the current study, antimicrobial resistance was more frequent in the O25b/ST131 clones than non-O25b/ST131 isolates and high resistance to all four generations of cephalosporins was detected. Furthermore, two studies in the United Kingdom and Iran reported the same cephalosporins with resistance rates of 68% and 49.5%, respectively [37, 38]. Moreover, there are several reports in recent years pointed out the importance of ST131 as a major clone for extraintestinal E. coli infections [8, 39,40,41].
Johnson et al. reported that 42.51% of investigated E. coli isolates were ST131, of which 67–69% were resistant to extended-spectrum cephalosporins or fluoroquinolones [39]. Furthermore, our study demonstrated 84% of isolates belonged to O25b/ST131, and 89.8% were resistant to extended-spectrum cephalosporins. In addition, other studies confirmed the close association between ST131 and ESBL production also, a recent meta-analysis study demonstrated the high prevalence of broadly disseminated ST131 clone among ESBLs isolates in the western Asia region. Additionally, Iran reported with highest MDR-ST131 isolates in this region, which is similar to our results [35, 36, 42].
The frequency of ESBLs genes, especially the blaCTX-M-15 has posed a serious threat to public health [43, 44]. Our result indicated that the frequency of ESBLs genes was higher in O25b/ST131 clone type than non-O25b/ST131 isolates, especially in blaCTX-M genes. ST131 clone type is known worldwide for its role in the dissemination of ESBLs genes, especially blaCTX-M-15 [36].
Shin et al. reported that the existence of the plasmid harboring blaCTX-M could be a major factor related to the emerging and dissemination of pandemic multi-resistant E. coli ST131. In addition, they demonstrated that isolates with plasmids harboring blaCTX-M-14 or blaCTX-M-15 showed raise to cephalosporin MICs, in comparison to susceptible hosts. Additionally, they showed high MICs of ampicillin, aztreonam, gentamicin, and piperacillin/tazobactam [45].
Our study showed similar results with the prevalence of blaCTX-M-15 was 96.2% in O25b/ST131 clone type isolates and high resistance to ceftriaxone, cefotaxime, cefuroxime, cefazolin and ampicillin was observed. Furthermore, the similar study conducted in Iran reports a 95.5% prevalence of blaCTX-M-15 gene among ESBL positive O25b/ST131 isolates [46].
In addition, a significant prevalence difference in other CTX genes between O25b/ST131 and non-O25b/ST131 isolates was observed. This result is in correlation with the Overdevest et al. study supporting the idea of O25/ST131 success are associated with the ESBL phenotype [47]. Moreover, we selected 10 O25b/ST131 clone type isolates and 10 non-O25b/ST131 and although in vitro biofilm formation is strongly depending on the method, there was a weak association between O25b/ST131 clone type isolates and the level of the biofilm formation, which is correlated with other studies that confirmed the ST131 isolates carrying blaCTX-M-15 were unable to develop mature biofilm [48].
For genotyping of isolates, ERIC-PCR method was considered with 50% cut-off for cluster definition, which is faster and more cost-effective rather than other techniques. Also, ERIC-PCR showed a higher discriminatory ability in comparison to other quick-typing techniques [30, 49]. As a result of genotyping, the E. coli ST131 isolate was genetically diverse and heterogeneous expected; since the isolates were collected from two different hospitals over a 9 months. We detected 14 groups of E. coli from 84 isolates and the most of isolates were classified into 6 groups that showed similar profiles which can be explained as the clonal transmission of our isolates. Similarly, another study from Iran reported the high diversity among 230 E. coli isolates, collected from two selective hospitals [50].
This study has several limitations including, the isolates being collected from one city rather than the whole country, which limit the generalizability of our data. On the other, more multicenter study are needed in the future to determine the other features of this clone type. The suitable genotyping method is MLST and could be the next step in near future.
In summary, the current study demonstrated once again the importance of E. coli O25b/ST131 clone as a type of clone capable of rapidly spreading and disseminate antibiotic resistance genes. Moreover, the study of various mechanisms in this clone type is useful to prevent the transmission of antimicrobial resistance genes, especially with the increasing rate of resistance to colistin among this clone [51]. Our study detected O25b/ST131 with a high resistance rate among clinical isolates of E. coli and ESBLs genes among them.
Availability of data and materials
All data were included.
References
Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–84.
Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis Mon. 2003;49(2):53–70.
Alam MF, Cohen D, Butler C, Dunstan F, Roberts Z, Hillier S, et al. The additional costs of antibiotics and re-consultations for antibiotic-resistant Escherichia coli urinary tract infections managed in general practice. Int J Antimicrob Agents. 2009;33(3):255–7.
Mirkalantari S, Masjedian F, Irajian G, Siddig EE, Fattahi A. Determination of the frequency of β-lactamase genes (bla SHV, bla TEM, bla CTX-M) and phylogenetic groups among ESBL-producing uropathogenic Escherichia coli isolated from outpatients. J Lab Med. 2020;44(1):27–33.
Ghazvini H, Taheri K, Edalati E, Miri A, Sedighi M, Mirkalantari S. Virulence factors and antimicrobial resistance in uropathogenic Escherichia coli strains isolated from cystitis and pyelonephritis. Turkish J Med Sci. 2019;49(1):361–7.
Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000;181(5):1753–4.
Gupta K, Scholes D, Stamm WE. Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA. 1999;281(8):736–8.
Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66(1):1–14.
Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon J-M, Garry L, et al. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS ONE. 2012;7(9): e46547.
Ismail MD, Ali I, Hatt S, Salzman EA, Cronenwett AW, Marrs CF, et al. Association of Escherichia coli ST131 lineage with risk of urinary tract infection recurrence among young women. J Global Antimicrob Resist. 2018;13:81–4.
Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27(3):543–74.
Sherlock O, Schembri MA, Reisner A, Klemm P. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J Bacteriol. 2004;186(23):8058–65.
Totsika M, Beatson SA, Sarkar S, Phan M-D, Petty NK, Bachmann N, et al. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS ONE. 2011;6(10): e26578.
Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi MP, et al. A distinctive’microbial signature’in celiac pediatric patients. BMC Microbiol. 2010;10(1):175.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Wayne PA: Clinical and Laboratory Standards Institute; 2020.
Bahramian A, Khoshnood S, Shariati A, Doustdar F, Chirani AS, Heidary M. Molecular characterization of the pilS2 gene and its association with the frequency of Pseudomonas aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island. Infect Drug Resist. 2019;12:221.
Pourhajibagher M, Mokhtaran M, Esmaeili D, Bahador A. Antibiotic resistance patterns among Acinetobacter baumannii strains isolated from burned patients. Der Pharmacia Lettre. 2016;8(8):347–51.
Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother. 2009;64(2):274–7.
Shariati A, Razavi S, Ghaznavi-Rad E, Jahanbin B, Akbari A, Norzaee S, et al. Association between colorectal cancer and Fusobacterium nucleatum and Bacteroides fragilis bacteria in Iranian patients: a preliminary study. Infect Agents Cancer. 2021;16(1):41.
Hemati S, Azizi-Jalilian F, Pakzad I, Taherikalani M, Maleki A, Karimi S, et al. The correlation between the presence of quorum sensing, toxin-antitoxin system genes and MIC values with ability of biofilm formation in clinical isolates of Pseudomonas aeruginosa. Iranian J Microbiol. 2014;6(3):133.
Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin–antitoxin systems. Nat Rev Microbiol. 2011;9(11):779–90.
Ardakani MA, Ranjbar R. Molecular typing of uropathogenic E. coli strains by the ERIC-PCR method. Electron Physician. 2016;8(4):2291.
Duan H, Chai T, Liu J, Zhang X, Qi C, Gao J, et al. Source identification of airborne Escherichia coli of swine house surroundings using ERIC-PCR and REP-PCR. Environ Res. 2009;109(5):511–7.
Cambau E, Lascols C, Sougakoff W, Bebear C, Bonnet R, Cavallo J-D, et al. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002–2005. Clin Microbiol Infect. 2006;12(10):1013–20.
Memariani M, Najar Peerayeh S, Zahraei Salehi T, Shokouhi Mostafavi SK. Occurrence of SHV, TEM and CTX-M β-Lactamase genes among enteropathogenic Escherichia coli strains isolated from children with diarrhea. Jundishapur J Microbiol. 2015;8(4): e15620.
Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–5.
Chen C-M, Ke S-C, Li C-R, Chiou C-S, Chang C-C. Prolonged clonal spreading and dynamic changes in antimicrobial resistance of Escherichia coli ST68 among patients who stayed in a respiratory care ward. J Med Microbiol. 2014;63(11):1531–41.
Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, et al. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50(4):1178–82.
Hong SS, Kim K, Huh JY, Jung B, Kang MS, Hong SG. Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann Lab Med. 2012;32(5):359–61.
El-Badawy MF, Tawakol WM, Maghrabi IA, Mansy MS, Shohayeb MM, Ashour MS. Iodometric and molecular detection of ESBL production among clinical isolates of E. coli fingerprinted by ERIC-PCR: the first Egyptian report declares the emergence of E. coli O25b-ST131clone harboring bla GES. Microb Drug Resist. 2017;23(6):703–17.
Bhattacharjee A, Sen MR, Prakash P, Anupurba S. Role of β-lactamase inhibitors in enterobacterial isolates producing extended-spectrum β-lactamases. J Antimicrob Chemother. 2008;61(2):309–14.
Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 1991;19(24):6823–31.
Shaikh S, Fatima J, Shakil S, Rizvi SMD, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J Biol Sci. 2015;22(1):90–101.
Behrooozi A, Rahbar M, Jalil V. Frequency of extended spectrum beta-lactamase (ESBLs) producing Escherichia coli and Klebseilla pneumonia isolated from urine in an Iranian 1000-bed tertiary care hospital. Afr J Microbiol Res. 2010;4(9):881–4.
Banerjee R, Robicsek A, Kuskowski MA, Porter S, Johnston BD, Sokurenko E, et al. Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-β-lactamase-positive and-negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob Agents Chemother. 2013;57(12):6385–8.
Hojabri Z, Mirmohammadkhani M, Kamali F, Ghassemi K, Taghavipour S, Pajand O. Molecular epidemiology of Escherichia coli sequence type 131 and its H30/H30-Rx subclones recovered from extra-intestinal infections: first report of OXA-48 producing ST131 clone from Iran. Eur J Clin Microbiol Infect Dis. 2017;36(10):1859–66.
Ciesielczuk H, Doumith M, Hope R, Woodford N, Wareham DW. Characterization of the extra-intestinal pathogenic Escherichia coli ST131 clone among isolates recovered from urinary and bloodstream infections in the United Kingdom. J Med Microbiol. 2015;64(12):1496–503.
Rasoulinasab M, Shahcheraghi F, Feizabadi MM, Nikmanesh B, Hajihasani A, Sabeti S, et al. Distribution of pathogenicity island markers and h-antigen types of Escherichia coli O25b/ST131 isolates from patients with urinary tract infection in iran. Microb Drug Resist. 2021;27(3):369–82.
Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis. 2010;51(3):286–94.
Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol. 2013;34(4):361–9.
Cagnacci S, Gualco L, Debbia E, Schito GC, Marchese A. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25: H4-ST 131 and O15: K52: H1 causing community-acquired uncomplicated cystitis. J Clin Microbiol. 2008;46(8):2605–12.
Jafari A, Falahatkar S, Delpasand K, Sabati H, Sedigh E-S. Emergence of Escherichia coli ST131 causing urinary tract infection in Western Asia: a systematic review and meta-analysis. Microb Drug Resist. 2020;26(11):1357–64.
Pana ZD, Zaoutis T. Treatment of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBLs) infections: what have we learned until now? F1000Res. 2018;7.
Goudarzi H, Aghamohammad S, Hashemi A, Nikmanesh B, Noori M. Distribution of blaTEM, blaSHV and blaCTX-M genes among Escherichia coli isolates causing urinary tract infection in children. Arch Clin Infect Dis. 2013;8(3):e16207. https://doi.org/10.5812/archcid.16207.
Shin J, Ko KS. Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int J Antimicrob Agents. 2015;46(2):214–8.
Namaei MH, Yousefi M, Ziaee M, Salehabadi A, Ghannadkafi M, Amini E, et al. First report of prevalence of CTX-M-15-producing Escherichia coli O25b/ST131 from Iran. Microb Drug Resist. 2017;23(7):879–84.
Overdevest I, Bergmans A, Verweij J, Vissers J, Bax N, Snelders E, et al. Prevalence of phylogroups and O25/ST131 in susceptible and extended-spectrum β-lactamase-producing Escherichia coli isolates, the Netherlands. Clin Microbiol Infect. 2015;21(6):570.e4-4.
Novais Â, Pires J, Ferreira H, Costa L, Montenegro C, Vuotto C, et al. Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother. 2012;56(7):3973–6.
Meacham KJ, Zhang L, Foxman B, Bauer RJ, Marrs CF. Evaluation of genotyping large numbers of Escherichia coli isolates by enterobacterial repetitive intergenic consensus-PCR. J Clin Microbiol. 2003;41(11):5224–6.
Ramazanzadeh R, Zamani S, Zamani S. Genetic diversity in clinical isolates of Escherichia coli by enterobacterial repetitive intergenic consensus (ERIC)-PCR technique in Sanandaj hospitals. Iran J Microbiol. 2013;5(2):126–31.
Moghadam MT, Mirzaei M, Moghaddam MFT, Babakhani S, Yeganeh O, Asgharzadeh S, et al. The challenge of global emergence of novel colistin-resistant Escherichia coli ST131. Microb Drug Resist. 2021. https://doi.org/10.1089/mdr.2020.0505.
Acknowledgements
We would like to thank the Antimicrobial Resistance Research Center of Iran University of Medical Sciences for supporting us during this study.
Funding
This research was supported by Grant No: 97-01-134-33238 from Iran University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
SHM, MTM, and AD conceived and designed the study. AKH and RGZ contributed in comprehensive research and sample collection. AKH, RGZ, and MTM wrote the paper. SHM and AD participated in manuscript editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
It was obtained from the ethics committee of Iran University of Medical Science. Reference Number: IR.IUMS.REC.1397.219.
Consent for publication
Not applicable.
Competing interests
None declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Khoshbayan, A., Golmoradi Zadeh, R., Taati Moghadam, M. et al. Molecular determination of O25b/ST131 clone type among extended spectrum β-lactamases production Escherichia coli recovering from urinary tract infection isolates. Ann Clin Microbiol Antimicrob 21, 35 (2022). https://doi.org/10.1186/s12941-022-00526-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-022-00526-2