Abstract
Background
The infection and prevalence of extended-spectrum β-lactamases (ESBLs) is a worldwide problem, and the presence of ESBLs varies between countries. In this study, we investigated the occurrence of plasmid-mediated ESBL/AmpC/carbapenemase/aminoglycoside resistance gene expression in Escherichia coli using phenotypic and genotypic techniques.
Methods
A total of 58 E. coli isolates were collected from hospitals in the city of Makkah and screened for the production of ESBL/AmpC/carbapenemase/aminoglycoside resistance genes. All samples were subjected to phenotypic and genotypic analyses. The antibiotic susceptibility of the E. coli isolates was determined using the Vitek-2 system and the minimum inhibitory concentration (MIC) assay. Antimicrobial agents tested using the Vitek 2 system and MIC assay included the expanded-spectrum (or third-generation) cephalosporins (e.g., cefoxitin, cefepime, aztreonam, cefotaxime, ceftriaxone, and ceftazidime) and carbapenems (meropenem and imipenem). Reported positive isolates were investigated using genotyping technology (oligonucleotide microarray-based assay and PCR). The genotyping investigation was focused on ESBL variants and the AmpC, carbapenemase and aminoglycoside resistance genes. E. coli was phylogenetically grouped, and the clonality of the isolates was studied using multilocus sequence typing (MLST).
Results
Our E. coli isolates exhibited different levels of resistance to ESBL drugs, including ampicillin (96.61%), cefoxitin (15.25%), ciprofloxacin (79.66%), cefepime (75.58%), aztreonam (89.83%), cefotaxime (76.27%), ceftazidime (81.36%), meropenem (0%) and imipenem (0%). Furthermore, the distribution of ESBL-producing E. coli was consistent with the data obtained using an oligonucleotide microarray-based assay and PCR genotyping against genes associated with β-lactam resistance. ST131 was the dominant sequence type lineage of the isolates and was the most uropathogenic E. coli lineage. The E. coli isolates also carried aminoglycoside resistance genes.
Conclusions
The evolution and prevalence of ESBL-producing E. coli may be rapidly accelerating in Saudi Arabia due to the high visitation seasons (especially to the city of Makkah). The health authority in Saudi Arabia should monitor the level of drug resistance in all general hospitals to reduce the increasing trend of microbial drug resistance and the impact on patient therapy.
Similar content being viewed by others
Background
Gram-negative bacteria that produce β-lactamases are a major concern in healthcare due to their ability to spread globally [1]. β-lactamases are produced by bacteria to hydrolyze the β-lactam ring in antibiotics, which results in ineffective drug treatment. Extended-spectrum β lactamases (ESBLs) are a major group of enzymes that confer resistance to several generations of β-lactam antibiotics, including third-generation cephalosporins [2, 3]. ESBL-producing Gram-negative bacteria are thought to be an important reason for cephalosporin therapy failure [4, 5]. Therefore, these bacteria should be monitored and reported by clinical laboratories to minimize their impact on patient therapy. ESBL resistance genes are primarily carried by plasmids. Plasmids may also carry genes encoding resistance to other antibiotic classes, such as ampA, ampC, aminoglycosides, chloramphenicol, macrolides, or quinolone. Therefore, treatment options are limited for bacteria that produce ESBLs due to the multiple resistance genes encoded in the plasmids. ESBL genes that are primarily encoded in plasmids include TEM, SHV, and CTX-M. Multiple variants of each gene are produced by altering the configuration of amino acids within the β-lactamase active site. Enterobacteriaceae, such as Klebsiella pneumoniae and Escherichia coli, are the major ESBL producers frequently isolated in clinical laboratories [6]. Acinetobacter baumannii is also an important ESBL-producing bacteria that has been reported globally [7]. Newer antibiotic classes, such as carbapenems, have been introduced by the pharmaceutical industry as treatment options for infections caused by ESBL-producing bacteria. Nevertheless, carbapenemase-producing bacteria have also been documented [8, 9]. Due to the increasing threat of multidrug-resistant bacteria, laboratory personnel, physicians, and clinical practitioners should implement a program to detect and report ESBLs as part of their infection control to limit the therapeutic failures caused by ESBL-producing bacteria. Molecular genotyping has been used concurrently with phenotyping techniques to detect and confirm antimicrobial drug resistance data and to detect Gram-negative ESBL producing bacteria [10]. PCR, multiplex PCR and oligonucleotide microarray-based assays have been developed and used to monitor the emergence of ESBLs and many other drug-resistant genes from E. coli, K. pneumonia and A. baumannii [11–15]. Strain characterization by multilocus sequence typing (MLST) is the method of choice in many clinical and research laboratories due to its high discriminatory power [16, 17]. The increased prevalence of ESBLs is being monitored and reported globally. Due to a lack of a solid data regarding the emergence of ESBLs from major Saudi general hospitals in the region of Makkah, this study reports the characterization of drug resistance genes for 58 E. coli isolates from an in-patient ward using phenotypic and genotypic approaches. Understanding the phenotypic and molecular nature of Enterobacteriaceae during the busy visitation Hajj season (pilgrimage season) in the city of Makkah, Saudi Arabia, is essential to reducing ESBL-strains prevalence.
Methods
Bacteriological culture
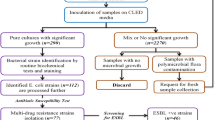
A total of 58 bacterial isolates were collected from two different general hospitals in the city of Makkah during the 2014–2015 Hajj (pilgrimage) season from patients with urinary tract infections. The bacterial isolates were phenotypically and genotypically investigated in microbiology laboratories at the King Abdulaziz City for Science and Technology (KACST). Single pure E. coli colonies were obtained from the all isolates. The bacteria were isolated from urine specimens using the clean-catch midstream urine sampling technique. Urine samples were inoculated using a calibrated 0.01 mL urine plastic loop on 5% sheep blood agar and MacConkey agar plates. The plates were incubated at 37 °C for 24 h. Urine samples were considered positive for UTIs if the number of colonies equaled or exceeded 105 CFU/mL. Gram staining was performed to identify urine specimens that contained more than 105 colony forming units (CFU)/mL of bacteria. A drop of well-mixed urine was fixed on a glass slide, stained, and examined under oil immersion (1000×) for the presence of at least one organism per oil immersion field.
Bacterial identification and antibiotic susceptibility testing
Bacterial identities were confirmed with a Vitek 2 GN ID card using the Vitek 2 system (bioMérieux, Marcy I’Etoile, France). Antibiotic susceptibility testing (AST) was completed using Vitek 2 cards (AST-N292) according to the manufacturer’s recommendations (bioMérieux, Marcy I’Etoile, France). The Vitek ESBL susceptibility tests were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) criteria using the Vitek system. E. coli ATCC 25922 was included in each Vitek testing step for quality control. The minimum inhibitory concentrations (MICs) were determined in Mueller–Hinton broth using microdilution plates [18]. The MICs of cefoxitin, ciprofloxacin, aztreonam, ceftazidime, meropenem, cefepime, cefotaxime, imipenem and ampicillin for all E. coli isolates were interpreted according to previous protocols (CLSI, 2014) [18]. Serial dilutions of the nine drugs were prepared (0.5, 1, 2, 4, 8, 16, 32, 64, 128, 256, 512 and 1024 µg/mL) in Mueller–Hinton broth and added to 96-well plates. Then, the bacterial suspensions were added to each well to achieve a final inoculum of 5×105 CFU/mL. The endpoint MIC was read visually and by spectrophotometer at 600 nm. The lowest concentration of antibiotic that inhibited visible bacterial growth after 24 h at 37 °C was defined as the MIC.
Amplification and sequencing of the 16S rRNA gene
The E. coli identity was confirmed using PCR and sequencing of the 16S rDNA gene [19–21]. Illustra PuReTaq Ready-To-Go PCR beads were used in the PCR reaction (GE Health Biosciences, USA). The 25-μL reaction was set up as follows: 2 μL of 10 pmol of each forward and reverse primer (IDT, Integrated DNA Technologies) were used with the 8-forward primer (AGA GTT TGA TCC TGG CTC AG) and 805-reverse primer (GAC TAC CAG GGT ATC TAA TC) [22]. Exactly 1 μL of DNA template was added to the beads, and the reaction was completed using 22 μL of nuclease-free ddH2O (Promega). The amplification cycling conditions were 3 min for the initial incubation at 95 °C, followed by 35 cycles of 1 min at 95 °C, 1 min at 55 °C, and 2 min at 72 °C and a final extension at 72 °C for 7 min. All PCR amplicons were fractionated using 1% agarose gel electrophoresis prior to staining with ethidium bromide. Images were obtained with a gel documentation system under a UV transilluminator. The PCR amplicons were purified and sequenced in an ABI 3130 Genetic Analyzer (Life Technologies, Carlsbad, CA, USA) using the 16S rDNA forward primer with the ABI BigDye terminator cycle sequencing ready reaction kit according to the manufacturer’s recommendations (Applied Biosystem, Foster City, CA, USA). The sequencing data were analyzed using the basic local alignment search tool BLAST-n (http://www.ncbi.nlm.nih.gov/BLAST) or the RDP database [23]. Unequivocal identification was obtained using 16S ribosomal DNA sequences.
Genetic background grouping of E. coli strains
The genetic background grouping of the E. coli isolates was explored. The grouping was performed based on Clermont phylo-typing method [24]. The majority of the pathogenic extra-intestinal E. coli strains belonged to groups B2. In contrast, the commensal E. coli strains belonged to groups A, B1, C, and F. The strains were subsequently phylogenetically inferred based on the presence or absence of the gene markers as follows: For group A, arpA+, chuA−, yjaA−, and TspE4.C2−, for group B1, arpA+, chuA−, yjaA−, and TspE4.C2+, for group F, arpA−, chuA+, yjaA−, and TspE4.C2−, for group B2, arpA−, chuA+, yjaA+, and TspE4.C2−, for group A or C, arpA+, chuA−, yjaA+, and TspE4.C2−, for group D or E, arpA+, chuA+, yjaA−, and TspE4.C2−. A standard 25-µL PCR reaction was used to investigate the genetic background of the E. coli strains used in our study. Illustra PuReTaq Ready-To-Go PCR beads were used in the PCR reaction (GE Health Biosciences, USA) as described by Clermont to amplify the arpA (400 bp) ChuA (288 bp), YjaA (211 bp), TspE4C2 (152 bp), and an internal control trpA (489 bp). Primers and PCR cycling conditions were described in (Table 1).
Detection of ESBL, ampC and carbapenemase genes
Global genotyping utilizing an oligonucleotide microarray-based assay and PCR genotyping were applied to identify and confirm the presence of drug resistance genes. For the microarray DNA analysis (Alere Technologies, Jena, Germany) [15], the ESBL/AmpC/carbapenemase genes were evaluated based on the manufacturer’s protocols (bla ACC , bla ACT , bla CMY , bla KHM , bla MOX-CMY9 , bla CTX-M-1 , bla CTX-M-15 , bla CTX-M 2 , bla CTX-M-8 , bla CTX-M-9 , bla CTX-M-26 , bla DHA-1 , bla FOX , bla GES-1 , bla G1M-1 , bla MI-3 , bla IMP , bla KPC , bla LAP-1 , bla LEN-1 , bla MOX , bla OXA-1 , bla OXA-2 , bla OXA-7 , bla OXA-9 , bla OXA-23 , bla OXA-23 , bla OXA-40 , bla OXA-48 , bla OXA-51 , bla OXA-54 , bla OXA-55 , bla OXA-58 , bla OXA-60 , bla PER1 , bla PER2 , bla PSE-1 , bla SHF-1 , bla SHV , bla SEM-1 , bla SPM-1 , bla TEM , bla VEB-1 , and bla VIM ). For the PCR analysis, DNA templates were obtained by boiling 500 µL of bacterial cells (OD600 = 1.0) in a sterile single 1.5 mL Eppendorf tube in a water bath at 100 °C. The DNA lysate was diluted 1:10 with ddH2O and frozen at −20 °C before use for bacterial genotyping confirmation with gene-specific primers (Table 2) [10]. The primers detected the β-lactamase genes and their variants (TEM, SHV, and CTX-M groups 1, 15, 2, 8, 9, and 26). The PCRs were performed using a 9800 Thermal Cycler (Applied Biosystem, USA) in a total volume of 25 µL containing 10 pmol of primers, 25 µmol of dNTPs, 10 µL of gDNA/plasmid lysate, 2.5 µL of 10Χ Taq buffer, 2.5 mM MgCl2 and 2.5 U of Taq polymerase. The cycling conditions used for the PCR were an initial denaturation at 94 °C for 10 min, 35 cycles of 94 °C for 40 s, 60 °C for 40 s and 72 °C for 1 min and a final extension step at 72 °C for 7 min [10]. All PCR amplicons were fractionated by 1% agarose gel electrophoresis prior to staining with ethidium bromide. Images were obtained with a gel documentation system under a UV transilluminator.
Bacterial genotyping by multilocus sequence typing
Fifty-eight E. coli isolates were subjected to multilocus sequence typing (MLST) as previously described [25]. The MLST schemes used to type E. coli were conducted using internal fragments of the following seven housekeeping genes: Adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), mdh (malate dehydrogenase), purA (adenylosuccinate dehydrogenase), icd (isocitrate/isopropylmalate dehydrogenase), recA (ATP/GTP binding motif) (ID Genomics Inc, Seattle, WA, USA). The primers used for amplification and sequencing are illustrated in (Table 3) [25]. The PCR amplifications were performed using a 9800 Thermal Cycler (Applied Biosystem, USA) with the following conditions: initial denaturation at 94 °C for 7 min, followed by 35 cycles of denaturation at 94 °C for 30 s and an annealing temperature of 56 °C for 30 s, extension at 72 °C for 2 min and a final 7-min extension at 72 °C. The PCR amplicons were checked with 1% agarose gel electrophoresis prior to purification for sequencing. Forward and reverse sequencing reactions were performed for every isolate. Different allele sequences were assigned for each locus with an arbitrary allele number for identification. Each bacterial isolate was characterized by a pattern of numbers defining its sequence type (ST). The sequencing data of the MLST genes were analyzed using the E. coli MLST Database (Warwick Medical School, Coventry, UK database; http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) [25]. The similarities of the allelic profiles were assessed using the Molecular Evolutionary Genetics Analysis (MEGA 6) software.
Phylogenetic analysis
Phylogenetic trees based on the concatenated alleles of seven MLST genes were constructed using the Molecular Evolutionary Genetics Analysis (Mega6) software for the E. coli dataset. The maximum likelihood trees were based on neighbor-joining (NJ) starting trees with Nearest-Neighbor Interchange branch swapping. The tree stability was assessed using the bootstrap method [26].
Results
Bacterial identification and antibiotic susceptibility testing
All obtained clinical E. coli isolates (n = 58) were phenotypically studied using the Vitek system and the microbroth dilution method to determine the MIC values against various antimicrobial drugs (cefoxitin, ciprofloxacin, aztreonam, ceftazidime, meropenem, cefepime, cefotaxime, imipenem and ampicillin), (Table 4). The greatest number of E. coli isolates was resistant to ampicillin (96.61%), followed by cefotaxime (76.27%), cefepime (74.58%), ceftazidime (81.36%), aztreonam (89.83%), and cefoxitin (15.25%). Meropenem and imipenem had rates of 0%, and the non-β-lactam ciprofloxacin had a rate of 79.66%. All E. coli isolates were sensitive to meropenem and imipenem (Fig. 1).
ESBL-producing E. coli genotyping
The genotyping of ESBL-producing E. coli isolates by the global analysis of an oligonucleotide microarray-based assay and PCR identified a high proportion of β-lactamase genes among the E. coli isolates. The blaCTX-M1, blaCTX-M15, blaOXA1 and blaTEM genotypes were more frequently identified and were more predominant among the E. coli isolates than the blaCTX-M, blaOXA and blaSHV variants. The prevalence was 46.7% for blaCTX-M1 and blaCTX-M15, 48.3% for blaOXA1, and 38.7% for blaTEM. The genotypic prevalence of blaSHV was 3.2% among all isolates, which was considerably lower than the prevalence for blaCTX-M1, blaCTX-M15, blaOXA1 and blaTEM. No carbapenem-resistant E. coli isolate was detected (Fig. 2).
Distribution of ESBL genes in E. coli
The β-lactam genes were studied to evaluate the distribution of the genes among our isolates. We found that 13.7% (8/58) of the E. coli isolates harbored four major ESBL genes (blaCTX-M1, blaCTX-M15, blaOXA1, and blaTEM) and 17.2% (10/58) contained three ESBL genes (blaCTX-M1, blaCTX-M15, and blaOXA1). Additionally, two positive ESBL genotypes (3.4%; 2/58) contained blaOXA1 and blaTEM and two isolates contained only blaSHV (3.4%; 2/58). No carbapenemase genes were detected among the isolates according to the phenotypic and genotypic testing.
The phylogenetic grouping of E. coli strains
The genetic background grouping of the E. coli isolates was assigned based on the Clermont method which is very specific for E. coli phylo-typing groups [24]. It has shown that 46.55% (27) of the ESBL-producing E. coli isolate collection belonged to group B2, and 6.9% (4) belonged to group D or E, while 12% (7) belonged to group B1, 3.4% (2) belonged to group A, 24% (14) belonged to group A or C and 6.9% (4) belonged to group F. The majority of the pathogenic extra-intestinal E. coli strains belonged to groups B2 (Fig. 3).
a Quadruples PCR profiles of 58 E. coli MDR isolates. Isolate IDs shown under each lane. Data assignment were performed according to the presence or absence of signals of the following gene order arpA, chuA, yjaA, TspE4.C2. Row 1, lanes 1, 12—group A (+−−−); lanes 2–11, 13, 15—groups A or C (+−+−), lanes 16–22—group B1 (+−+), lane 14—group B2 (−+++), lanes 23–24—group F (−+−); Row 2, lanes 1–2—group F (−+−), lanes 3–24—group B2 (−+++) or (−++−) in lane 7; Row 3, lanes 1–4—group B2 (−+++), lanes 5–8—groups E or D (++−+), lanes 9–10—groups A or C (+−+−). b Groups A/C differentiation. In all lanes, both bands for internal control and Group C specific trpA fragment (219 bp) are present. c Groups D/E differentiation. Lanes 1, 3 and 4—both bands for internal control and Group E specific arpA fragment (301 bp) are present. Lane 2—only internal control present
Prevalence of genes associated with aminoglycoside resistance in the E. coli isolates
The prevalence of genes associated with aminoglycoside resistance among the 58 E. coli isolates was investigated. A total of 44.8% of the E. coli isolates carried the aac6 gene, 43% harbored aac6Ib, 42% contained aadA4, and 36% contained strB; these prevalences represented high proportions and were more common than the other aminoglycoside genes (15% for aadA1, 12% for aadA2, 4% for aadB, 4% for ant2, 12% for aphA and 1% for strA) (Fig. 4).
Bacterial genotyping by MLST
In this study, 58 E. coli isolates were assigned sequence types (ST) and ST complexes (STc) using a standard multi-locus sequence typing method (Fig. 5) [25]. Of the 58 E. coli isolates, 7 isolates (12%) belonged to STc 10, 20 isolates (34.5%) to STc 131, and 4 isolates (7%) to STc 648 and STc 38. Other ST complexes (46, 448, 156, 155, 23, 12 and 73) were represented by 1–3 isolates (2–5%). The ST complex could not be determined for 14 isolates (24%). A new allele of the recA gene was identified in isolate ID 4. This allele had one nucleotide substitution compared to allele 2 at position 40 from the beginning of the allele (C substituted by T). The allelic profile of one isolate (isolate ID 12) was not previously reported (Fig. 5).
Phylogenetic tree of 58 MDR E. coli strains constructed using the maximum likelihood algorithm in the MEGA6 software [27] and based on the concatenated alleles of 7 housekeeping genes according to Achtman’s scheme (columns 4–10). The numerals on the branches represent bootstrap values. The phylogenetic analysis identified 24 ST (column 11) that corresponded to 11 ST complexes (STc, column 12). Phylogenetic groups in column 2 were identified using phylo-typing method [24] by quadruplex PCR using four target genes (arpA±, chuA±, yjaA±, TspE4.C2±). Strains incorrectly assigned using the extended quadruplex method are indicated in red. The phylo-group memberships of two isolates (1097, 1125) were ambiguous. Notably, E. coli Clone ST131, ST410, and ST1193, which are disseminated globally, were among the identified STs in this study
Phylogenetic analysis
We conducted a phylogenetic analysis to examine the clonal diversity and phylogenetic relationships among the E. coli isolates. The E. coli isolates clustered in several monophyletic clades that corresponded to known phylogenetic groups. The majority of the E. coli isolates fell into the phylogenetic groups B2: ST131 (20 isolates), ST1193 (3 isolates) [27], ST73 (1 isolate), ST12 (1 isolate) and ST127 (1 isolate). In this study, four E. coli isolates appeared to belong to group E: ST38 (3 isolates) [27] ST315 (1 isolate), of which all isolates were from STc 38. Of the rest, one isolate belonged to phylogroup A, thirteen to phylogroup C, eight to phylogroup B1, two to phylogroup C, and four to phylogroup F. Most of the known virulent extraintestinal strains primarily belong to group B2 and to a lesser extent to group D or E (4). Group A together with group B1, F, and C are considered predominant among commensal E. coli strains (5).
Discussion
During congested visitation seasons, especially during the religious pilgrimage (Hajj), the risk of spreading multidrug-resistant microbes increases substantially [28]. Therefore, this study investigated the prevalence of ESBL-producing E. coli in local general hospitals in Makkah during Hajj and the correlation between the ESBL phenotype and antimicrobial drug resistance using genotyping approaches. This study also explored multilocus sequence typing (MLST) to characterize these isolates and to enhance our understanding of the epidemiology of ESBL-producing E. coli in our region. Our data had demonstrated that our E. coli isolates exhibited varying levels of resistance to ESBL drugs and carried genes associated with aminoglycoside resistance. No carbapenem resistance genes were identified in any of the collected and investigated E. coli isolates (n = 58). All uropathogenic E. coli isolates in this study are classified as multidrug-resistant (MDR) according to the criteria by Magiorakos [29].
The distribution of ESBL-producing E. coli was consistent with the data obtained from an oligonucleotide microarray-based assay and PCR genotyping against genes associated with β-lactam resistance. The dominant sequence type lineage of the isolates was ST131, which is the most uropathogenic E. coli lineage. The most commonly isolated bacterium from patients with urinary tract infections is E. coli. These isolates usually harbor different plasmids. Therefore, they have a tendency to acquire multidrug-resistant phenotypes and are difficult to treat.
Consistent with our study, studies have associated ST131/CTX-M15 human uropathogenic E. coli with ST410/CTX-M15, and ST648/CTX-M15. However, it was shown that ST410/CTX-M15, and ST648/CTX-M15 were also isolated from uropathogenic E. coli of cats and dogs (feline and canine) in Switzerland [30, 31]. These circulating STs may suggest that disease transmission between companion animals and humans may occur by direct contact. Furthermore, three isolates of E. coli ST1193 were evident in this study. E. coli ST1193 is associated with urinary tract infection and fluoroquinolone-resistance, namely ciprofloxacin-resistance (Figs. 1, 5) [32, 33]. E. coli ST1193 may also be transmitted through household contact as suggested by a study from Japan [34].
It was demonstrated that multidrug-resistance (MDR) E. coli clone ST131 was globally disseminated in six different geographical countries. The rapid spread and emergence among countries may be due to high virulence factor gene-content, the presence of ESBL CTX-M-15, and fluoroquinolones resistance. Notably, the E. coli clone ST131 fluoroquinolone resistance phenotype was the most prevalent geographically and was detected in this current study in Saudi Arabia (Figs. 1, 5) [35].
Studies have shown that pathogenicity island acquisition by uropathogenic E. coli may worsen urinary tract infections and aid in evasion of the host immune response, resulting in spreading in the bloodstream. Gene acquisition by intestinal pathogenic bacteria promotes their colonization in different intestinal regions; these genes may also alter the mechanism of bacterial-host interactions, causing distinct gastrointestinal pathology [27]. It was shown that a urinary sepsis niche for E. coli ST131 compared to bacteremia in non-ST131 E. coli clones [32, 36]. However, this niche could not be linked to the clinical manifestation of renal tract infections in humans. No clinical focus to other sites of infection, such as intra-abdominal abscess, ascitic fluid, bones/joints, respiratory tract or bacteremia, has been established for the ST131 clone [37, 38]. There is evidence for a direct human-to-human route of transmission for ST131. For instance, an elderly father with pyelonephritis transmitted ST131 E. coli to his daughter, which initiated a similar illness in her. Similarly, an identical clone was identified from a young child with osteoarticular infection and a fecal sample from his mother [39, 40]. ST131 may contain genes for multiple antimicrobial resistance mechanisms, which may make therapy difficult. Carbapenems alone or in combination with amikacin were successfully used to treat infections caused by clones harboring CTX-M genes [32, 39]. A study demonstrated ESBL-producing E. coli acquisition among Makkah visitors after the Hajj (pilgrimage) season, especially CTX-M genes [28].
Molecular epidemiological studies have focused on the characterization and spread of the ST131 clone; however, global studies on the distribution and spread between humans and animals are limited, particularly in the developing world. There is a high frequency of infection by the drug-resistant ST131 clone in developing countries and a lack of timely countermeasures resulting in high morbidity and mortality rates [41–43].
Conclusion
In this study, 58 E. coli isolates were collected from patients with urinary tract infections from two local hospitals in the city of Makkah during the 2014–2015 Hajj (pilgrimage) season and screened for the production of ESBL/AmpC/carbapenemase/aminoglycoside resistance genes using phenotypic and genotypic analyses. Our data had demonstrated that our isolates primarily carried various ESBL and aminoglycoside resistance genes. The dominant sequence type lineage of the isolates was ST131, which was the most uropathogenic E. coli lineage. A genetic analysis of multiple isolates from the Middle Eastern regions may contribute to the development of rapid molecular detection methods and may lead to new therapies. Furthermore, controlling the endemicity of emerging MDR and decreasing the occurrence and prevalence of drug-resistant uropathogenic E. coli during heavy visitation seasons in the city of Makkah may involve the implementation of a stringent antidrug treatment policy.
Abbreviations
- MDR:
-
multi drug resistance
- ESBL:
-
extended-spectrum-β-lactamase
- AmpC:
-
AmpC beta-lactamases
- MIC:
-
minimum inhibitory concentration
- MLST:
-
multilocus sequence typing
- ST:
-
sequence typing
- STc:
-
sequence typing complex
References
Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–66.
Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11(6):315–7.
Philippon A, Labia R, Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989;33(8):1131.
Drawz SM, Papp-Wallace KM, Bonomo RA. New beta-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother. 2014;58(4):1835–46.
Rawat D, Nair D. Extended-spectrum beta-lactamases in Gram Negative Bacteria. J Glob Infect Dis. 2010;2(3):263–74.
Livermore DM. Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis. 2003;36(Suppl 1):S11–23.
Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292–301.
Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53(1):60–7.
Kanj SS, Kanafani ZA. Current concepts in antimicrobial therapy against resistant gram-negative organisms: extended-spectrum β-lactamase—producing Enterobacteriaceae, carbapenem-resistant Enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clin Proc. 2011;86(3):250–9.
Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–5.
Trung NT, Hien TT, Huyen TT, Quyen DT, Binh MT, Hoan PQ, Meyer CG, Velavan TP, le Song H. Simple multiplex PCR assays to detect common pathogens and associated genes encoding for acquired extended spectrum betalactamases (ESBL) or carbapenemases from surgical site specimens in Vietnam. Ann Clin Microbiol Antimicrob. 2015;14(1):23.
Valverde A, Turrientes MC, Norman F, San Martín E, Moreno L, Pérez-Molina JA, López-Vélez R, Cantón R. CTX-M-15-non-ST131 Escherichia coli isolates are mainly responsible of faecal carriage with ESBL-producing Enterobacteriaceae in travellers, immigrants and those visiting friends and relatives. Clin Microbiol Infect. 2015;21(3):252.e251–4.
Tawfik AF, Alswailem AM, Shibl AM, Al-Agamy MH. Prevalence and genetic characteristics of TEM, SHV, and CTX-M in clinical Klebsiella pneumoniae isolates from Saudi Arabia. Microb Drug Resist. 2011;17(3):383–8.
Alyamani EJ, Khiyami MA, Booq RY, Alnafjan BM, Altammami MA, Bahwerth FS. Molecular characterization of extended-spectrum beta-lactamases (ESBLs) produced by clinical isolates of Acinetobacter baumannii in Saudi Arabia. Ann Clin Microbiol Antimicrob. 2015;14(1):38.
Braun SD, Monecke S, Thurmer A, Ruppelt A, Makarewicz O, Pletz M, Reibetaig A, Slickers P, Ehricht R. Rapid identification of carbapenemase genes in gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS ONE. 2014;9(7):e102232.
Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53(8):3365–70.
Salerno A, Deletoile A, Lefevre M, Ciznar I, Krovacek K, Grimont P, Brisse S. Recombining population structure of Plesiomonas shigelloides (Enterobacteriaceae) revealed by multilocus sequence typing. J Bacteriol. 2007;189(21):7808–18.
CLSI. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement (M100-S24). Wayne, PA: Clinical and Laboratory Standard Institute; 2014.
Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703.
Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28(9):1942–6.
James G. Universal bacterial identification by PCR and DNA sequencing of 16S rRNA gene. In: Schuller M, Sloots TP, James GS, Halliday CL, Carter IWJ, editors. PCR for clinical microbiology: an Australian and international perspective. New York: Springer; 2010. p. 209–14.
Srinivasan R, Karaoz U, Volegova M, MacKichan J, Kato-Maeda M, Miller S, Nadarajan R, Brodie EL, Lynch SV. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE. 2015;10(2):e0117617.
Maidak BL, Olsen GJ, Larsen N, Overbeek R, McCaughey MJ, Woese CR. The ribosomal database project (RDP). Nucl Acids Res. 1996;24(1):82–5.
Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58–65.
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–51.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S: MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Welch RA, Burland V, Plunkett G 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99(26):17020–4.
Leangapichart T, Dia NM, Olaitan AO, Gautret P, Brouqui P, Rolain JM. Acquisition of extended-spectrum beta-lactamases by Escherichia coli and Klebsiella pneumoniae in gut microbiota of pilgrims during the Hajj Pilgrimage of 2013. Antimicrob Agents Chemother. 2016;60(5):3222–6.
Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, Harbarth S, Hindler J, Kahlmeter G, Olsson-Liljequist B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Huber H, Zweifel C, Wittenbrink MM, Stephan R. ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Vet Microbiol. 2013;162(2–4):992–6.
Giufrè M, Graziani C, Accogli M, Luzzi I, Busani L, Cerquetti M: Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother. 2012;67(4):860–7.
Johnson JR, Johnston B, Clabots C, Kuskowski MA, Pendyala S, DebRoy C, Nowicki B, Rice J. Escherichia coli sequence type ST131 as an emerging fluoroquinolone-resistant uropathogen among renal transplant recipients. Antimicrob Agents Chemother. 2010;54(1):546–50.
Rubin JE, Pitout JD. Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet Microbiol. 2014;170(1):10–8.
Kojima Y, Harada S, Aoki K, Ishii Y, Sawa T, Hasegawa K, Saji T, Yamaguchi K, Tateda K. Spread of CTX-M-15 extended-spectrum β-lactamase-producing Escherichia coli isolates through household contact and plasmid transfer. J Clin Microbiol. 2014;52(5):1783–5.
Petty NK, Zakour NLB, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan M-D, Moriel DG, Peters KM, Davies M. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci. 2014;111(15):5694–9.
Pitout JDD, Gregson DB, Campbell L, Laupland KB. Molecular characteristics of extended-spectrum-β-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob Agents Chemother. 2009;53(7):2846–51.
Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother. 2009;63(1):72–9.
Bert F, Johnson JR, Ouattara B, Leflon-Guibout V, Johnston B, Marcon E, Valla D, Moreau R, Nicolas-Chanoine MH. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J Clin Microbiol. 2010;48(8):2709–14.
Lee MY, Choi HJ, Choi JY, Song M, Song Y, Kim SW, Chang HH, Jung SI, Kim YS, Ki HK, et al. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J Infect. 2010;60(2):146–53.
Ender PT, Gajanana D, Johnston B, Clabots C, Tamarkin FJ, Johnson JR. Transmission of an extended-spectrum-beta-lactamase-producing Escherichia coli (sequence type ST131) strain between a father and daughter resulting in septic shock and emphysematous pyelonephritis. J Clin Microbiol. 2009;47(11):3780–2.
Manges AR, Johnson JR, Foxman B, O’Bryan TT, Fullerton KE, Riley LW. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Engl J Med. 2001;345(14):1007–13.
Schaeffer AJ. Global molecular epidemiology of the O15:K52:H1 extraintestinal pathogenic Escherichia coli clonal group: evidence of distribution beyond Europe. J Urol. 2003;169(4):1612.
Oteo J, Pérez-Vázquez M, Campos J. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis. 2010;23(4):320–6.
Authors' contributions
EA and AK contributed to the study design. RB, FB, MM, and ER collected the samples, and all authors contributed equally to the laboratory experiments. All authors contributed to the data interpretation. EA and AK drafted and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
All authors wish to acknowledge the Life Science & Environment Research Institute and the National Center for Biotechnology at KACST for supporting and facilitating this work.
Competing interests
All authors declare that they have no competing interests.
Availability of data and material
All data and material are available.
Consent for publication
All authors approve the publication of this work.
Ethics approval and consent to participate
Ethical approval and consent were not required for this project because no human or animal subjects were used.
Funding
This work was funded by an internal grant from the Life Science & Environment Research Institute and from the National Center for Biotechnology at KACST.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Alyamani, E.J., Khiyami, A.M., Booq, R.Y. et al. The occurrence of ESBL-producing Escherichia coli carrying aminoglycoside resistance genes in urinary tract infections in Saudi Arabia. Ann Clin Microbiol Antimicrob 16, 1 (2017). https://doi.org/10.1186/s12941-016-0177-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-016-0177-6