Abstract
Background
The occurrence of cyanobacterial blooms in freshwater presents a threat to human health. However, epidemiological studies on the association between cyanobacterial blooms in drinking water sources and human health outcomes are scarce. The objective of this study was to evaluate if cyanobacterial blooms were associated with increased emergency room visits for gastrointestinal (GI), respiratory and dermal illnesses.
Methods
Satellite-derived cyanobacteria cell concentrations were estimated in the source of drinking water for the Greater Boston area, during 2008–2011. Daily counts of hospital emergency room visits for GI, respiratory and dermal illnesses among drinking water recipients were obtained from an administrative record database. A two-stage model was used to analyze time-series data for an association between cyanobacterial blooms and the occurrence of illnesses. At the first stage, predictive autoregressive generalized additive models for Poisson-distributed outcomes were fitted to daily illness count data and daily predictive variables. At the second stage, residuals from the first stage models were regressed against lagged categorized cyanobacteria concentration estimates.
Results
The highest cyanobacteria concentration (above the 75th percentile) was associated with an additional 4.3 cases of respiratory illness (95% confidence interval: 0.7, 8.0, p = 0.02, n = 268) compared to cyanobacteria concentrations below the 50th percentile in a two-day lag. There were no significant associations between satellite derived cyanobacterial concentrations and lagged data on GI or dermal illnesses.
Conclusion
The study demonstrated a significant positive association between satellite-derived cyanobacteria concentrations in source water and respiratory illness occurring 2 days later. Future studies will require direct measures of cyanotoxins and health effects associated with exposure to cyanobacteria-impacted drinking water sources.
Similar content being viewed by others
Introduction
Cyanobacteria are widely distributed in the environment and are problematic in aquatic systems when they create dense assemblages (blooms) or produce potent secondary metabolites (cyanotoxins) that are harmful to animals, plants, and humans [1, 2]. Driven by climate and anthropogenic alterations, cyanobacterial blooms may change in frequency [3], extent [4], and magnitude [5] in many areas, which pose a threat to the sustainability of aquatic ecosystems as well as to human health [2, 6–8].
Humans can be exposed to cyanobacterial harmful algal blooms (cyanoHABs) via drinking water [6, 9–11]. CyanoHABs and cyanotoxins are found in source waters worldwide [6, 12–14], posing a global health risk. The most common reported acute health effect following cyanotoxin exposure through drinking water is gastrointestinal (GI) illness [15]. Recreational water exposure to cyanobacteria and cyanotoxins can result in multiple non-specific acute illnesses such as GI, respiratory, dermal, otic, neurological, musculoskeletal, and other signs and symptoms such as fever and anorexia [16, 17].
Measurement and ascertainment of cyanoHAB exposure is an important element during evaluation of the risk of cyanoHABs to human health. Standard exposure assessment may include in situ water sampling, cyanobacteria cell count, and quantifying toxin concentrations. These methods are often time-consuming, costly, and only feasible on a small geographic or temporal scale. Although satellite observations cannot detect toxins [18], they can assist in quantifying cyanobacteria abundance near the water surface. The use of remote sensing is a potentially attractive, low cost approach to characterizing the risk of human exposure to cyanoHABs [19]. Satellite remote sensing has been used to detect harmful algal blooms in large inland water bodies such as the Great Lakes in North America [20–22]. Recently, the use of remote sensing for cyanoHABs identification in smaller lakes using satellites has improved [23]. Satellite, sensor, and algorithm improvements now allow for the study of numerous inland lakes, improving our ability to remotely assess cyanoHABs in water bodies that serve as sources of drinking water or as recreational venues [3]. Satellite images acquired by the European Space Agency’s (ESA’s) MEdium Resolution Imaging Spectrometer (MERIS) can measure phytoplankton spectral signatures [24], and are now used to estimate cyanobacteria abundance in water [5, 25, 26].
The objective of this study is to explore the use of satellite-derived cyanobacteria abundance for exposure assessment and analysis of potential associations with human health effects. We used emergency room (ER) visit data to detect temporally- and spatially-associated acute illnesses in a human population served by a cyanoHAB-impacted drinking water source. We analyzed the three main classes of acute illnesses that could result from exposure to cyanobacterial blooms: GI, respiratory, and dermal illnesses [16, 27, 28].
Methods
Study area and water distribution system
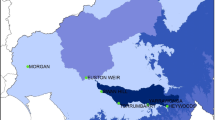
Our study area included the city of Boston and other cities and towns in the Boston Metropolitan Area that receive municipal drinking water from the Massachusetts Water Resources Authority (MWRA) sourced from the Wachusett Reservoir located in Middlesex county, Massachusetts (Fig. 1). The Wachusett Reservoir is the second largest water body in the state. It serves as a source of drinking water for nearly 3 million people in the metropolitan Boston area (https://www.mass.gov/locations/wachusett-reservoir). The water is treated at the John J. Carroll Water Treatment Plant. The water is unfiltered; ozone is used as a primary disinfectant and chloramine as a secondary disinfectant (http://www.mwra.com/04water/html/watsys.htm). Twenty-two towns, located within 20–35 miles (32–56 km) of the reservoir intake, that also receive drinking water only from Wachusett Reservoir, were selected for inclusion in this study. The distance limits were used in order to ensure similar water residence times of approximately 2 days in the distribution system [29].
Health data
Data on ER visits for GI, respiratory, and dermal illnesses during 2008–2011 were obtained from the Massachusetts Center for Health Information Analysis (CHIA) (http://www.chiamass.gov/), which accepts the submission of health data from approximately 70 Massachusetts hospitals. These anonymous, publicly-available, administrative health data were determined not to be personally identifiable human subjects data by the Human Subjects Research Protocol Officer for the U.S. Environmental Protection Agency (USEPA), and therefore are exempt from review by the USEPA’s Institutional Review Board of record. Administrative data included diagnostic code (International Classification of Diseases, Ninth Revision (ICD-9)), registration date, age, and the patients’ residential Zone Improvement Plan (ZIP) code. Patient residence in the study area determined their inclusion in the analysis. Eligible patients may have visited hospitals outside of the study area. We extracted the records on residents of the 22 towns included in the study using their ZIP codes. GI illnesses included ICD-9 codes 001–009.9, 558.9, 787.91, 787.01, and 787.03. Respiratory illnesses included ICD-9 codes 460, 461, 465.9, 493, 786.2, 786.05, and 786.07. Dermal illnesses included ICD-9 codes 782.1, 136.9, 686.9, 692, and 691.8 (see the description for these codes in Table S1). The response variable was the total daily number of patient visits for each category of illness in the study area.
Cyanobacterial bloom data
Cyanobacteria concentration at the Wachusett Reservoir water intake were estimated using satellite images from MEdium Resolution Imaging Spectrometer (MERIS), an ESA sensor onboard the Envisat satellite which offers 300-m spatial resolution at nadir, where nadir is defined as the point on Earth’s surface directly below the satellite. The MERIS data archive includes a consistent time series over the study region for the years 2008–2011 with an image collected approximately every 3 days, and a usable image collected every 5 days on average when taking into account cloud, snow, and ice cover, which confound the satellite signal. Image processing was performed following the previous reported method [25, 30] and estimation of the cyanobacteria index (CI_cyano) followed Lunetta et al. 2015 [31]. Multiple inland water satellite algorithms could be considered to monitor cyanobacteria, however the updated CI_cyano from Lunetta et al. 2015 [31] was used here because it was validated across six New England states, including Wachusett Reservoir and several other lakes throughout Massachusetts. Additional validation efforts include agreement with state health advisories [32], state reported toxins and cell counts [33], and agreement against expected seasonality [25]. Only pixels within a 900-m buffer of the water intake that fell completely within the reservoir and excluding mixed land and water pixels along the shoreline were included in this analysis. CI_cyano was then converted into cyanobacteria concentration in cells/mL (cyan C) using the following equation: cyan C = CI*108 following Lunetta et al. 2015 [31]. At a 300-m spatial resolution, five satellite pixels fell within a 900-m buffer of the drinking water intake. For each satellite image, cyanobacteria concentrations were calculated as the maximum cyan C value of these five pixels.
Estimated maximum cyanobacteria concentration data were classified into three categories defined using the sample distribution of cyan C values. Category 1 included observations with cyanobacteria concentrations <= 10,000 cells/mL, which is the assumed detection limit of the sensor; this level contained 50 % of the observations. Category 2 included those above the estimated detection limit but below the 3rd quartile of the distribution (cyanobacteria concentrations <= approximately 109,468 cells/mL); this category contained about 25 % of the observations. Category 3 included those above the 3rd quartile of the distribution up to the maximum satellite-derived cyanobacteria concentrations of 575,440 cells/mL and contained about 25 % of the observations.
Covariates
Daily mean temperature and precipitation data for the Boston area were obtained from the PRISM Climate Group (http://www.prism.oregonstate.edu/explorer/). We selected one grid (4 km × 4 km, longitude: 42.3312, latitude: − 71.0761) to represent the weather conditions in the study area. Air pollution data were obtained from USEPA’s outdoor air quality website (https://www.epa.gov/outdoor-air-quality-data/download-daily-data). These data included daily concentrations of carbon monoxide (CO), nitrogen dioxide (NO2), sulfur dioxide (SO2), ozone (O3), and fine particulate matter (PM2.5) from a single air quality monitoring site (AQS) (ID: 250250042) in the study area. Given that the spatial variability of weather and air pollution data is relatively small, we assumed that these parameters were representative of the entire study area. ER visit counts differed from school vacation to school year and among days of the week and holidays [34, 35]. We created two binary variables: school days (all days in the school year including weekends) and holidays. In addition, we created dummy variables for each day of the week.
Statistical analysis
We conducted exploratory data analyses, including plotting distributions of major variables, visually assessing temporal patterns, conducting correlation analysis of potential associations between temporal factors, such as day of the week, calendar month, holidays, meteorological variables and health data, and autocorrelation analysis of the health data. Time-series analysis of data was employed to assess associations between cyanobacteria concentration and lagged daily counts of illness as illness count data and meteorological data were daily while valid satellite images suitable for estimating cyanobacteria data were available at a lower frequency (on average, once per 5 days and often at irregular intervals). In order to fully use the available daily data in a time-series analysis, we conducted a regression analysis in two stages. First, we developed a predictive autoregressive generalized additive model (GAM) for each Poisson-distributed health outcome (respiratory, GI and dermal illness counts). All models included the spline function of time to account for seasonality and an autoregressive component (lagged daily counts of ER visits) at a 1-day lag. Other covariates tested included daily weather variables, air quality variables and indicator variables for weekdays, school days, and holidays as predictors. We used Akaike information criterion (AIC) to select the predictors for final stage one models [36].
Residuals from the first stage model were lacking temporal trends, seasonal patterns and serial autocorrelation. They were generally homoscedastic and normally distributed (Fig. 3). This suggests that the first stage analysis removed the effects of potential time-varying confounders, such as seasonality.
Next, residuals from the first stage models were regressed against original categorized cyanobacteria concentration data at lags from 0 to 6 days using univariable models for normally distributed (Gaussian) outcomes. As exploratory analysis of data demonstrated that cyanobacteria data for the winter period included many missing observations or values below the detection limit of the sensor, the second stage regression models excluded data for December, January and February. It has been shown previously that remotely sensed cyanobacterial bloom ascertainment is less available in winter due to spectral interference by ice or snow, particularly in northern latitude states such as Massachusetts [25].
Results
The daily counts of GI, respiratory, and dermal illnesses during 2008–2011 are illustrated in Fig. 2. Visits for GI and respiratory illnesses included in this study generally peaked in the winter and their seasonal curves largely overlapped, although counts of respiratory illnesses had larger temporal variability. Counts of ER visits for GI illness averaged 72.6 (±14.9) per day, and count of respiratory illness visits averaged 79.1 (±30.0) per day (Table 1). In contrast, the daily count of dermal illness visits was much smaller, with a mean value of 24.2 (±7.4) per day; dermal illness visits were slightly higher in the summer compared to the winter.
The estimated daily counts of remotely sensed cyanobacteria concentration near the drinking water intake during 2008–2011 are shown in Fig. 3. Shaded areas on this graph indicate winter months (December, January, and February) when the time-series of cyanobacteria concentrations contained many missing data due to unfavorable meteorological conditions (e.g., snow and ice cover). These data were excluded from stage 2 analysis.
Estimates of cyanobacteria concentrations at the Wachusett drinking water intake during 2008–2011. Cyanobacteria concentrations were calculated as the maximum value of the five satellite pixels within a 900 m buffer of the drinking water intake. The shadowed data in December, January and February were not used in the statistical analysis
Daily counts of GI illness had a significant positive correlation with school days, the concentrations of NO2, and cyanobacteria concentration, but a significant negative correlation with O3, PM2.5, and daily mean temperature. Similarly, the daily count of respiratory illness visits had a significant positive correlation with school days, the concentrations of NO2, CO, and cyanobacteria concentration, but a significant negative correlation with O3, PM2.5, and daily mean temperature. Daily counts of dermal illness were negatively correlated with school days, SO2, NO2, CO, precipitation, and cyanobacteria concentration, but positively correlated with holidays, O3, PM2.5, and daily mean temperature (Table 2).
The results from the first stage model for the three types of illnesses (GI, respiratory, dermal) are shown in Table 3. Days of the week and autoregression components, as well as spline functions of time, were significant predictors of daily counts of illness in all three models. Different meteorological and air pollution variables were retained in final stage 1 models for respiratory, dermal, and GI illnesses. The model residuals from the first stage models for the three types of illnesses are shown in Fig. 4. All residuals are approximately homoscedastic and normally distributed, lacking temporal trends and seasonal patterns.
The results from the second stage models show that there were no significant associations between cyanobacteria concentrations and daily visits for GI illness at all time lags analyzed (Table 4). There was a significant positive association between satellite-derived cyanobacteria abundance and visits for respiratory illnesses at the two-day lag (Table 5). The adjusted effect associated with the highest category of cyanobacteria concentration was 4.3 additional cases with the 95% CI: 0.7–8.0 cases (n = 268). Associations at all other lags were not significant. Visits for dermal illnesses were not significantly associated with the level of cyanobacteria concentrations (Table 6). In addition, we divided the population of water consumers in different age groups (children, adults, and the elderly) and conducted stratified analysis using the same statistical models for each stratum. We did not find significant associations in individual groups.
Discussion
This study evaluated acute illness among consumers of municipal drinking water associated with exposure to cyanobacterial blooms estimated using remotely sensed imagery. We investigated the association between satellite-derived cyanobacteria concentration in Wachusett Reservoir in Massachusetts during 2008–2011 and ER visits for three types of acute illnesses among residents of towns that receive drinking water from this reservoir.
To account for potential time-varying confounding factors, the two-stage modeling approach enabled utilization of daily data on health outcomes, weather, and air pollution. Regression analysis of detrended and normally distributed residuals from stage one autoregression GAM against cyanobacteria data demonstrated a lagged increase in the number of ER visits for respiratory illnesses associated with the highest category of cyanobacteria concentrations (> 109,468 cells/mL). It is important to highlight that the only significant association was detected at a 2-day lag corresponding to the average water residence time in the distribution system. This result suggests that drinking water from source water impacted by cyanobacterial blooms might be associated with respiratory symptoms following consumption. However, in contrast to other studies on effects associated with drinking water exposures to cyanobacteria, we did not observe significant associations between cyanobacteria in source water and ER visits for GI or dermal illnesses.
We are not aware of previously published reports of respiratory illness associated with exposure to cyanobacteria-impacted drinking water. However, several studies have examined the associations between respiratory illnesses and exposure to cyanobacteria in recreational water [16, 17, 27, 37–40]. Some of the previous reports describe severe respiratory illnesses such as pneumonia that resulted in hospital visits [27, 39, 40].
Hilborn et al. 2014 [16] reported that individuals recreating in or at cyanobacteria-impacted waters generally reported illness (including cough, wheeze, congestion and tight chest) on the day of exposure. In our study, the significant effect was observed at a two-day lag, which is the average time interval between water entering the intake at Wachusett Reservoir and water receipt by households in the study area. Fortunately, exposure to cyanobacteria via drinking water is not commonly reported because of the general effectiveness of drinking water treatment. If contamination does occur, there are multiple potential respiratory exposure pathways, including dishwashing, bathing, lawn watering, or other uses of tap water where a water aerosol is created [41].
Reports of GI and dermal illnesses associated with cyanobacterial blooms in drinking water have previously been published; however, findings from other studies were inconsistent. El Saadi et al. 1995 [42] found that the risk of GI symptoms was significantly increased for people drinking chlorinated water derived from a cyanobacteria-impacted river, and the risks of GI and dermal symptoms were increased for people using untreated water from the same river for bathing and dishwashing. However, the odds for developing GI and dermal symptoms was not significant for those exposed to recreational river water. Lévesque et al. 2014 [28] reported that exposure to cyanobacteria and microcystins in source water was linked to subsequent GI, muscle pain, skin, and ear symptoms, but symptoms were reported to be mild and did “not require a medical consultation.”
In this study, we did not observe a significant association between cyanobacteria and GI or dermal illnesses while some previous studies detected significant associations. A possible explanation for these discrepant results is that the exposure to cyanobacteria and health effects were defined in different ways in various studies. For example, Levesque et al. 2014 [28] counted cyanobacterial cells at multiple depths at sample sites in the drinking water source, and also measured cyanotoxin at those sampling sites. In their study, health effects among drinking water consumers were self-reported; therefore, they could capture mild symptoms which did not result in an ER visit. It is possible that any exposure to cyanobacteria in tap water in the Greater Boston area was associated with mild symptoms among water consumers which our study could not detect. This may also be because Levesque et al. 2014 [28] measured cyanotoxins in water while we used satellite data, which have two important limitations. First, satellite images cannot be used to measure toxins; and second, satellite images provide information for the surface layer of a waterbody. The satellite only measures the water’s surface, penetrating up to a depth of about 2 m in clear water [43] and less than 2 m in more turbid waters [9, 44]. This means that, depending on the depth of the drinking water intake, cyanobacteria concentrations at the water intake depth may not have been assessed in this study [3].
Beaudeau et al. 2014 [45] studied the same public water supply system, MWRA, but during a different time frame (1998–2008). They reported an association between the presence of cyanobacteria at the drinking water intake and hospital admissions for GI illness among the elderly 23–27 days later. Their study did not evaluate respiratory or dermal illnesses. Beaudeau et al. 2014 [45] also observed a decline in hospital admissions for GI illness after the water utility started treating drinking water with ozone in 2005 (prior to that, they used only chlorine for disinfection). Our study was wholly conducted after the introduction of ozone treatment. Therefore, it is possible that exposure to cyanobacteria via drinking water was diminished after 2005, as ozone is more effective at removing cyanobacterial toxins than chlorine [13].
Satellite remote sensing has a few additional limitations for human exposure assessment to cyanobacterial blooms. First, the availability and quality of satellite data is subject to the influence of cloud cover, snow and ice as well as other factors [3, 26]. Therefore, cyanobacterial bloom data were not available for every time the satellite passed over the study area. As our analysis was conducted in two stages, we were able to include full-year daily data on health, weather and air quality in stage one models. Thus, seasonality analysis was conducted using a full health dataset enabling us to properly characterize and remove seasonal patterns from health data. We had to exclude cyanobacteria data for December, January, and February from our stage two regression models because the presence of ice and snow or dense cloud cover could impact the accuracy of estimating cyanobacterial cell concentrations using satellite images. This limitation was due to the location of our study site in a northern part of the country where ice cover is formed on surface water bodies in winter.
Additionally, satellite-derived cyanobacteria concentration represents the maximum value within a 900-m buffer around the intake while actual cyanobacteria concentrations may be heterogenous spatially. Surface cyanobacteria concentrations may not accurately represent water quality at the actual drinking water intake depth.
This study has other inherent limitations. As an observational study, it could only demonstrate statistical associations but not causality. Although statistical analysis involved adjusting for multiple time-varying covariates and spline function of time, there could still be residuals from unmeasured time-varying factors correlated with cyanobacteria occurrence and daily disease counts. Furthermore, there could be ingestion exposure misclassification due to some individuals not drinking tap water. That might explain the lack of observed association between cyanobacteria and GI illness as such exposure misclassification would have biased the observed effect towards the null.
Despite the mentioned limitations, results from this study add to the existing literature of merging satellite technology for water quality with human health [46]. Our study has a few strengths. First, we simultaneously investigated three types of acute illnesses previously identified as being associated with cyanobacterial blooms, where previous studies have been largely restricted to the evaluation of gastrointestinal effects. A restricted a priori focus limits the ability to characterize health risks associated with exposure to drinking water impacted by cyanobacterial blooms. Second, we used daily health outcome data, which allowed us to characterize lagged effects of cyanobacterial blooms. Third, we analyzed associations between cyanobacteria concentration categories and illness counts at various lags from zero to 6 days and detected an association only at the two-day lag corresponding to the water residence time in the distribution system. Previous studies of recreational exposure demonstrated that illness symptoms occur on the day of exposure to cyanobacteria [16, 47]. We conducted statistical analysis at all 7 days in order to rule out confounding effects.
Conclusions
We found a significant positive association between respiratory illnesses in the Greater Boston area and satellite-derived cyanobacteria concentration in source water at a time lag consistent with the water residence time in the distribution system. Because in situ monitoring data at the drinking water intake were not analyzed, this novel finding needs to be confirmed in future studies of health effects associated with exposure to cyanobacteria-impacted drinking water sources.
Availability of data and materials
The health data used in this study are available from the Massachusetts Center for Health Information Analysis but restrictions apply to the availability of these data, which were used under data use agreement for the current study. The other data will be available from USEPA ScienceHub as the paper is published.
Abbreviations
- AIC:
-
Akaike information criterion
- AQS:
-
Air quality monitoring site
- CO:
-
Carbon monoxide
- ER:
-
Emergency room
- cyanoHABs:
-
Cyanobacterial harmful algal blooms
- GAM:
-
Generalized additive model
- GI:
-
Gastrointestinal
- ICD-9:
-
International Classification of Diseases, Ninth Revision
- MERIS:
-
MEdium Resolution Imaging Spectrometer
- MWRA:
-
Massachusetts Water Resources Authority
- NO2 :
-
Nitrogen dioxide
- O3 :
-
Ozone
- PM2.5 :
-
Fine particulate matter
- SO2 :
-
Sulfur dioxide
- USEPA:
-
U.S. Environmental Protection Agency
References
Hilborn ED, Beasley VR. One health and cyanobacteria in freshwater systems: animal illnesses and deaths are sentinel events for human health risks. Toxins. 2015;7(4):1374–95. https://doi.org/10.3390/toxins7041374.
Huisman J, Codd GA, Paerl HW, Ibelings BW, Verspagen JM, Visser PM. Cyanobacterial blooms. Nat Rev Microbiol. 2018;16(8):471–83. https://doi.org/10.1038/s41579-018-0040-1.
Clark JM, Schaeffer BA, Darling JA, Urquhart EA, Johnston JM, Ignatius AR, et al. Satellite monitoring of cyanobacterial harmful algal bloom frequency in recreational waters and drinking water sources. Ecol Indic. 2017;80:84–95. https://doi.org/10.1016/j.ecolind.2017.04.046.
Paerl HW, Paul VJ. Climate change: links to global expansion of harmful cyanobacteria. Water Res. 2012;46(5):1349–63. https://doi.org/10.1016/j.watres.2011.08.002.
Mishra S, Richard P, Stumpf RP, Schaeffer BA, WERdell PJ, Loftin KA, Meredith A. Measurement of cyanobacterial bloom magnitude using satellite remote sensing. Sci Rep. 2019;9(1):18310. https://doi.org/10.1038/s41598-019-54453-y.
Falconer I, Humpage A. Health risk assessment of cyanobacterial (blue-green algal) toxins in drinking water. Int J Environ Res Public Health. 2005;2(1):43–50. https://doi.org/10.3390/ijerph2005010043.
O’neil J, Davis T, Burford M, Gobler C. The rise of harmful cyanobacteria blooms: the potential roles of eutrophication and climate change. Harmful Algae. 2012;14:313–34. https://doi.org/10.1016/j.hal.2011.10.027.
Paerl HW, Otten TG. Harmful cyanobacterial blooms: causes, consequences, and controls. Microb Ecol. 2013;65(4):995–1010. https://doi.org/10.1007/s00248-012-0159-y.
Hoeger SJ, Hitzfeld BC, Dietrich DR. Occurrence and elimination of cyanobacterial toxins in drinking water treatment plants. Toxicol Appl Pharmacol. 2005;203(3):231–42. https://doi.org/10.1016/j.taap.2004.04.015.
Qin B, Zhu G, Gao G, Zhang Y, Li W, Paerl HW, et al. A drinking water crisis in Lake Taihu, China: linkage to climatic variability and lake management. Environ Manag. 2010;45(1):105–12. https://doi.org/10.1007/s00267-009-9393-6.
Ueno Y, Nagata S, Tsutsumi T, Hasegawa A, Watanabe MF, Park H-D, et al. Detection of microcystins, a blue-green algal hepatotoxin, in drinking water sampled in Haimen and Fusui, endemic areas of primary liver cancer in China, by highly sensitive immunoassay. Carcinogenesis. 1996;17(6):1317–21. https://doi.org/10.1093/carcin/17.6.1317.
de Figueiredo DR, Azeiteiro UM, Esteves SM, Gonçalves FJ, Pereira MJ. Microcystin-producing blooms—a serious global public health issue. Ecotoxicol Environ Saf. 2004;59(2):151–63. https://doi.org/10.1016/j.ecoenv.2004.04.006.
He XX, Liu YL, Conklin A, Westrick J, Weavers LK, Dionysiou DD, et al. Toxic cyanobacteria and drinking water: impacts, detection, and treatment. Harmful Algae. 2016;54:174–93. https://doi.org/10.1016/j.hal.2016.01.001.
McCarty CL, Nelson L, Eitniear S, Zgodzinski E, Zabala A, Billing L, et al. Community needs assessment after microcystin toxin contamination of a municipal water supply - Lucas County, Ohio, September 2014. Morb Mortal Wkly Rep. 2016;65(35):925–9. https://doi.org/10.15585/mmwr.mm6535a1.
Cheung MY, Liang S, Lee J. Toxin-producing cyanobacteria in freshwater: A review of the problems, impact on drinking water safety, and efforts for protecting public health. J Microbiol. 2013;51(1):1–10. https://doi.org/10.1007/s12275-013-2549-3.
Hilborn ED, Roberts VA, Backer L, DeConno E, Egan JS, Hyde JB, et al. Algal bloom–associated disease outbreaks among users of freshwater lakes—United States, 2009–2010. Morb Mortal Wkly Rep. 2014;63(1):11–5.
Stewart I, Webb PM, Schluter PJ, Shaw GR. Recreational and occupational field exposure to freshwater cyanobacteria–a review of anecdotal and case reports, epidemiological studies and the challenges for epidemiologic assessment. Environ Health. 2006;5(1):6. https://doi.org/10.1186/1476-069X-5-6.
Stumpf RP, Davis TW, Wynne TT, Graham JL, Loftin KA, Johengen TH, et al. Challenges for mapping cyanotoxin patterns from remote sensing of cyanobacteria. Harmful Algae. 2016;54:160–73. https://doi.org/10.1016/j.hal.2016.01.005.
Hunter PD, Tyler AN, Gilvear DJ, Willby NJ. Using remote sensing to aid the assessment of human health risks from blooms of potentially toxic cyanobacteria. Environ Sci Technol. 2009;43(7):2627–33. https://doi.org/10.1021/es802977u.
Bosse KR, Sayers MJ, Shuchman RA, Fahnenstiel GL, Ruberg SA, Fanslow DL, et al. Spatial-temporal variability of in situ cyanobacteria vertical structure in Western Lake Erie: implications for remote sensing observations. J Great Lakes Res. 2019;45(3):480–9. https://doi.org/10.1016/j.jglr.2019.02.003.
Malayeri HZ, Twardowski M, Sullivan J, Moore T, Choi H. Correlation of cyanobacterial harmful bloom monitoring parameters: A case study on western Lake Erie. AIMS Environ Sci. 2018;5(1):24–34. https://doi.org/10.3934/environsci.2018.1.24.
Stumpf RP, Johnson LT, Wynne TT, Baker DB. Forecasting annual cyanobacterial bloom biomass to inform management decisions in Lake Erie. J Great Lakes Res. 2016;42(6):1174–83. https://doi.org/10.1016/j.jglr.2016.08.006.
Greb S, Dekker A, Binding C, Bernard S, Brockmann C, DiGiacomo P, et al. Earth observations in support of global water quality monitoring; 2018.
Kutser T. Passive optical remote sensing of cyanobacteria and other intense phytoplankton blooms in coastal and inland waters. Int J Remote Sens. 2009;30(17):4401–25. https://doi.org/10.1080/01431160802562305.
Coffer MM, Schaeffer BA, Urquhart EA, Darling JA, Salls WB. Quantifying national and regional cyanobacterial occurrence in US lakes using satellite remote sensing. Ecol Indic. 2020;111:105976. https://doi.org/10.1016/j.ecolind.2019.105976.
Urquhart EA, Schaeffer BA, Stumpf RP, Loftin KA, Werdell PJ. A method for examining temporal changes in cyanobacterial harmful algal bloom spatial extent using satellite remote sensing. Harmful Algae. 2017;67:144–52. https://doi.org/10.1016/j.hal.2017.06.001.
Giannuzzi L, Sedan D, Echenique R, Andrinolo D. An acute case of intoxication with cyanobacteria and cyanotoxins in recreational water in Salto Grande dam, Argentina. Marine Drugs. 2011;9(11):2164–75. https://doi.org/10.3390/md9112164.
Lévesque B, Gervais M-C, Chevalier P, Gauvin D, Anassour-Laouan-Sidi E, Gingras S, et al. Prospective study of acute health effects in relation to exposure to cyanobacteria. Sci Total Environ. 2014;466:397–403.
Lin C. Drinking water quality and human health: impact of harmful algae and water pipe breaks. Chapel Hill: University of North Carolina; 2018.
Urquhart EA, Schaeffer BA. Envisat MERIS and Sentinel-3 OLCI satellite Lake biophysical water quality flag dataset for the contiguous United States. Data in Brief. 2019;28:104826.
Lunetta RS, Schaeffer BA, Stumpf RP, Keith D, Jacobs SA, Murphy MS. Evaluation of cyanobacteria cell count detection derived from MERIS imagery across the eastern USA. Remote Sens Environ. 2015;157:24–34. https://doi.org/10.1016/j.rse.2014.06.008.
Schaeffer BA, Bailey SW, Conmy RN, Galvin M, Ignatius AR, Johnston JM, et al. Mobile device application for monitoring cyanobacteria harmful algal blooms using Sentinel-3 satellite ocean and land colour instruments. Environ Model Softw. 2018;109:93–103. https://doi.org/10.1016/j.envsoft.2018.08.015.
Mishra S, Stumpf RP, Schaeffer B, Werdell PJ, Loftin KA, Meredith A. Evaluation of a satellite-based cyanobacteria bloom detection algorithm using field-measured microcystin data. Sci Total Environ. 2021;774:145462. https://doi.org/10.1016/j.scitotenv.2021.145462.
Holleman DR, Bowling RL, Gathy C. Predicting daily visits to a waik-in clinic and emergency department using calendar and weather data. J Gen Intern Med. 1996;11(4):237–9. https://doi.org/10.1007/BF02642481.
Noble JH, LaMontagne ME, Bellotti C, Wechsler H. Variations in visits to hospital emergency care facilities: ritualistic and meteorological factors affecting supply and demand. Med Care. 1971;9(5):415–27. https://doi.org/10.1097/00005650-197109000-00005.
Akaike H. A new look at the statistical model identification. In: selected papers of Hirotugu Akaike. Springer. 1974;19(6):714–23.
Lin CJ, Wade TJ, Sams EA, Dufour AP, Chapman AD, Hilborn ED. A prospective study of marine phytoplankton and reported illness among recreational beachgoers in Puerto Rico, 2009. Environ Health Perspect. 2016;124(4):477–83. https://doi.org/10.1289/ehp.1409558.
Pilotto LS, Douglas RM, Burch MD, Cameron S, Beers M, Rouch GJ, et al. Health effects of exposure to cyanobacteria (blue–green algae) during recreational water–related activities. Aust N Z J Public Health. 1997;21(6):562–6. https://doi.org/10.1111/j.1467-842X.1997.tb01755.x.
Trevino-Garrison I, DeMent J, Ahmed F, Haines-Lieber P, Langer T, Ménager H, et al. Human illnesses and animal deaths associated with freshwater harmful algal blooms—Kansas. Toxins. 2015;7(2):353–66. https://doi.org/10.3390/toxins7020353.
Turner PC, Gammie AJ, Hollinrake K, Codd GA. Pneumonia associated with contact with cyanobacteria. BMJ. 1990;300(6737):1440–1. https://doi.org/10.1136/bmj.300.6737.1440.
Facciponte DN, Bough MW, Seidler D, Carroll JL, Ashare A, Andrew AS, et al. Identifying aerosolized cyanobacteria in the human respiratory tract: A proposed mechanism for cyanotoxin-associated diseases. Sci Total Environ. 2018;645:1003–13. https://doi.org/10.1016/j.scitotenv.2018.07.226.
Saadi OE, Esterman AJ, Cameron S, Roder DM. Murray River water, raised cyanobacterial cell counts, and gastrointestinal and dermatological symptoms. Med J Aust. 1995;162(3):122–5. https://doi.org/10.5694/j.1326-5377.1995.tb138473.x.
Mishra DR, Narumalani S, Rundquist D, Lawson M. Characterizing the vertical diffuse attenuation coefficient for downwelling irradiance in coastal waters: implications for water penetration by high resolution satellite data. ISPRS J Photogramm Remote Sens. 2005;60(1):48–64. https://doi.org/10.1016/j.isprsjprs.2005.09.003.
Wynne TT, Stumpf RP, Tomlinson MC, Dyble J. Characterizing a cyanobacterial bloom in western Lake Erie using satellite imagery and meteorological data. Limnol Oceanogr. 2010;55(5):2025–36. https://doi.org/10.4319/lo.2010.55.5.2025.
Beaudeau P, Schwartz J, Levin R. Drinking water quality and hospital admissions of elderly people for gastrointestinal illness in eastern Massachusetts, 1998–2008. Water Res. 2014;52:188–98. https://doi.org/10.1016/j.watres.2014.01.005.
Stroming S, Robertson M, Mabee B, Kuwayama Y, Schaeffer B. Quantifying the human health benefits of using satellite information to detect cyanobacterial harmful algal blooms and manage recreational advisories in US Lakes. GeoHealth. 2020;4(9):e2020GH000254.
Billings WH. Water-associated human illness in northeast Pennsylvania and its suspected association with blue-green algae blooms. In: Carmichael WW, editor. The Water Environment Environmental Science Research. Boston: Springer; 1981. p. 243–55.
Acknowledgements
The views expressed in this report are those of the individual authors and do not necessarily reflect the views and policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Funding
This work was supported the NASA Ocean Biology and Biogeochemistry Program/Applied Sciences Program (proposal 14-SMDUNSOL14–0001 and SMDSS20–000) and by U.S. EPA, NOAA, U.S. Geological Survey Toxic Substances Hydrology Program. This research was also supported in part by an appointment to the Research Participation Program at the US EPA, Office of Research and Development (ORD), administered by the Oak Ridge Institute for Science and Education (ORISE) through interagency agreement between the US Department of Energy and EPA.
Author information
Authors and Affiliations
Contributions
EDH conceived the study and its design, coordinated data collection and analysis. JW conducted the data analysis and drafted the initial manuscript. BAS, EU, MMC and CJL participated in the data collection and provided technical input to the data analyses. AIE participated in data analysis and result interpretation. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
N/A
Consent for publication
N/A
Competing interests
The authors declare that they have no conflict of interests with this work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, J., Hilborn, E.D., Schaeffer, B.A. et al. Acute health effects associated with satellite-determined cyanobacterial blooms in a drinking water source in Massachusetts. Environ Health 20, 83 (2021). https://doi.org/10.1186/s12940-021-00755-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12940-021-00755-6