Abstract
Background
Spatio-temporal models are increasingly being used to predict exposure to ambient outdoor air pollution at high spatial resolution for inclusion in epidemiological analyses of air pollution and health. Measurement error in these predictions can nevertheless have impacts on health effect estimation. Using statistical simulation we aim to investigate the effects of such error within a multi-level model analysis of long and short-term pollutant exposure and health.
Methods
Our study was based on a theoretical sample of 1000 geographical sites within Greater London. Simulations of “true” site-specific daily mean and 5-year mean NO2 and PM10 concentrations, incorporating both temporal variation and spatial covariance, were informed by an analysis of daily measurements over the period 2009–2013 from fixed location urban background monitors in the London area. In the context of a multi-level single-pollutant Poisson regression analysis of mortality, we investigated scenarios in which we specified: the Pearson correlation between modelled and “true” data and the ratio of their variances (model versus “true”) and assumed these parameters were the same spatially and temporally.
Results
In general, health effect estimates associated with both long and short-term exposure were biased towards the null with the level of bias increasing to over 60% as the correlation coefficient decreased from 0.9 to 0.5 and the variance ratio increased from 0.5 to 2. However, for a combination of high correlation (0.9) and small variance ratio (0.5) non-trivial bias (> 25%) away from the null was observed. Standard errors of health effect estimates, though unaffected by changes in the correlation coefficient, appeared to be attenuated for variance ratios > 1 but inflated for variance ratios < 1.
Conclusion
While our findings suggest that in most cases modelling errors result in attenuation of the effect estimate towards the null, in some situations a non-trivial bias away from the null may occur. The magnitude and direction of bias appears to depend on the relationship between modelled and “true” data in terms of their correlation and the ratio of their variances. These factors should be taken into account when assessing the validity of modelled air pollution predictions for use in complex epidemiological models.
Similar content being viewed by others
Background
The lack of accurate measurements of a subject’s short (e.g. day to day) or long-term (e.g. year to year) exposure to ambient outdoor air pollution, leads to estimated health effects of such exposure in epidemiological studies that are prone to bias and / or reduced statistical power with the extent of these problems depending on the magnitude of the imprecision or measurement error and its type [1]. In the past most studies estimated individual-level exposure to air pollutants based on the nearest monitor(s) to subject residence or an area average of monitor measurements. However more recently spatio-temporal models have been used facilitating the estimation of daily pollutant concentrations at high spatial resolution. While these models increase the precision of address-level exposure estimation, they are not free of measurement error: classical/classical-like error due to model parameter estimation and Berkson/Berkson-like error due to spatial smoothing [2]. While classical error tends to bias health effect estimates towards the null, both error types but particularly Berkson error results in reduced statistical power [3]. Various simulation studies have investigated the effects of measurement error in different scenarios involving different epidemiological models and evaluating different approaches to the estimation of ambient air pollution concentrations [2, 4,5,6,7,8,9,10,11,12,13]. In one such study we investigated the use of outputs from the EMEP-WRF chemistry transport model in a time-series analysis [5]. In this paper we extend the methodology previously applied by giving our “true” pollution data a more representative distribution spatially (i.e. allowing for the spatial correlation of long-term pollutant means as well as the spatial correlation of day to day pollutant concentrations) and by investigating the effects of measurement error in a multi-level analysis for the joint estimation of the health effects of both short and long-term pollutant exposure [14]. We simulate scenarios in which we specify a) the spatial and the temporal correlation between “true” and model data and b) the ratio of the variance in model data to the variance in “true” data (which we also assume is the same both temporally and spatially). For each scenario we run 500 simulations and report on the impact in terms of bias in estimation, coverage of 95% confidence intervals (CIs) and statistical power.
Methods
Data analysis
Our simulation of “true” exposure and outcome data were informed by an analysis of 63,865 daily mean NO2 measurements and 48,151 daily mean PM10 measurements from 47 (1 suburban and 46 urban) and 37 (2 suburban and 35 urban) background monitoring sites respectively, and covering the period 2009–13. The monitoring data were sourced from: Air Quality England [15] and the London Air Quality Network, [16] which included data from the Automatic Urban and Rural Network (AURN) [17]. All sites were operated to comparable international QA/QC standards, [18] and were situated within the confines of the London M25 circular road network.
The mean (variance) of the site-specific 5-year means was 36.52 μg/m3 (76.200 (μg/m3)2) for NO2 and 20.17 μg/m3 (8.715 (μg/m3)2) for PM10; the average within-site variance was 274.608 (μg/m3)2 for daily mean NO2 and 104.815 (μg/m3)2 for daily mean PM10; the average within-site variance of the 5-year means was 0.237 (μg/m3)2 for NO2 and 0.094 (μg/m3)2 for PM10. A full description of the analysis for NO2 is given in Additional file 1.
Simulation set-up
Based on London’s extensive monitoring network we initially simulated daily “true” concentrations for each pollutant over a period of 5 years in 1000 locations. We consequently simulated: total mortality data from the “true” exposure series through previously identified effect estimates; and then modelled exposure data from the “true” series under several measurement error scenarios. The section below briefly describes the steps involved and these will be illustrated using results from our NO2 analysis (Additional file 1).
Step 1
Our simulation study sample consisted of 1000 sites. Each site was assumed to represent the centroid of a Lower Super Output Area (LSOA) and was defined by a pair of easting (E) and northing (N) co-ordinates. An LSOA is a small area with an average population of approximately 1500 subjects [19]. The co-ordinate variables E and N (i.e. (ei, ni), i = 1, …, 1000) were sampled at random from a multivariate normal distribution with means (528, 182), variances (172.544, 51.260) and covariance (9.097).
Step 2
For each site i (i = 1, …, 1000) and each day t (t = 1, …1826) we simulated “true” mean daily concentrations xi,t as follows:
The systematic component of spatial variation ui in (1) was estimated from modelling the long-term average pollutant measurements as a function of co-ordinates e.g. for NO2
The spatial variance covariance matrix S in (2) was estimated by fitting a model with exponential covariance function to a semivariogram of the residuals e.g. for NO2
where di,j is the Euclidian distance between sites i.e.
The temporal variance covariance matrix Λ in (3) was informed by the mean of the within-site variances and a linear regression line linking Pearson correlations over-time between site-pairs (i, j) with their corresponding Euclidean between-site distances (di,j) e.g. for NO2.
Λ (i, j) = 274.608 × (0.7999 − (0.0016 × di,j)).
Step 3
We simulated outcome data yi,t for site i on day t from the “true” pollutant data xi,t based on the average crude death rate per day in a London LSOA in 2011 (i.e. 0.0264), which we estimated using data from the Office of National Statistics, [20, 21] and pre-specified concentration response functions (CRF) for deaths associated with both short-term and long-term exposure, as follows:
where \( {\overline{x}}_i \) is the average site-specific “true” concentration over the 5-year study period, β1 is the short-term estimate, β2 the long-term estimate and ei ~N(0, 1).
For NO2, we assumed a short-term CRF (β1in eq. (4)) of loge(1.0071)/10 = 0.000707 per 1 μg/m3, [22] and a long-term CRF (β2 in equation (4)) of loge(1.023)/10 = 0.00227 per 1 μg/m3, [23] (personal communication) and for PM10 short and long-term CRFs of loge(1.0051)/10 = 0.000509 per 1 μg/m3, [24] and loge(1.07)/10 = 0.00677 per 1 μg/m3, [23] respectively.
Step 4
Next we simulated “pseudo” model data zi,t from the “true” pollutant data setting the temporal correlation between “true” and model data to αt; the spatial correlation between “true” and model 5-year means to αs; the ratio of model versus “true” variances temporally (variance of daily data within site) to γt; and the ratio of model versus “true” variances spatially (variance of 5-year means across sites) to γs.
The following formula is an extension of that used in Butland et al., [5] and has its origins in an approach by Reeves et al., [25] and a generalisation of second-order regression as outlined in Cox and Hinkley [26]. Our choice of a constant term here was arbitrary (we used 3.5 μg/m3 for both NO2 and PM10). Further details are contained in Additional file 2.
In the above, ϵ2 represents the variance of “true” daily data within-site which is assumed to be the same across all sites. Thus for NO2: \( \mathit{\operatorname{var}}\left({\overline{x}}_i\right)=76.200-0.237= \)75.963 and ϵ2 = 274.608 × 0.7999 = 219.659.
Step 5
Finally we analysed the association between outcomes yi,t and modelled short (zi,t) and long-term \( \left({\overline{z}}_i\right) \) exposures using a simplified version of the statistical model proposed by Kloog et al., [14] i.e.
The aim, to obtain coefficient estimates and their standard errors i.e., \( \hat{\beta_1} \), \( se\left(\hat{\beta_1}\right),\kern0.5em \hat{\beta_2} \), \( se\left(\hat{\beta_2}\right) \).
Step 6
Steps 2–5 were then repeated 500 times and summary statistics calculated for the coefficient estimates and their standard errors.
Defining the different scenarios
We simplified our scenarios by setting γt = γs=λ and αt = αs = τ but allowed λ to take values (2, 1.25, 1, 0.75, 0.5) and τ to take values (0.5, 0.6, 0.7, 0.8, 0.9). It is worth noting that based on standard measurement error theory pure classical error would produce a value of λ > 1 and pure Berkson error a value of λ < 1 [1]. All simulations were run in R versions 3.3.2 and 3.4.3, [27] using the packages MASS, [28] Hmisc, [29] and lme4 [30]. Each scenario was run serially with a different 9 digit starting seed chosen at random from published tables of random numbers [31, 32].
Results
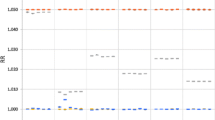
From Tables 1 and 2, it would appear that in general the health effect estimates were biased toward the null and to a similar degree for both short and long-term exposures. This bias tended to become more negative as the correlation coefficient decreased from 0.9 to 0.5 and the ratio of variances (model versus “true”) increased from 0.5 to 2.0 (Fig. 1).
At the extreme scenario under which the correlation coefficient was 0.5 and variance ratio was 2.0, attenuation was 65% for short-term exposure to NO2 and 74% for long-term exposure, while for PM10 the corresponding figures were 65% and 66%. However for high correlation of 0.9 combined with a low variance ratio of 0.5 bias away from the null was observed for both pollutants reaching 27% and 40% for short and long-term exposure to NO2 and 31% and 34% for short and long-term exposure to PM10. For both pollutants the standard errors of the health effect estimates appeared to be attenuated for variance ratios> 1 but inflated for variance ratios< 1 and these effects appeared to be independent of the correlation coefficient.
For effect estimates associated with short-term exposure, particularly those in Table 1 the coverage of 95% CIs appeared to depend on both the correlation coefficient and the variance ratio, reducing as the former got smaller and the latter increased. This can be seen graphically in Additional file 3: Figure S3.1. At the extreme scenario within which the correlation was 0.5 and a variance ratio was 2, the coverage probability fell to an estimated 19% for short-term exposure to NO2 (suggesting that only in 95 of our 500 simulated samples did the 95% CI contain the true value of β1), but a far less marked 72.8% for short-term exposure to PM10. For effect estimates associated with long-term exposure the 95% coverage probability exhibited comparatively little change across the various scenarios never falling below 84%.
For both pollutants the statistical power to detect an association with short-term exposure appeared to fall as the correlation between model and monitor data decreased, although for long-term exposure there was some slight tendency for power to decrease with both an increase in the variance ratio and a decrease in the correlation (see in Additional file 3: Figure S3.2 ).
Discussion
Based on our simulations we demonstrated downward biases in the health effect estimates associated with both long and short-term pollutant exposure, the magnitude of which depended on the correlation between modelled and true pollutant concentrations and the ratio of their variances (the lower the correlation coefficient and the higher the variance ratio of model versus “true” data the greater the attenuation). However for high correlation combined with a low variance ratio we observed some bias away from the null which at the extreme (i.e. correlation of 0.9 and variance ratio of 0.5) was non-trivial. The standard error of the simulated effect estimate appeared to depend on the variance ratio, with ratios >1 resulting in attenuation and those <1 in inflation. Marked attenuation in the coverage probability was observed for short-term exposures to NO2 when the temporal correlation between modelled and “true” data was low and the model exposure variance was greater than the “true”; and reductions in statistical power were observed for short-term exposures to both pollutants as the correlation coefficient decreased. Overall, statistical power for short-term exposure effects was higher for NO2 than PM10 (Additional file 3) but this may be attributed at least in part to the different CRFs driving their respective scenarios.
The aim of our methodology was to introduce measurement error of both types (i.e. classical / classical-like and Berkson / Berkson-like) by simulating “pseudo” model data which had on average a pre-specified correlation with the “true” data and a pre-specified variance ratio both spatially and temporally. The importance of the correlation coefficient (τ) and the variance ratio (λ) is clear simply from a consideration of the standard formula for total measurement error between model (Z) and true (X) data i.e.
The correlation coefficient between modelled and monitored data is often used as a measure of model validity, [33] and while a correlation of 0.8 would seem reasonably high, using outputs from such a model as exposure metrics in an epidemiological analysis may result in bias in the health effect estimate. Within our simulations assuming a correlation of 0.8 and a variance ratio of 2 we observed negative biases in the health effect estimates of between 42% and 46%. Increasing the correlation to 0.9 still resulted in a 32–37% negative bias in the health effect estimates emphasizing that measurement error adjustment is important in cohort studies as well as time-series and panel studies.
Gryparis et al.,[7] suggest that the smoothing inherent in spatio-temporal models effectively converts classical error into Berkson error, so that the latter is more of a concern. Thus for modelled pollution data a more realistic scenario maybe one where the overall variance of the model predictions is less than that of the “true” exposures (λ < 1); and under the scenarios of, λ = 0.5 and λ = 0.75, (Fig. 1) attenuation in the health effect estimate appeared to be less marked than for λ = 1, λ = 1.25 or λ = 2.0. However, for λ = 0.5 combined with a high correlation coefficient of 0.9, bias away from the null was observed for both short and long-term exposure ranging from 27% to 40%. In trying to explain these findings we note that the scenario effectively sets the covariance between the model and “true” data equal to 1.27 times (i.e.\( \frac{0.9}{\sqrt{0.5}} \)) the variance of the model data. This relationship is indicative of positive bias (based on simple regression calibration) [10, 25] but may only occur in practice if there is a lack of independence between the Berkson component of measurement error and the modelled data [9, 10]. While, in general Berkson error is not thought to introduce bias into the health effect estimate, some studies have shown that bias away from the null can occur due to Berkson error if additive on a log scale [9, 10].
Error (both classical and Berkson-like) can be introduced into an epidemiological analysis due to the use of model predictions that are misaligned in space from the observed data on which the model is based. In a simulation study and in the context of a linear regression analysis of cohort data, Szpiro et al., [4] investigated the impact of such error and reported only minimal bias in estimating the health effect estimate associated with long-term exposure. This is in contrast to our findings where negative bias in the health effect estimate was pronounced when the spatial correlation between “true” and modelled exposures was low, even for λ = 0.75 (Fig. 1). Low correlation may arise due to spatial misalignment but also model misspecification (i.e. the omission from LUR and/or kriging models of an important spatial covariate). Alexeef et al., 2016, [8] in the context of a linear regression analysis investigated the effects of model miss-specification for long-term exposures and, in common with our findings, their simulations illustrated a downward bias in the health effect estimate. However Szpiro et al., [2] demonstrated scenarios in which the use of a correctly specified model compared to a miss-specified model though resulting in more precise long-term exposure prediction did not result in improved health effect estimation. They concluded that more accurate exposure prediction does not necessarily improve the estimation of health effects as the additional parameter estimation involved may increase the classical-like error. It is therefore important, as illustrated by our own simulations, to consider both the correlation and the variance ratio when assessing the validity of modelled air pollutant outputs for use in epidemiological analyses.
The fact that bias in the standard error depends on the variance ratio is not unexpected. Indeed the pattern in standard errors observed across values of λ, is in line with the error inflation we might expect under a Berkson error model (λ < 1) and the bias in standard error estimation which can be in either direction (here attenuation) that we might expect under a classical error model (λ > 1) [3]. However it is not so clear why the standard error should not be influenced by the magnitude of the correlation coefficient.
Our simulations were based on 1000 sites (assumed to be the centroids of 1000 LSOAs) and therefore each simulated dataset was based on 1000 LSOAs × 1826 days =1,826,000 observations. Nevertheless, given the very small concentration response functions [22,23,24] this implies that statistical power to detect associations with both short-term exposure and particularly for long-term exposure were low. Indeed our simulations suggest that the power of our study set-up would be around 85% and 34% for short-term exposures to NO2 and PM10 respectively and 13% for long-term exposures. This combined with the use of only 500 simulations may have obscured any patterns in statistical power across the different scenarios for long-term exposure. However, despite this we did observe some reductions in power for long-term exposure, with some suggestion of greater attenuation with decreasing correlation and increasing variance ratio.
In terms of 95% CIs we observed some under-coverage for short-term exposures, especially for NO2 and for low correlation / high variance ratio scenarios. However coverage probabilities for long-term exposures varied little across all scenarios. This is likely due to the fact that within our simulations, as in real studies of the type considered here, health effect estimates associated with short-term exposures were based on larger numbers of observations and were therefore estimated with more precision as illustrated by their smaller standard errors. Thus for short-term exposures it only takes a small bias in the health effect estimate to move the 95% confidence interval so that it no longer contains the “true” value. The follow-on from this is that given a more powerful study any reduction in coverage probability may be more extreme and observed for both pollutants and both health effect estimates.
Simulation studies are limited in that they only inform you about the scenario in which they are set. It is therefore important that the scenario resembles to some extent a real world situation [34]. To this end we have simulated “true” pollutant data incorporating both temporal and spatial variation informed by real measurements from a large number of monitors situated within the London area. This is particularly important as two previous simulation studies [7, 12], suggest that the adverse effects of measurement error on health effect estimation may be moderated if there is high spatial correlation in the underlying true exposure surface. Nevertheless as in all simulation studies we have made various assumptions which may hold to a lesser or greater degree. In a real world setting for example the temporal and spatial correlation coefficients (model versus “true”) may not be the same and similarly variance ratios may differ. However the aim of our study was to present generalised scenarios rather than those that may be specific to any particular air pollution model, although our methods can easily be adapted to a more tailored approach if required. We also assume that modelled data are linearly related to the “true” exposures both over time and space. In other words the daily data are linearly related within site and the 5-year means are linearly related across sites. Given that the aim of pollution modelling is to provide an accurate representation of “true” pollutant values this does not seem to be unreasonable. The way in which we incorporated error into our “true” data in order to simulate “pseudo” model data is based on second-order regression equations [25, 26], and does not allow for the possibility that the classical components of measurement error may be spatially correlated. For modelled air pollution data output from spatio-temporal models based on LUR and / or universal kriging, it has been shown that classical type error resulting from parameter estimation will tend to be spatially correlated and heteroscedastic.[4] While we acknowledge this as a limitation of our approach, the aim of our simulations was to produce “pseudo” model datasets with given temporal and spatial correlations to the “true” and with a given variance ratio and it is often these measures that are used as markers of model performance particularly in terms of performance in epidemiological models [1, 25]. The success of incorporating these correlations and variance ratios into our “pseudo” modelled data was assessed by checks within our simulation programs. While overall these checks were reassuring they did suggest that in terms of the spatial variance ratio, the actual value introduced might be slightly higher than intended. However across all the scenarios in Tables 1 and 2 estimates of this bias (to 2 decimal places) were never more than 0.02 (e.g. spatial variance ratio 2.02 rather than 2.00).
It should also be appreciated that our hypothesized correlations between modelled and “true” exposures assume that the latter have had additive classical instrument error removed. While the assumption that monitor measurements are accurate (i.e. with no instrument error) may not be so important for long-term exposure estimation [7] it is not trivial in terms of short-term daily exposures [9]. Another point to consider is that our analysis is based at the level of a London LSOA, which is an area containing roughly 1500 subjects, [19] and was chosen in order to provide adequate numbers of events under the epidemiological model considered. Thus underlying our simulations is the assumption that monitor data (bar instrument error) accurately reflects the average exposure of residents within an LSOA and that we can ignore the Berkson error introduced by this effective averaging. Finally when simulating our “true” pollutant data we did not incorporate any seasonal pattern or time trend. This was done for simplicity and to avoid any corresponding adjustment in the multi-level Poisson regression analyses and thus any unforeseen effects of such an adjustment on our findings.
While our simulation study is designed to provide some insight into the effects of measurement error due to the use of modelled air pollution data in a complex epidemiological analysis, our results may also be informative to multi-level health analysis of other spatially distributed exposures.
Conclusions
Our results illustrate that measurement error in modelled air pollutant exposures can lead to non-trivial bias in health effect estimation. Although in general this bias is towards the null, under certain conditions bias away from the null may occur. In order to assess the magnitude and direction of this bias we need to consider both variance ratios and correlation coefficients. By allowing these factors to differ spatially and temporally, as outlined in Additional file 2, statistical simulation can be used to compare the performance (in terms of bias, coverage probability and power) of different pollutant modelling approaches (e.g. LUR, dispersion, satellite-based etc.) in order to find the best model or combination of models for use in a multi-level analysis of air pollution and health.
Abbreviations
- AURN:
-
Automatic urban and rural network
- CI:
-
Confidence interval
- CRF:
-
Concentration response function
- LSOA:
-
Lower super output area
- LUR:
-
Land use regression
References
Armstrong B. Effect of measurement error on epidemiological studies of environmental and occupational exposures. Occup Environ Med. 1998;55:651–6.
Szpiro AA, Paciorek CJ, Sheppard L. Does more accurate exposure prediction necessarily improve health effect estimates? Epidemiology. 2011;22:680–5.
Sheppard L, Burnett RT, Szpiro AA, Kim S-Y, Jerrett M, Pope III CA, Brunekreef B. Confounding and exposure measurement error in air pollution epidemiology. Air Qual Atmos Health. 2012;5:203–16.
Szpiro AA, Sheppard L, Lumley T. Efficient measurement error correction with spatially misaligned data. Biostatistics. 2011;12:610–23.
Butland BK, Armstrong B, Atkinson RW, Wilkinson P, Heal MR, Doherty RM, Vieno M. Measurement error in time-series analysis: a simulation study comparing modelled and monitored data. BMC Med Res Methodol. 2013;13:136.
Szpiro AA, Paciorek CJ. Measurement error in two-stage analyses, with application to air pollution epidemiology. Environmetrics. 2013;24:501–17.
Gryparis A, Paciorek CJ, Zeka A, Schwartz J, Coull BA. Measurement error caused by spatial misalignment in environmental epidemiology. Biostatistics. 2009;10:258–74.
Alexeeff SE, Carroll RJ, Coull B. Spatial measurement error and correction by spatial SIMEX in linear regression models when using predicted air pollution exposures. Biostatistics. 2016;17:377–89.
Strickland MJ, Gass KM, Goldman GT, Mulholland JA. Effects of ambient air pollution measurement error on health effect estimates in time series studies: a simulation-based analysis. J Expo Sci Environ Epidemiol. 2015;25:160–6.
Goldman GT, Mulholland JA, Russell AG, Strickland MJ, Klein M, Waller LA, Tolbert PE. Impact of exposure measurement error in air pollution epidemiology: effect of error type in time-series studies. Environ Health. 2011;10:61.
Dionisio KL, Chang HH, Baxter LK. A simulation study to quantify the impacts of exposure measurement error on air pollution health risk estimates in copollutant time-series models. Environ Health. 2016;15:114.
Kim S-Y, Sheppard L, Kim H. Health effects of long-term air pollution: influence of exposure prediction methods. Epidemiology. 2009;20:3.
Alexeeff SE, Schwartz J, Kloog I, Chudnovsky A, Koutrakis P, Coull BA. Consequences of kriging and land use regression for PM2.5 predictions in epidemiologic analyses: insights into spatial variability using high-resolution satellite data. J Expo Sci Environ Epidemiol. 2015;25:138–44.
Kloog I, Coull BA, Zanobetti A, Koutrakis P, Schwartz JD. Acute and chronic effects of particles on hospital admissions in New-England. PLoS One. 2012;7:e34664.
Air Quality England. Ricardo Energy and Environment http://www.airqualityengland.co.uk/. Accessed 1 Mar 2017.
London Air Quality Network. King’s college, London http://www.londonair.org.uk/. Accessed 1 Mar 2017.
Automatic Urban and Rural Network (AURN) Data Archive. © Crown 2017 copyright Defra via uk-air.defra.gov.uk, licenced under the Open Government Licence (OGL) v2.0. http://www.nationalarchives.gov.uk/doc/open-government-licence/version/2/. Accessed 1 Mar 2017.
Department for Environment Food and Rural Affairs. The Air Quality Validation and Ratification Process. https://uk-air.defra.gov.uk/assets/documents/Data_Validation_and_Ratification_Process_Apr_2017.pdf. Accessed 26 Feb 2018.
Department of Communities and Local Government. English Indices of Deprivation – LSOA level. https://data.gov.uk/dataset/english-indices-of-deprivation-2015-lsoa-level. Accessed 25 Sept 2017. Licenced under the Open Government Licence (OGL) v3.0. http://www.nationalarchives.gov.uk/doc/open-government-licence/version/3/.
Office for National Statistics‚ National Records of Scotland‚ Northern Ireland Statistics and Research Agency. Mortality Statistics: Deaths registered by area of usual residence, 2011 registrations. https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/deaths/datasets/deathsregisteredbyareaofusualresidenceenglandandwales. Accessed 21 Aug 2017. The data are © Crown Copyright 2013, licenced under the Open Government Licence (OGL) v3.0. http://www.nationalarchives.gov.uk/doc/open-government-licence/version/3/.
Office for National Statistics. 2011 Census: Usual residents by resident type, and population density, number of households with at least one usual resident and average household size, Output Areas (OAs) in London. https://www.ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/datasets/2011censuspopulationandhouseholdestimatesforwardsandoutputareasinenglandandwales. Accessed 22 Aug 2017. The data are © Crown Copyright 2012, licenced under the Open Government Licence v3.0. http://www.nationalarchives.gov.uk/doc/open-government-licence/version/3/.
Mills IC, Atkinson RW, Kang S, Walton H, Anderson HR. Quantitative systematic review of the associations between short-term exposure to nitrogen dioxide and mortality and hospital admissions. BMJ Open. 2015;5:e006946.
Carey IM, Atkinson RW, Kent AJ, van Staa T, Cook DG, Anderson HR. Mortality associations with long-term exposure to outdoor air pollution in a national English cohort. Am J Respir Crit Care Med. 2013:187:1226–33.
Anderson HR, Atkinson RW, Bremner SA, Carrington J, Peacock J. Quantitative systematic review of short term associations between ambient air pollution (particulate matter, ozone, nitrogen dioxide, sulphur dioxide and carbon monoxide), and mortality and morbidity. Department of Health. 2007. https://www.gov.uk/government/publications/quantitative-systematic-review-of-short-term-associations-between-ambient-air-pollution-particulate-matter-ozone-nitrogen-dioxide-sulphur-dioxide-and-carbon-monoxide-and-mortality-and-morbidity. Accessed 3 Oct 2017.
Reeves GK, Cox DR, Darby SC, Whitley E. Some aspects of measurement error in explanatory variables for continuous and binary regression models. Statist Med. 1998;17:2157–77.
Cox DR, Hinkley DV. Appendix 3 second-order regression for arbitrary random variables. In: Theoretical statistics. London: Chapman and Hall; 1974. p. 475–7.
Core Team R. R: a language and environment for statistical computing. Vienna: R Foundation for statistical computing. 2016 and 2017. https://www.R-project.org/.
Venables WN, Ripley BD. Modern applied statistics with S. 4th ed. New York: Springer; 2002.
Harrell Jr FE, with contributions from Dupont C and many others. Hmisc: Harrell miscellaneous. 2016 and 2018. R package versions 4.0–2 and 4.1–1. https://CRAN.R-project.org/package=Hmisc.
Bates D, Maechler M, Bolker B, Walkers S. Fitting linear mixed-effects models using lme4. J Statist Software. 2015;67:1–48.
Machin D, Campbell MJ. Statistical tables for the design of clinical trials. Oxford: Blackwell Scientific Publication; 1987. p. 200–2.
Armitage P. Statistical methods in medical research. Oxford: Blackwell Scientific Publications; 1971. p. 470–3.
Thunis P, Pederzoli A, Pernigotti D. Performance criteria to evaluate air quality modelling applications. Atmos Environ 2012;59:476–82.
Burton A, Altman DG, Royston P, Holder RL. The design of simulation studies in medical statistics. Statist Med. 2006;25:4279–92.
Acknowledgements
We acknowledge use of monitored pollutant data from: “Air Quality England” operated by Ricardo Energy and Environment (http://www.airqualityengland.co.uk/) and the “London Air Quality Network” operated by King’s College London (http://www.londonair.org.uk/) and which includes data from the Automatic Urban and Rural Network (AURN) Data Archive, © Crown 2017 copyright Defra via uk-air.defra.gov.uk, licenced under the Open Government Licence v2 (http://www.nationalarchives.gov.uk/doc/open-government-licence/version/2/).
Funding
Research in this paper as part of the STEAM project was funded under the MRC UK Grant ref.: MR/N014464/1.
Availability of data and materials
All monitoring data used in our study are available publically via data download tools available at websites listed in [15,16,17].
Author information
Authors and Affiliations
Contributions
BKB analysed the monitoring data, conducted the simulations and took the lead in drafting the paper. BB constructed the monitoring dataset. BKB, ES, RWA and KK were involved in the study design. All authors contributed to the drafting of the paper, and read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
There are no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Full set of results from the analysis of NO2 monitor data. (DOCX 1330 kb)
Additional file 2:
Further details of equations (5–7) used to express “pseudo” model data in terms of spatial and temporal correlations (αs and αt) and variance ratios (γs and γt). (DOCX 16 kb)
Additional file 3:
Additional graphs for coverage probabilities and statistical power. (DOCX 413 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
About this article
Cite this article
Butland, B.K., Samoli, E., Atkinson, R.W. et al. Measurement error in a multi-level analysis of air pollution and health: a simulation study. Environ Health 18, 13 (2019). https://doi.org/10.1186/s12940-018-0432-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12940-018-0432-8