Abstract
Accurate segmentation of multiple organs in the head, neck, chest, and abdomen from medical images is an essential step in computer-aided diagnosis, surgical navigation, and radiation therapy. In the past few years, with a data-driven feature extraction approach and end-to-end training, automatic deep learning-based multi-organ segmentation methods have far outperformed traditional methods and become a new research topic. This review systematically summarizes the latest research in this field. We searched Google Scholar for papers published from January 1, 2016 to December 31, 2023, using keywords “multi-organ segmentation” and “deep learning”, resulting in 327 papers. We followed the PRISMA guidelines for paper selection, and 195 studies were deemed to be within the scope of this review. We summarized the two main aspects involved in multi-organ segmentation: datasets and methods. Regarding datasets, we provided an overview of existing public datasets and conducted an in-depth analysis. Concerning methods, we categorized existing approaches into three major classes: fully supervised, weakly supervised and semi-supervised, based on whether they require complete label information. We summarized the achievements of these methods in terms of segmentation accuracy. In the discussion and conclusion section, we outlined and summarized the current trends in multi-organ segmentation.
Similar content being viewed by others
Introduction
Accurate segmentation of multiple organs in medical images is essential for various medical applications such as computer-aided diagnosis, surgical planning, navigation, and radiotherapy treatment [1, 2]. For instance, radiation therapy is a common treatment option for cancer patients, where tumor masses and high-risk microscopic areas are targeted [3]. However, radiation therapy can pose a significant risk to normal organs adjacent to the tumor, which are called organs at risk (OARs). Therefore, precise segmentation of both tumor and OARs contours is necessary to minimize the risk of radiation therapy [4, 5].
The early segmentation process relies heavily on manual labeling by physicians, which is labour-intense and time-consuming. For example, mapping 24 OARs in the head and neck region takes over 3 h, resulting in potential long waits for patients, especially in cases of patient overload [6]. Due to a shortage of experienced doctors, the mapping process becomes even more time-consuming, potentially delaying the patient's treatment process and missing the optimal treatment window [7]. Furthermore, the labeling results obtained by different physicians or hospitals exhibit significant variability [8,9,10,11]. Therefore, there is a pressing requirement for accurate and automated multi-organ segmentation methods in clinical practice.
Traditional methods [12,13,14,15] usually utilize manually extracted image features for image segmentation, such as the threshold method [16], graph cut method [17], and region growth method [18]. Limited by numerous manually extracted image features and the selection of non-robust thresholds or seeds, the segmentation results of these methods are usually unstable, and often yield only a rough segmentation result or only apply to specific organs. Knowledge-based methods leverage labeled datasets to automatically extract detailed anatomical information for various organs, reducing the need for manual feature extraction. This method can enhance the accuracy and robustness of multi-organ segmentation techniques, such as multi-atlas label fusion [19, 20] and statistical shape models [21, 22]. The method based on multi-atlas uses image alignment to align predefined structural contours to the image to be segmented. But this method typically includes multiple steps, therefore, the performance of this method may be influenced by various relevant factors involved in each step. Moreover, due to the use of fixed atlases, it is challenging to manage the anatomical variation of organs between patients. In addition, it is computationally intensive and takes a long time to complete an alignment task. The statistical shape model uses the positional relationships between different organs, and the shape of each organ in the statistical space as a constraint to regularize the segmentation results. However, the accuracy of this method is largely dependent on the reliability and extensibility of the shape model, and the model based on normal anatomical structures has very limited effect in the segmentation of irregular structures [23].
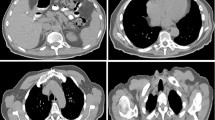
Compared to traditional methods that require manual feature extraction, deep learning can automatically learn the parameters of the model from a large number of data samples, enabling the model to learn complex features and patterns from the data. Recently, deep learning-based methods have gained considerable attention in several image processing applications such as image classification [24], object detection [25], image segmentation [26, 27], image fusion [28], image registration [29] due to their ability to extract features automatically. Methods based on deep learning have become a mainstream in the field of medical image processing. However, there are still several major challenges in multi-organ segmentation tasks. Firstly, there are significant variations in organ sizes, as illustrated by the head and neck in Fig. 1, the chest in Fig. 2, the abdomen in Fig. 3, and the organ size statistics in Fig. 4. Such size imbalances can lead to poor segmentation performance of the trained network for small organs. Secondly, the inherent noise and low contrast in CT images often result in ambiguous boundaries between different organs or tissue regions, thereby reducing the accuracy of organ boundary segmentation achieved by segmentation networks. Finally, due to safety and ethical concerns, many hospitals do not disclose their datasets, as a result, datasets used to train multiple organ segmentation models are very limited, and many segmentation methods are trained and validated on private datasets, making it difficult to compare with other methods. Consequently, there is an increasing demand for the development of multi-organ segmentation techniques that can accurately segment organs of different sizes, as shown in Fig. 5.
Schematic diagram of the organs of the head and neck, where the numbers are arranged in order: (1) brainstem, (2) left eye, (3) right eye, (4) left lens, (5) right lens, (6) left optic nerve, (7) right optic nerve, (8) Optical chiasm, (9) left temporal lobe, (10) right temporal lobe, (11) pituitary gland, (12) left parotid gland, (13) right parotid gland, (14) left temporal bone rock, (15) right temporal bone rock, (16) left temporal bone, (17) right temporal bone, (18) left mandibular condyle, (19) right mandibular condyle, (20) spinal cord, (21) left mandible, (22) right mandible. The segmentations and images are from the Automatic Radiotherapy Planning Challenge (StructSeg) in 2019 (https://structseg2019.grand-challenge.org/Dataset/)
Schematic diagram of the abdominal organs, where the numbers are arranged in order: (1) liver, (2) kidney, (3) spleen, (4) pancreas, (5) aorta, (6) inferior vena cava, (7) stomach, (8) gallbladder, (9) esophagus, (10) right adrenal gland, (11) left adrenal gland, and (12) celiac artery. The segmentations and images are from the Multi-Atlas Labelling Beyond the Cranial Vault (BTCV) by MICCAI [34]
Illustration of the percentage of voxels in each organ of the head and neck (a), abdomen (b), and chest (c), respectively, which is calculated based on the BTCV data set [34]
Recently, only a few comprehensive reviews have provided detailed summaries of existing multi-organ segmentation methods. For example, Fu et al. [30] summarized literature of deep learning-based multi-organ segmentation methods up to 2020, providing a comprehensive overview of developments in this field; Vrtovec et al. [31] systematically analyzed 78 papers published between 2008 and 2020 on the automatic segmentation of OARs in the head and neck. However, these reviews encounter certain issues. Firstly, with the rapid development of technology, many novel methods such as transformer architecture [32], foundation models [33] have emerged for addressing multi-organ segmentation, and more public datasets have also been introduced. However, these reviews only encompassed literature up to 2020; secondly, they categorized methods solely based on network design, without categorizing and summarizing specific solutions unique to the challenges of multi-organ segmentation; thirdly, the majority of these reviews primarily covered fully supervised methods and did not provide a summary of papers related to weakly supervised and semi-supervised; lastly, they did not provide a comprehensive summary of the segmentation accuracy for each organ, making it difficult for readers to assess the current segmentation precision for each organ and knew which organs have reached a mature stage of segmentation and which organs still pose challenges.
In this review, we have summarized around the datasets and methods used in multi-organ segmentation. Concerning datasets, we have provided an overview of existing publicly available datasets for multi-organ segmentation and conducted an analysis of these datasets. In terms of methods, we categorized them into fully supervised, weakly supervised, and semi-supervised based on whether complete pixel-level annotations are required. Within the fully supervised methods, we organized the methods according to the network architectures used, input image dimensions, segmentation modules specifically designed for multi-organ segmentation, and the loss functions employed. For weakly supervised and semi-supervised methods, we summarized the latest papers in each subcategory. Detailed information on the datasets and network architectures used in each paper, along with the segmentation accuracy achieved for each organ, has been provided to enable readers to quickly understand the current segmentation accuracy of each organ on the respective datasets. In the discussion section, we have summarized the existing methods in this field and, in conjunction with the latest technologies, discussed future trends in the field of multi-organ segmentation.
The structure of this review is as follows. The first section elaborates on the mathematical definition of multi-organ segmentation and the corresponding evaluation metrics. The second section describes how we conducted literature research and screening based on PRISMA [35]. The third section presents the literature analysis we retrieved, categorized into two main sections: data and methods. In the data section, we summarize existing public datasets and conduct analysis. In the methods section, we divide into three categories: supervised methods, weakly and semi-supervised methods. In the fourth section, we discuss existing methods and their future prospects, while in the fifth section, we summarize the entire paper.
Definition and evaluation metrics
Let \({\varvec{X}}\) represent the union of input images, \({\varvec{G}}\) represent the union of ground truth labels, \({\varvec{P}}\) represent the union of predicted labels, f represents the neural network, and \({\varvec{\theta}}\) represents its parameters, where \({\varvec{P}}={\varvec{f}}({\varvec{X}};\boldsymbol{ }{\varvec{\theta}})\).
Given a multi-organ segmentation task, \({\varvec{\Psi}}\) represents the class set of organs to be segmented. \({\left\{{\varvec{x}}\right\}}_{\boldsymbol{*}}\) represents the set of organs annotated in \({\varvec{x}}\). According to the available annotations, multi-organ segmentation can be implemented according to three learning paradigms, as shown in Fig. 6: fully supervised learning, weakly supervised learning, and semi-supervised learning. Fully supervised learning means that the labels of all organ are given, which indicates that \(\forall {\varvec{x}}\in {\varvec{X}},\boldsymbol{ }{\left\{{\varvec{x}}\right\}}_{\boldsymbol{*}}={\varvec{\Psi}}\). Weakly supervised learning often means that the data come from \({\varvec{n}}\) different datasets. However, each dataset provides the annotations of one or more organs but not all organs, which means that \({\varvec{X}}={{\varvec{X}}}_{1}\cup {{\varvec{X}}}_{2}\cup \cdots \cup {{\varvec{X}}}_{n},\boldsymbol{ }\boldsymbol{ }\forall \boldsymbol{ }{{\varvec{x}}}_{k,i}\in {{\varvec{X}}}_{k}, k=\mathrm{1,2},\dots n,\boldsymbol{ }\boldsymbol{ }{\left\{{{\varvec{x}}}_{k,i}\right\}}_{\boldsymbol{*}}\subseteq{\varvec{\Psi}}\),\(\bigcup_{k=1}^{n}{\left\{{{\varvec{x}}}_{k,i}\right\}}_{\boldsymbol{*}}={\varvec{\Psi}}\). Here, \({{\varvec{x}}}_{{\varvec{k}},{\varvec{i}}}\) denotes the ith image in \({{\varvec{X}}}_{{\varvec{k}}}\). Semi-supervised learning indicate that some of the training datasets are fully labeled and others are unlabelled, \({\varvec{X}}={{\varvec{X}}}_{{\varvec{l}}}\cup {{\varvec{X}}}_{{\varvec{u}}}\). \({{\varvec{X}}}_{{\varvec{l}}}\) represents the fully labeled dataset, \({{\varvec{X}}}_{{\varvec{u}}}\) represents the unlabelled dataset, which indicates that \(\forall {{\varvec{x}}}_{{\varvec{l}}}\in {{\varvec{X}}}_{{\varvec{l}}},\boldsymbol{ }{\left\{{{\varvec{x}}}_{{\varvec{l}}}\right\}}_{\boldsymbol{*}}={\varvec{\Psi}}\) and \(\forall {{\varvec{x}}}_{{\varvec{u}}}\in {{\varvec{X}}}_{{\varvec{u}}},\boldsymbol{ }{\left\{{{\varvec{x}}}_{{\varvec{u}}}\right\}}_{\boldsymbol{*}}={\varvec{\phi}}\), which represents the empty set, and the size of \({{\varvec{X}}}_{{\varvec{l}}}\) is far less than the one of \({{\varvec{X}}}_{{\varvec{u}}}\).
General overview of the learning paradigms reviewed in this paper. (The images presented in this figure are sourced from the MICCAI Multi-Atlas Labelling Beyond the Cranial Vault (BTCV) data set [34].)
The performance of the segmentation methods is typically evaluated using metrics such as the Dice Similarity Coefficient (DSC), 95% Hausdorff Distance (HD95) and Mean Surface Distance (MSD). DSC is a measure of the volume overlap between the predicted outputs and ground truth, HD95 and MSD are measures of the surface distance between them:
where \({P}^{c}\) and \({G}^{c}\) represent the set of predicted pixels and the set of real pixels of the \(c\) class organ, respectively; \({P}_{s}^{c}\) and \({G}_{s}^{c}\) represent the set of predicted pixels and the set of real pixels of the surface of the \(c\) class organ, respectively; and \(d\left({p}_{s}^{c},{G}_{s}^{c}\right)={min}_{{g}_{s}^{c}\in {G}_{s}^{c}}{||{p}_{s}^{c}-{g}_{s}^{c}||}_{2}\) represents the minimal distance from point \({p}_{s}^{c}\) to surface \({G}_{s}^{c}\). The review reports various methods based on DSC values.
Search protocol
This paper adopts the method proposed by the PRISMA guidelines [35] to determine the articles included in the analysis. The articles were primarily obtained through Google Scholar. Using the keywords “multi-organ segmentation” and “deep learning”, the search covered the period from January 1, 2016, to December 31, 2023, resulting in a total of 327 articles. We focused on highly cited articles, including those published in top conferences (such as NeurIPS, CVPR, ICCV, ECCV, AAAI, MICCAI, etc.) and top journals (such as TPAMI, TMI, MIA, etc.). Two researchers independently reviewed these articles to determine their eligibility. Among them, 67 articles did not meet the inclusion criteria based on the title and abstract, and 45 complete manuscripts were evaluated separately. In the end, we included 195 studies for analysis.
Result
Datasets
Public datasets
To obtain high-quality datasets for multi-organ segmentation, numerous research teams have collaborated with medical organizations. A summary of commonly used datasets for validating multi-organ segmentation methods in the head and neck, thorax, and abdomen regions can be found in Table 1, with references in [34, 36,37,38,39,40,41,42,43,44,45,46,47,48,49]. The table also reveals that the amount of annotated data available for deep learning studies remains insufficient.
Datasets analysis
Data play a crucial role in improving model performance. In certain cases, such as lung segmentation, the key issue has shifted from algorithm complexity to dataset quality. Accurate lung segmentation does not necessarily require complex techniques [50]. Even with simple network architectures, superior results can be achieved with more extensive and heterogeneous private data. The lack of diversity in training data is considered one of the primary obstacles to building robust segmentation models.
Therefore, acquiring large-scale, high-quality, and diverse multi-organ segmentation datasets has become an important direction in current research. Due to the difficulty of annotating medical images, existing publicly available datasets are limited in number and only annotate some organs. Additionally, due to the privacy of medical data, many hospitals cannot openly share their data for training purposes. For the former issue, techniques such as semi-supervised and weakly supervised learning can be utilized to make full use of unlabeled and partially labeled data. Alternatively, human-in-the-loop [51] techniques can combine human knowledge and experience with machine learning to select samples with the highest annotation value for training. For the latter issue, federated learning [52] techniques can be applied to achieve joint training of data from various hospitals while protecting data privacy, thus fully utilizing the diversity of the data.
Dataset size
Incorporating unannotated data into training or integration; existing partially labeled data can be fully utilized to enhance model performance, as detailed in Section of Weakly and semi-supervised methods.
Annotation quality
Human-in-the-loop integration of human knowledge and experience minimizes the cost of training accurate predictive models [51]. By closely collaborating, humans and machines leverage each other’s primary strengths to maximize efficiency. Human-in-the-loop primarily consists of two categories: active learning [53] and interactive segmentation [54]. Active learning selects the next batch of annotated samples through algorithms to maximize model performance, presenting an economically effective method for expanding training datasets. Another category, interactive segmentation, expedites the annotation process by allowing expert annotators to interactively correct initial segmentation masks generated by the model.
Wang et al.[55] comprehensively reviewed core methods of deep active learning, including informative assessment, sampling strategies, integration with other techniques such as semi-supervised and self-supervised learning, and customized active learning works specifically for medical image analysis. Recently, Qu et al.[56] proposed a novel and systematically effective active learning-based organ segmentation and labeling method. They annotated spleen, liver, kidney, stomach, gallbladder, pancreas, aorta, and inferior vena cava in 8,448 CT volumes. The proposed active learning process generated an attention map, highlighting areas that radiologists need to modify, reducing annotation time from 30.8 years to 3 weeks and accelerating the annotation process by 533 times.
Interactive segmentation in medical imaging typically involves a sequential interactive process, where medical professionals iteratively improve annotation results until the desired level of accuracy is achieved [57]. In recent years, many deep learning-based interactive segmentation methods have been proposed. Recent advancements in natural image segmentation have witnessed the emergence of segmentation-agnostic models like the Segmentation Anytime Model (SAM) [58, 59], demonstrating remarkable versatility and performance in various segmentation tasks. Various large models for medical interactive segmentation have also been proposed, providing powerful tools for generating more high-quality annotated datasets.
Dataset diversity
One significant reason for the limited availability of data for multi-organ segmentation is the issue of data privacy. Many institutions are unable to share their data for training due to privacy concerns. The emergence of federated learning addresses this problem precisely. Federated learning is a distributed learning approach in machine learning aimed at training models across multiple devices or data sources without centralizing the dataset in a single location. In federated learning, model training occurs on local devices, and then locally updated model parameters are sent to a central server, where they are aggregated to update the global model [52]. This distributed learning approach helps protect user privacy because data do not need to leave devices for model training.
In federated learning, the heterogeneity of statistical data is a crucial research issue. FedAvg is one of the pioneering works to address this issue, using weighted averaging of local weights based on local training scale and has been widely recognized as a baseline for federated learning [60]. Recently, several federated learning algorithms have been proposed for medical image segmentation tasks. For example, FedSM [61] employs a model selector to determine the model or data distribution closest to any testing data. Studies [62] have shown that architectures based on self-attention exhibit stronger robustness to distribution shifts and can converge to better optimal states on heterogeneous data.
Federated learning enables data from multiple sites to participate in training simultaneously without requiring hospitals to disclose their data, thereby enhancing dataset diversity and training more robust segmentation models.
Methods
Fully supervised methods
The fully supervised methods require complete annotation of all organs involved in the multi-organ segmentation task. The existing methods can be analyzed from four parts: network architecture, network dimension, image segmentation modules, and network loss function. The network architecture is further divided into single network, cascade network and step-by-step segmentation networks; while network dimension categorizes methods based on the image dimension used (2D, 3D, or multi-view); image segmentation modules refer to modules that are frequently used in multi-organ segmentation to improve segmentation performance, and network loss function summarizes the innovative use of common loss functions for multi-organ segmentation.
Network architecture
Multi-organ segmentation methods can be categorized based on their network architecture, which can be divided into three types: single network, cascade network, and step-by-step segmentation network, which is shown in Fig. 7. Tables 2, 3, 4 summarize the literature related to methods for the segmentation of multi-organ in the head and neck, abdomen and chest based on DSC metrics. Since there are so many organs in the head and neck as well as the abdomen, this paper mainly reports on 9 organs in the head and neck and 7 organs in the abdomen. Tables 5, 6 summarize the DSC values of other organs.
Single network
CNN-based methods
CNN can automatically extract features from input image. Multiple neurons are connected to each neuron in next layer, where each layer can perform tasks such as convolution, pooling or loss computation [63]. CNNs have been successfully applied to medical images, such as brain [64, 65] and pancreas [66] segmentation tasks.
Early CNN-based methods
Earlier CNN-based methods mainly utilized convolutional layers for feature extraction, followed by pooling layers and fully connected layers for final prediction. In the work of Ibragimov and Xing [67], deep learning techniques were employed for the segmentation of OARs in head and neck CT images for the first time. They trained 13 CNNs for 13 OARs and demonstrated that the CNNs outperformed or were comparable to advanced algorithms in accurately segmenting organs such as the spinal cord, mandible and optic nerve. However, they did not perform well in segmenting organs such as the optical chiasm. Fritscher et al. [68] incorporated shape location and intensity information with CNN for segmenting the optic nerve, parotid gland, and submandibular gland. Moeskops et al. [69] investigated whether a single CNN can be used for segmenting multiple tissues across different modalities, including six tissues in brain MR images, pectoral muscles in breast MR images, and coronary arteries in heart CTA images. Their results demonstrated that a single CNN can effectively segment multiple organs across different imaging modalities.
FCN-based methods
Early methods based on CNN showed some improvement in segmentation accuracy compared to traditional methods. However, CNN involves multiple identical computations of overlapping voxels during the convolution operation, which may cause some performance loss. Moreover, the final fully connected network layer in CNN can introduce spatial information loss to the image. To overcome these limitations, Shelhamer et al. [70] proposed the Fully Convolutional Network (FCN), which utilized transposed convolutional layers to achieve end-to-end segmentation while preserving spatial information. Wang et al. [71] used FCN with a novel sample selection strategy to segment 16 organs in the abdomen, while Trullo et al. [72] employed a variant of FCN called SharpMask [73] to enhance the segmentation performance of 5 organs in the thorax compared to standard FCN.
U-Net-based methods
The U-Net architecture, proposed by Ronneberger et al. [74], builds upon the FCN framework and consists of an encoder and a decoder, connecting them layer by layer with skip connections that allow for multiscale feature fusion. U-Net has become a widely adopted architecture in multi-organ segmentation [75,76,77,78,79,80,81,82,83,84,85,86,87,88,89]. For example, Roth et al. [79] employed U-Net to segment 7 organs in the abdomen with an average Dice value of 0.893. Lambert et al. [45] proposed a simplified U-Net for segmenting the heart, trachea, aorta, and esophagus of the chest, which improved performance by adding dropout and bilinear interpolation. Apart from U-Net, V-Net [90] introduced a volumetric, fully convolutional neural network for 3D image segmentation [91,92,93]. Gibson et al. [91] used dense V-Networks to segment 8 organs in the abdomen, while Xu et al. [92] proposed a probabilistic V-Net model with a conditional variational autoencoder (cVAE) and hierarchical spatial feature transform (HSPT) for abdominal organs segmentation. The nnU-Net [94] is a novel framework based on U-Net architecture with adaptive pre-processing, data enhancement, and postprocessing techniques, which has demonstrated state-of-the-art performance in various biomedical segmentation challenges [95,96,97,98]. Podobnik et al. [95] reported successful results in segmenting 31 OARs in the head and neck using nnU-Net, with both CT and MR images being employed.
GAN-based methods
GAN [99] usually comprises a pair of competitive networks: generators and discriminators. The generator attempts generate synthetic data that can deceive the discriminator, while the discriminator strives to accurately distinguish between real and generated data. After iterative optimization training, the performance of both networks can be improved. In recent years, several GAN-based multi-organ segmentation methods have been proposed and achieved high segmentation accuracy [100,101,102,103,104,105,106,107].
Dong et al. [102] employed a GAN framework with a set of U-Nets as the generator and a set of FCNs as the discriminator to segment the left lung, right lung, spinal cord, esophagus and heart from chest CT images. The results showed that the adversarial networks enhanced the segmentation performance of most organs, with average DSC values of 0.970, 0.970, 0.900, 0.750, and 0.870 for the above five organs. Tong et al. [100] proposed a Shape-Constraint GAN (SC-GAN) for automatic segmentation of head and neck OARs from CT and low-field MR images. It used DenseNet [108], a deep supervised fully convolutional network, to segment organs for prediction and uses a CNN as the discriminator network to correct the prediction errors. The results showed that combining GAN and DenseNet could further improve the segmentation performance of CNN by incorporating original shape constraints.
While GAN can enhance accuracy with its adversarial losses, training a GAN network is challenging and time-consuming since the generator must achieve Nash equilibrium with the discriminator [99]. Moreover, its adversarial loss, as a shape modifier, can only achieve higher segmentation accuracy when segmenting organs with regular and distinctive shapes (e.g., liver and heart) but may not work well for irregular or tubular structures (such as the pancreas and aorta) [109].
Transformer-based methods
CNN-based methods have demonstrated impressive effectiveness in segmenting multiple organs across various tasks. However, a significant limitation arises from the inherent shortcomings of the limited perceptual field within the convolutional layers. Specifically, these limitations prevent CNNs from effectively modeling global relationships. This constraint impairs the models' overall performance by limiting their ability to capture and integrate broader contextual information which is critical for accurate segmentation. The self-attention mechanism of transformer [32] can overcome the long-term dependency problem and achieve superior results compared to CNNs in several tasks, including natural language processing and computer vision. In recent studies, it has been demonstrated that medical image segmentation networks employing transformers can achieve comparable or superior accuracy compared to current state-of-the-art methods [110,111,112,113].
For instance, Cao et al. [114] incorporated the transformer into a U-shaped network, named Swin-UNet, to investigate the effectiveness of the pure transformer model in abdominal multi-organ segmentation, which showed promising segmentation accuracy. However, this method requires initializing the network encoder and decoder with the training weights of the Swin transformer on ImageNet. Huang et al. [115] introduced MISSFormer, a novel architecture for medical image segmentation that addresses convolution's limitations by incorporating an Enhanced Transformer Block. This innovation enables effective capture of long-range dependencies and local context, significantly improving segmentation performance. Furthermore, in contrast to Swin-UNet, this method can achieve comparable segmentation performance without the necessity of pre-training on extensive datasets. Tang et al.[116] introduce a novel framework for self-supervised pre-training of 3D medical images. This pioneering work includes the first-ever proposal of transformer-based pre-training for 3D medical images, enabling the utilization of the Swin Transformer encoder to enhance fine-tuning for segmentation tasks.
While transformer-based methods can capture long-range dependencies and outperform CNNs in several tasks, they may struggle with the detailed localization of low-resolution features, resulting in coarse segmentation results. This concern is particularly significant in the context of multi-organ segmentation, especially when it involves the segmentation of small-sized organs [117, 118].
Hybrid networks
CNNs are proficient at detecting local features but frequently struggle to capture global features effectively. In contrast, transformers can capture long-range feature dependencies but may lose local feature details and result in poor segmentation accuracy for small organs. To overcome the limitations, researchers have explored hybrid methods that combine CNN and transformer frameworks [111, 119,120,121,122,123].
For example, Suo et al. [124] proposed the I2-Net, a collaborative learning network that combines features extracted by CNNs and transformers to accurately segment multiple abdominal organs. This method resulted in an enhancement of the segmentation accuracy for small organs by 4.19%, and for medium-sized organs by a range of 1.83% to 3.8%. Kan et al. [125] proposed ITUnet, which added transformer-extracted features to the output of each block of the CNN-based encoder, obtaining segmentation results that leveraged both local and global information. ITUnet demonstrated better accuracy and robustness than other methods, especially on difficult organs such as the lens. Chen et al. [126] introduced TransUNet, a network architecture that utilized transformers to build stronger encoders and competitive results for head and neck multi-organ segmentation. Similarly, Hatamizadeh et al. [127] introduced UNETR and Swin UNETR [128], which employed transformers (Swin transformer) as encoders and CNNs as decoders. This hybrid method captured both global and local dependencies, leading to improved segmentation accuracy.
In addition to the methods combining CNN and transformer, there are some other hybrid architectures. For example, Chen et al. [129] integrated U-Net with long short-term memory (LSTM) for chest organ segmentation, and the DSC values of all five organs were above 0.8. Chakravarty et al. [130] introduced a hybrid architecture that leveraged the strengths of both CNNs and recurrent neural networks (RNNs) to segment the optic disc, nucleus, and left atrium. The hybrid methods effectively merge and harness the advantages of both architectures for accurate segmentation of small and medium-sized organs, which is a crucial research direction for the future.
Cascade network
Segmenting small organs in medical images is challenging because most organs occupy only a small volume in the images, making it difficult for segmentation models to accurately identify them. To address this constraint, researchers have proposed cascade multi-stage methods, which can be categorized into two types. One is coarse-to-fine-based method [131,132,133,134,135,136,137,138,139,140,141], where the first network is utilized to acquire a coarse segmentation, followed by the second network that refines the coarse outcomes for improved accuracy. The other is localization and segmentation-based method [105, 142,143,144,145,146,147,148,149,150,151,152,153], where registration methods or localization networks are used to identify candidate boxes for the location of each organ, which are then input into the segmentation network, which is shown in Fig. 7 (B). Additionally, the first network can provide other information, including organ shape, spatial location, or relative proportions, to enhance the segmentation accuracy of the second network.
Coarse-to-fine-based methods
The coarse-to-fine-based methods first input the original image and its corresponding labels into the first network to obtain probability map. This probability map will multiply the original image and be input into the second network to refine the coarse segmentation, as illustrated in Fig. 7(A). Over the years, numerous methods utilizing the coarse-to-fine method have been developed for multi-organ segmentation, with references in [131,132,133,134,135,136,137,138,139,140,141].
Trullo et al. [72] proposed 2 deep architectures that work synergistically to segment several organs such as the esophagus, heart, aorta, and trachea. In the first stage, probabilistic maps were obtained to learn anatomical constrains. Then, four networks were trained to distinguish each target organ from the background in separate refinements. Zhang et al. [133] developed a new cascaded network model with Block Level Skip Connections (BLSC) between two networks, allowing the second network to benefit from the features learned by each block in the first network. By leveraging these skip connections, the second network can converge more quickly and effectively. Xie et al. [134] proposed a new framework named the Recurrent Saliency Transformation Network (RSTN) which used coarse segmentation masks as spatial weights in the fine stage, effectively guiding the network's attention to important regions for accurate segmentation. Moreover, by enabling gradients to be backpropagated from the loss layer to the entire network, the RSTN facilitates joint optimization of the two stages. Ma et al. [154] presented a comprehensive coarse-to-fine segmentation model for automatic segmentation of multiple OARs in head and neck CT images. This model used a predetermined threshold to classify the initial results of the coarse stage into large and small OARs, and then designed different modules to refine the segmentation results.
This coarse-to-fine method efficiently simplifies the background and enhances the distinctiveness of the target structures. By dividing the segmentation task into two stages, this method achieves better segmentation results for small organs compared to the single-stage method. Nevertheless, it is essential to acknowledge that this method entails certain limitations, including heightened memory usage and extended training times attributed to the necessity of train at least two networks.
Localization and segmentation-based methods
In the localization and segmentation-based method, the first network provides location information and generates a candidate frame, which is then used to extract the Region of Interests (ROIs) from the image. This extracted region, free from interference of other organs or background noise, serves as the input for the second network. By isolating the targeted organ, the segmentation accuracy is improved. The process is illustrated in Fig. 7(B). The organ location in the first stage can be obtained through registration or localization network, with reference in [105, 142,143,144,145,146,147,148,149,150,151,152,153].
Wang et al. [142], Men et al. [143], Lei et al. [149], Francis et al. [155], and Tang et al. [144] used neural networks in both stages. In the first stage, networks were used to localize the target OARs by generating bounding boxes. In the second stage, the target OARs were segmented within the bounding boxes. Among them, Wang et al. [142] and Francis et al. [155] utilized 3D U-Net in both stages, while Lei et al. [149] used Faster RCNN to automatically locate the ROI of organs in the first stage. Furthermore, FocusNet [105, 147] presented a novel neural network that effectively addresses the challenge of class imbalance in the segmentation of head and neck OARs. The small organs are first localized using the organ localization network, and then high-resolution features of small organs are fed into the segmentation network. Liang et al. [146] introduced a multi-organ segmentation framework that utilizes multi-view spatial aggregation to integrate the learning of both organ localization and segmentation subnetworks. This framework mitigates the impact of neighboring structures and background regions in the input data, and the proposed fine-grained representation based on ROIs enhances the segmentation accuracy of organs with varying sizes, particularly small organs.
Larsson et al. [152], Zhao et al. [153], Ren et al. [156], and Huang et al. [150] utilized registration-based methods to localize organs, while CNN was employed for accurate segmentation. Ren et al. [156] used interleaved cascades of 3D-CNNs to segment each organ, exploiting the high correlation between adjacent tissues. Specifically, the initial segmentation results of a particular tissue can improve the segmentation of its neighboring tissues. Zhao et al. [153] proposed a flexible knowledge-assisted framework that synergistically integrated deep learning and traditional techniques to improve segmentation accuracy in the second stage.
Localization and segmentation-based methods have proven to enhance the accuracy of organ segmentation by reducing background interference, particularly for small organs. However, this method requires considerable memory and training time, and the accuracy of segmentation is heavily reliant on the accuracy of organ localization. Therefore, improving the localization of organs and enhancing segmentation accuracy are still areas of research that need further exploration in the future.
Other cascade methods
In addition to probability maps and localization information, the first network can also provide other types of information that can be used to improve segmentation accuracy, such as scale information and shape priors. For instance, Tong et al. [157] combined FCNN with a shape representation model (SRM) for head and neck OARs segmentation. The SRM serves as the first network for learning highly representative shape features in head and neck organs, which are then used to improve the accuracy of the FCNN. The results from comparing the FCNN with and without SRM indicated that the inclusion of SRM greatly raised the segmentation accuracy of 9 organs, which varied in size, morphological complexity, and CT contrasts. Roth et al. [158] proposed two cascaded FCNs, where low-resolution 3D FCN predictions were upsampled, cropped, and connected to higher-resolution 3D FCN inputs.
Step-by-step segmentation network
In the context of multi-organ segmentation, step-by-step segmentation refers to sequentially segmenting organs in order of increasing complexity, starting with easier-to-segment organs before moving on to more challenging ones, which is shown in Fig. 7(C). The fundamental assumption is that segmenting more challenging organs (e.g., those with more complex shapes and greater variability) can benefit from the segmentation results of simpler organs processed earlier [159]. Step-by-step segmentation has been demonstrated to be highly effective for segmenting some of the most challenging organs, such as the pancreas (Hammon et al. [160]), utilizing surrounding organs (such as the liver and spleen) as supportive structures.
In recent years, many deep learning-based step-by-step segmentation methods have emerged. For example, Zhao et al. [161] first employed the nnU-Net to segment the kidneys and then to segment kidney tumors based on the segmentation results of the kidneys. Similarly, Christ et al.[136] first segment the liver, followed by the segmentation of liver tumors based on the segmentation results of the liver. In [162], organs susceptible to segmentation errors, such as the lungs, are segmented first, followed by the segmentation of less susceptible organs, such as airways, based on lung segmentation. Guo et al. [163] proposed a method called Stratified Organ at Risk Segmentation (SOARS), which categorizes organs into anchor, intermediate, and small and hard (S&H) categories. Each OAR category uses a different processing framework. Inspired by clinical practice, anchor organs are utilized to guide the segmentation of intermediate and S&H category organs.
Network dimension
Considering the dimension of input images and convolutional kernels, multi-organ segmentation networks can be divided into 2D, 2.5D and 3D architectures, and the differences among three architectures will be discussed in follows.
2D- and 3D-based methods
The 2D multi-organ segmentation network takes as input slices from a three-dimensional medical image, and the convolution kernel is also two-dimensional. Several studies, including those by Men et al. [89], Trullo et al. [72], Gibson et al. [91], Chen et al. [164], Zhang et al. [78], and Chen et al. [165], have utilized 2D networks for multi-organ segmentation. 2D architectures can reduce the GPU memory burden. But CT or MR images are inherently 3D, slicing images into 2D tends to ignore the rich information in the entire image voxel, so 2D models are insufficient for analyzing the complex 3D structures in medical images.
3D multi-organ segmentation networks can extract features directly from 3D medical images by using 3D convolutional kernels. Some studies, such as Roth et al.[79], Zhu et al. [75], Gou et al. [77], and Jain et al. [166], have employed 3D network for multi-organ segmentation. However, since 3D network requires a large amount of GPU memory, they may face computationally intensive and memory shortage problems. As a result, most 3D network-based methods use sliding windows acting on patches. To overcome the constraints of GPU memory, Zhu et al. [75] proposed a model called AnatomyNet, which took full-volume of head and neck CT images as inputs and generated masks for all organs to be segmented at once. To balance GPU memory usage and network learning capability, they employed a down-sampling layer solely in the first encoding block, which also preserved information of small anatomical structures.
Multi-view-based methods
Accurate medical image segmentation requires effective use of spatial information among image slices. Inputting 3D images directly to the neural network can lead to high memory usage, while converting 3D images to 2D slices results in the loss of spatial information between slices. As a solution, multi-view-based methods have been proposed, which include using 2.5D neural networks with multiple 2D slices or combining 2D and 3D convolutions. This method can reduce memory usage while maintaining the spatial information between slices, improving the accuracy of medical image segmentation.
The 2.5D-based method uses 2D convolutional kernels and takes in multiple slices as input. The slices can either be a stack of adjacent slices using interslice information [167, 168], or slices along three orthogonal directions (axial, coronal, and sagittal) [67, 68, 148, 169], which is shown in Fig. 8. Zhou et al. [170] segmented each 2D slice using FCN by sampling a 3D CT case on three orthogonally oriented slices and then assembled the segmented output (i.e., 2D slice results) back into 3D. Chen et al. [165] developed a multi-view training method with a majority voting strategy. Wang et al. [171] used a statistical fusion method to combine segmentation results from three views. Liang et al. [148] performed context-based iterative refinement training on each of the three views and aggregated all the predicted probability maps to obtain final segmentation results. These methods have shown improved segmentation results compared to the three separate views.
Tang et al. [172] proposed a novel method which combines the strengths of 2D and 3D models. This method utilized high-resolution 2D convolution for accurate segmentation and low-resolution 3D convolution for extracting spatial contextual information. A self-attention mechanism controlled the corresponding 3D features to guide 2D segmentation, and experiments demonstrated that this method outperforms both 2D and 3D models. Similarly, Chen et al. [164] devised a novel convolutional neural network, OrganNet2.5D, that effectively processed diverse planar and depth resolutions by fully utilizing 3D image information. This network combined 2D and 3D convolutions to extract both edge and high-level semantic features.
Some studies only used 2D images to avoid memory and computation problems, but they did not fully exploit the potential of 3D image information. Although 2.5D methods can make better use of multiple views, their ability to extract spatial contextual information is still limited. Current 2.5D methods in multi-organ segmentation aggregate three perspectives at the outcome level, but the intermediate processes are independent of each other, and more effective use of intermediate learning processes is an area for further investigation. Pure 3D networks have a high parameter and computational burden, which limits their depth and performance. As for this reason, some people have begun researching lightweight 3D networks, Zhao et al.[173] proposed a novel framework based on lightweight network and Knowledge Distillation (KD) for delineating multiple organs from 3D CT volumes. Thus, finding better ways to combine multi-view information to achieve accurate multi-organ segmentation while considering memory and computational resources is a promising research direction.
Image segmentation modules
The design of network architecture is a crucial factor in improving the accuracy of multi-organ segmentation, but the process of designing such a network is quite intricate. In multi-organ segmentation tasks, various special mechanisms, such as dilation convolution module, feature pyramid module, and attention module, have been developed to enhance the accuracy of organ segmentation. These modules increase the perceptual field, combine features of different scales, and concentrate the network on the segmented region, thereby enhancing the accuracy of multi-organ segmentation. Cheng et al. [174] have explored the efficacy of each module in the network compared with the basic U-Net network for the head and neck segmentation task.
Shape prior module
Shape prior has been shown to be particularly effective for medical images due to the fixed spatial relationships between internal structures. As a result, incorporating anatomical priors in multi-organ segmentation task can significantly enhance the segmentation performance.
There are two main methods used for incorporating anatomical priors in multi-organ segmentation tasks. The first method is based on statistical analysis, which involves calculating the average distribution of organs in a fully labeled dataset. The segmentation network predictions are then guided to be as close as possible to this average distribution of organs [66, 68, 102, 175, 176]. The second method involves training a shape representation model that is pretrained using annotations from the training dataset. This model is used as a regularization term to constrain the predictions of the network during training [100, 157]. For example, Tappeiner et al.[177] propose that using stacked convolutional autoencoders as shape priors can enhance segmentation accuracy, both on small datasets and complete datasets. Recently, it has been demonstrated that generative models such as diffusion models [178, 179] can learn anatomical priors [180]. Therefore, utilizing generative models to obtain anatomical prior knowledge is a promising future research direction for improving segmentation performance.
Dilated convolutional module
In conventional CNN, down-sampling and pooling operations are commonly employed to expand the perception field and reduce computation, but these can cause spatial information loss and hinder image reconstruction. Dilated convolution (also referred to as "Atrous") introduces an additional parameter, expansion rate, to the convolution layer, which can allow for the expansion of the perception field without increasing computational cost. Dilated convolution is widely used in multi-organ segmentation tasks [66, 80, 168, 181, 182] to enlarge the sampling space and enable the neural network to extract multiscale contextual features across a wider receptive field. For instance, Li et al.[183] proposed a high-resolution 3D convolutional network architecture that integrates dilated convolutions and residual connections to incorporates large volumetric context. The effectiveness of this approach has been validated in brain segmentation tasks using MR images. Gibson et al. [66] utilized CNN with dilated convolution to accurately segment organs from abdominal CT images. Men et al. [89] introduced a novel Deep Dilated Convolutional Neural Network (DDCNN) for rapid and consistent automatic segmentation of clinical target volumes (CTVs) and OARs. Vesal et al. [182] integrated dilated convolution into the 2D U-Net for segmenting esophagus, heart, aorta, and thoracic trachea.
Multiscale module
Neural networks are composed of layers that progressively extract features from input data. The lower layers capture fine-grained geometric details with a smaller receptive field, providing high-resolution but weaker semantic representation. Conversely, higher layers have a larger receptive field and stronger semantic representation, but lower feature map resolution, which may cause information loss for small targets. To address this, multiscale fusion modules have been proposed, including bottom-up, top-down, and lateral feature pyramids (FPNs) [184], spatial pooling pyramids (ASPPs) [185] that combine dilated convolution and multiscale fusion. In multi-organ segmentation tasks, multiscale feature fusion is widely used because of the different sizes of organs. For example, Jia and Wei [80] introduced the feature pyramid into a multi-organ segmentation network using two opposite feature pyramids (top-down and bottom-up) to handle multiscale changes and improve the segmentation accuracy of small targets. Shi et al. [168] used the pyramidal structure of lateral connections between encoders and decoders to capture contextual information at multiple scales. Additionally, Srivastava et al. [186] introduced OARFocalFuseNet, a novel segmentation architecture that utilized a focal modulation scheme for aggregating multiscale contexts in a dedicated resolution stream during multiscale fusion.
Attention module
The attention module is a powerful tool that allows the network to dynamically weight important features. It can leverage the inherent self-attentiveness of the network and is especially useful for multi-organ segmentation tasks [101, 187]. There are several kinds of attention mechanisms, such as channel attention, spatial attention, and self-attention, which can be used to selectively emphasize the most informative features.
Squeeze-and-excitation (SE) module [188] is an effective channel attention technique that enables the network to emphasize important regions in an image. AnatomyNet [75] utilized 3D SE residual blocks to segment the OARs in the head and neck. This method enabled the extraction of 3D features directly from CT images and dynamically adjusted the mapping of residual features within each channel by generating a channel attention tensor. Liu et al. [189] proposed a novel network architecture, named Cross-layer Spatial Attention map Fusion CNN (CSAF-CNN), which could integrate the weights of different spatial attentional maps in the network, resulting in significant improvements in segmentation performance. In particular, the average DSC of 22 organs in the head and neck was 72.50%, which outperformed U-Net (63.9%) and SE-UNet (67.9%). Gou et al. [77] designed a Self-Channel-Spatial-Attention neural network (SCSA-Net) for 3D head and neck OARs segmentation. This network could adaptively enhance both channel and spatial features, and it outperformed SE-Res-Net and SE-Net in segmenting the optic nerve and submandibular gland. Lin et al. [190] proposed a variance-aware attention U-Net network that embedded variance uncertainty into the attention architecture to improve the attention to error-prone regions (e.g., boundary regions) in multi-organ segmentation. This method significantly improved the segmentation results of small organs and organs with irregular structures (e.g., duodenum, esophagus, gallbladder, and pancreas). Zhang et al. [78] proposed a novel network called Weaving Attention U-Net (WAU-Net) that combined the U-Net + + [191] with axial attention blocks to efficiently model global relationships at different levels of the network. This method achieved competitive performance in segmenting OARs of the head and neck.
Other modules
The dense block [108] can efficiently use the information of the intermediate layer, and the residual block [192] can prevent gradient disappearance during backpropagation. These two modules are often embedded in the basic segmentation framework. The convolution kernel of the deformable convolution [193] can adapt itself to the actual situation and better extract features. Heinrich et al. [194] proposed the OBELISK-Net, a 3D abdominal multi-organ segmentation architecture that incorporated sparse deformable convolutions with conventional CNNs to enhance segmentation of small organs with large shape variations such as the pancreas and esophagus. The deformable convolutional block proposed by Shen et al. [195] can handle shape and size variations across organs by generating specific receptive fields with trainable offsets. The strip pooling [196] module targets long strip structures (e.g., esophagus and spinal cord) by using long pooling instead of square pooling to avoid contamination from unrelated regions and capture remote contextual information. For example, Zhang et al. [197] utilized a pool of anisotropic strips with three directional receptive fields to capture spatial relationships between multiple organs in the abdomen. Compared to network architectures, network modules have gained widespread use due to their simple design process and ease of integration into various architectures.
Loss function
It is widely recognized that the choice of loss function is of vital importance in determining the segmentation accuracy. In multi-organ segmentation tasks, choosing an appropriate loss function can address the class imbalance issue and improve the segmentation accuracy of small organs. Jadon [198] has provided a comprehensive overview of commonly used loss functions in semantic segmentation; Ma et al.[199] systematically summarized common loss functions used in medical image segmentation and evaluated the effectiveness of each loss function across multiple datasets. In the context of multi-organ segmentation, commonly used loss functions include CE loss [200], Dice loss [201], Tversky loss [202], focal loss [203], and their combinations.
CE loss
The CE loss (cross-entropy loss) [200] is a widely used information theoretic measure that compares the predicted output labels with the ground truth. Men et al. [89], Moeskops et al. [95], and Zhang et al. [78] utilized CE loss for multi-organ segmentation. However, in situations where the background pixels greatly outnumber the foreground pixels, CE loss can result in poor segmentation outcomes by heavily biasing the model towards the background. To overcome this issue, the weighted CE loss [204] added weight parameters to each category based on CE loss, making it better suited for situations with unbalanced sample sizes. Since multi-organ segmentation often faces a significant class imbalance problem, using the weighted CE loss is a more effective strategy than using only the CE loss. As an illustration, Trullo et al. [72] used a weighted CE loss to segment the heart, esophagus, trachea, and aorta in chest images, while Roth et al. [79] applied a weighted CE loss for abdomen multi-organ segmentation.
Dice loss
Milletari et al. [90] proposed the Dice loss to quantify the intersection between volumes, which converted the voxel-based measure to a semantic label overlap measure, becoming a commonly used loss function in segmentation tasks. Ibragimov and Xing [67] used the Dice loss to segment multiple organs of the head and neck. However, using the Dice loss alone does not completely solve the issue that neural networks tend to perform better on large organs. To address this, Sudre et al. [201] introduced the weighted Dice score (GDSC), which adapted its Dice values considering the current class size. Shen et al. [205] assessed the impact of class label frequency on segmentation accuracy by evaluating three types of GDSC (uniform, simple, and square). Gou et al. [77] employed GDSC for head and neck multi-organ segmentation, while Tappeiner et al. [206] introduced a class-adaptive Dice loss based on nnU-Net to mitigate high imbalances. The results showcased the method's effectiveness in significantly enhancing segmentation outcomes for class-imbalanced tasks. Kodym et al. [207] introduced a new loss function named as the batch soft Dice loss function for training the network. Compared to other loss functions and state-of-the-art methods on current datasets, models trained with batch Dice loss achieved optimal performance.
Other losses
The Tversky loss [202] is an extension of the Dice loss and can be fine-tuned by adjusting its parameters to balance the rates of false positives and false negatives. The focal loss [203] was originally proposed for object detection to highlight challenging samples during training. Similarly, the focal Tversky loss [208] assigns less weight to easy-to-segment organs and focuses more on difficult organs. Berzoini et al. [81] applied the focal Tversky loss to smaller organs, which balances the performance between organs of different sizes and assigns more weight to hard-to-segment small organs, thus solving the class imbalance issue caused by kidneys and bladders. Inspired by the exponential logarithmic loss (ELD-Loss) [209], Liu et al. [189] introduced the top-k exponential logarithmic loss (TELD-Loss) to address the issue of class imbalance in head and neck OARs segmentation. Results indicate that the TELD-Loss is a robust method, particularly when dealing with mislabeling problems.
Combined loss
To address the advantages and disadvantages of different loss functions in multi-organ segmentation, researchers have proposed combining multiple loss functions for improved outcomes. The commonly employed method is a weighted sum of Dice loss and CE loss. Dice loss tackles class imbalance, while CE loss enhances curve smoothing. For instance, Isensee et al. [94] introduced a hybrid loss function that combines Dice loss and CE loss to calculate the similarity between predicted voxels and ground truth. Several other studies, including Isler et al. [181], Srivastava et al. [186], Xu et al. [92], Lin et al. [190], and Song et al. [210], have also adopted this weighted combination loss for multi-organ segmentation. Zhu et al. [75] specifically studied different loss functions for the unbalanced head and neck region and found that combining Dice loss with focal loss was superior to using the ordinary Dice loss alone. Similarly, both Cheng et al. [174] and Chen et al. [164] have used this combined loss function in their studies.
Conventional Dice loss may not effectively handle smaller structures, as even a minor misclassification can greatly impact the Dice score. Lei et al. [211] introduced a novel hardness-aware loss function that prioritizes challenging voxels for improved segmentation accuracy. Song et al. [212] proposed a dynamic loss weighting algorithm that dynamically assigns larger loss weights to organs that are classified as more difficult to segment based on data and network state, forcing the network to learn more from these organs, thereby maximizing segmentation performance. Designing an appropriate loss function is crucial for optimizing neural networks and significantly enhancing organ segmentation precision. This area of research remains essential and continues to be a critical focus for further advancements.
Weakly supervised methods
Obtaining simultaneous annotations for multiple organs on the same medical image poses a significant challenge in image segmentation. Existing datasets, such as LiTS [213], KiTS (p19) [214], and pancreas datasets [215], typically provide annotations for a single organ. How to utilize these partially annotated datasets to achieve a multi-organ segmentation model has arisen increasing interest.
Early methods involved training a segmentation model for each partially annotated dataset, and then combining the output of each model to obtain multi-organ segmentation results, referred to as multiple networks. Although this method is intuitive, it increases computational complexity and storage space. Later, Chen et al. [216] improved upon the multiple networks method by introducing a multi-head network. This network consists of a task-shared encoder and multiple task-specific decoders. When an image with annotations for a specific organ is input into the network, only the decoder parameters corresponding to that organ are updated, while the parameters for decoders corresponding to other organs are frozen. Though the multi-head network represents an improvement over multiple networks, this architecture is not flexible and cannot easily adapt to a newly annotated dataset. Recently, various methods have been proposed to use these partially annotated datasets, primarily falling into two categories: conditional network-based methods and pseudo-label-based methods.
Conditional network-based methods
Conditional network-based methods primarily involve embedding conditional information into the segmentation network, thus establishing a relationship between parameters of the segmentation model and the target segmented organs, which is shown in Fig. 9(a). Considering the way in which conditional information is incorporated into the segmentation network, methods based on conditional networks can be further categorized into task-agnostic and task-specific methods. Task-agnostic methods refer to cases where task information and the feature extraction by the encoder–decoder are independent. Task information is combined with the features extracted by the encoder and subsequently converted into conditional parameters introduced into the final layers of the decoder. Typical methods include DoDNet [217] and its variations [218], which utilized dynamic controllers to generate distinct weights for different tasks, and these weights were then incorporated into the final decoder layers to facilitate the segmentation of various organs and tumors.
Task-specific methods involve incorporating task information into the process of segmentation feature extraction by the encoder–decoder. For example, Dmitriev et al. [219] encoded task-related information into the activation layer between convolutional layers and nonlinear layers of decoder. Tgnet [220] adopted a task-guided method to design new residual blocks and attention modules for fusing image features with task-specific encoding. CCQ [221] embedded class relationships among multiple organs or tumors and utilizes learnable query vectors representing semantic concepts of different organs, achieving new state-of-the-art results on large partially annotated MOTS dataset.
However, currently, most methods based on conditional networks encode task information as one-hot labels, neglecting the prior relationships among different organs and tumors. Recently, foundation models [33] have seen significant development. Contrastive Language-Image Pretraining (CLIP) [222] can reveal the inherent semantics of anatomical structures by mapping similar concepts closer together in the embedding space. Liu et al. [223] was among the pioneers in applying CLIP to medical imaging. They introduced a CLIP-driven universal model for abdominal organ segmentation and tumor detection. This model achieved outstanding segmentation results for 25 organs based on 3D CT images and demonstrated advanced performance in detecting six types of abdominal tumors. The model ranked first on the MSD public leaderboard [41] and achieved state-of-the-art results on BTCV dataset [34]. However, since CLIP is predominantly trained on natural images, its capacity for generalization on medical images is constrained. Ye et al. [224] introduced a prompt-driven method that transformed organ category information into learnable vectors. While prompt-based methods could capture the intrinsic relationships between different organs, randomly initialized prompts may not fully encapsulate the information about a specific organ.
Pseudo-label-based methods
Pseudo-label-based methods initially train a segmentation model on each partially annotated dataset. Then, they utilize the trained models to generate pseudo labels for corresponding organs on other datasets, resulting in a fully annotated dataset with pseudo labels. A multi-organ segmentation model is subsequently trained using this dataset, which is shown in Fig. 9(b). Clearly, the performance of the final multi-organ segmentation model is closely tied to the quality of the generated pseudo-labels. In recent years, numerous methods have been proposed to enhance the quality of these pseudo-labels. Huang et al. [225] proposed a weight-averaging joint training framework that can correct the noise in the pseudo labels to train a more robust model. Zhang et al. [226] proposed a multi-teacher knowledge distillation framework, which utilizes pseudo labels predicted by teacher models trained on partially labeled datasets to train a student model for multi-organ segmentation. Lian et al. [176] improved pseudo-label quality by incorporating anatomical priors for single and multiple organs when training both single-organ and multi-organ segmentation models. For the first time, this method considered the domain gaps between partially annotated datasets and multi-organ annotated datasets. Liu et al. [227] introduced a novel training framework called COSST, which effectively and efficiently combined comprehensive supervision signals with self-training. To mitigate the impact of pseudo labels, they assessed the reliability of pseudo labels through outlier detection in latent space and excluded the least reliable pseudo labels in each self-training iteration.
Other methods
The issue of partially annotated can also be considered from the perspective of continual learning. Continual learning primarily addresses the problem of non-forgetting, where a model trained in a previous stage can segment several organs. After training, only the well-trained segmentation model is retained, and the segmentation labels and data become invisible. Next state, when new annotated organs become available, the challenge is how to ensure that the current model can both segment the current organs and not forget how to segment the previous organs. Inspired by [228], Liu et al. [229] first applied continual learning to aggregate partially annotated datasets in stages, which solved the problem of catastrophic forgetting and the background shift. Xu and Yan [230] proposed Federated Multi-Encoding U-Net (Fed-MENU), a new method that effectively uses independent datasets with different annotated labels to train a unified model for multi-organ segmentation. The model outperformed any model trained on a single dataset or on all datasets combined. Zhang et al. [231] proposed an innovative architecture specifically for continuous organ and tumor segmentation, in which a lightweight, class specific head was used to replace the traditional output layer, thereby improving flexibility in adapting to emerging classes. At the same time, CLIP was embedded into the heads of specific organs, which encapsulates the semantic information of each class through extensive image text collaborative training, this information would be an advantage for training new classes with pre-known class names. Ji et al.[232] introduced a novel CSS framework for the continual segmentation of a total of 143 whole-body organs from four partially labeled datasets. Utilizing a trained and frozen General Encoder alongside continually added and architecturally optimized decoders, this model prevents catastrophic forgetting while accurately segmenting new organs.
Others solved this problem from alternative perspectives. Zhou et al. [175] proposed a Prior-aware Neural Network (PaNN) that guided the training process based on partially annotated datasets by utilizing prior statistics obtained from a fully labeled dataset. Fang and Yan [233] and Shi et al. [234] trained uniform models on partially labeled datasets by designing new networks and proposing specific loss functions.
In multi-organ segmentation tasks, weak annotation not only includes partial annotation, but also includes other forms such as image-level annotation, sparse annotation, and noisy annotation [235]. For example, Kanavati et al. [236] proposed a weakly supervised method for the segmentation of liver, spleen, and kidney based on classification forests, where the organs were labeled through scribbles.
Semi-supervised methods
Semi-supervised methods are gaining popularity in organ segmentation due to their ability to enhance segmentation performance while reducing the annotation burden. These methods have found application in diverse medical image segmentation tasks, such as heart segmentation [237,238,239], pancreas segmentation [240], and tumor target region segmentation [241]. In a comprehensive review by Jiao et al. [242], the authors categorized semi-supervised learning methods in medical image segmentation into three paradigms: pseudo-label-based, consistency regularization-based, and knowledge prior-based methods. In this work, we specifically focus on exploring semi-supervised methods for multi-organ segmentation.
Ma et al. [39] proposed a semi-supervised method for abdominal multi-organ segmentation using pseudo-labeling. Initially, a teacher model was trained on labeled datasets to generate pseudo labels for unlabeled datasets. Subsequently, a student model was trained on both the labeled and pseudo-labeled datasets, and the student model replaced the teacher model for final training.
Semi-supervised multi-organ segmentation often employs multi-view methods to leverage information from multiple image planes and improve the reliability of pseudo-labels. Zhou et al. [243] proposed the DMPCT framework, which incorporated a multi-planar fusion module to iteratively update pseudo-labels for different configurations of unlabeled datasets in abdominal CT images. Xia et al. [244] proposed the uncertainty-aware multi-view collaborative training (UMCT) method, which employed spatial transformations to create diverse perspectives for training independent deep networks. Subsequently, these networks were collectively trained using multi-view consistency on unlabeled data, resulting in improved segmentation effectiveness.
Apart from collaborative training, consistency-based learning is another effective approach for multi-organ segmentation, given the diverse organ categories and their dense distribution. This method promotes the consistency of network outputs by using different parameters. For example, Lai et al. [245] proposed a semi-supervised DLUNet, which consisted of two lightweight U-Nets in the training phase. Additionally, for unlabeled data, the outputs from both networks were used to supervise each other, improving the segmentation accuracy of these unlabeled data. This method achieved an average DSC of 0.8718 for 13 organs in the abdomen. Chen et al. [246] proposed a novel teacher–student semi-supervised multi-organ segmentation model, called MagicNet, which normalized consistency training between teacher and student models by enhancing unlabeled data. MagicNet mainly included two data enhancement strategies, encouraging unlabeled images to learn relative organ semantics (cross-branch) from images and enhancing the segmentation accuracy of small organs (within-branch), Numerous experiments conducted on two common CT multi-organ datasets have demonstrated the effectiveness of MagicNet and were significantly superior to state-of-the-art semi-supervised medical image segmentation methods.
Furthermore, several other methods have been proposed for semi-supervised based method. For example, Lee et al. [247] developed a method that employed a discriminator module, which incorporated human-in-the-loop quality assurance (QA) to supervise the learning of unlabelled data. The QA scores were used as a loss function for the unlabelled data. Raju et al. [248] proposed an effective semi-supervised multi-organ segmentation method, CHASe, for liver and lesion segmentation. CHASe leverages co-training and hetero-modality learning within a co-heterogeneous training framework. This framework can be trained on a small single-phase dataset and can be adapted for label-free multi-center and multi-phase clinical data.
Discussion
This paper systematically summarizes the methods of multi-organ segmentation-based on deep learning, mainly from the aspects of data and methodology. In terms of data, it provides an overview of existing publicly available datasets and conducts an in-depth analysis of data-related issues. In terms of methodology, existing methods are categorized into fully supervised, weakly supervised, and semi-supervised based approaches. The proposal of these methods holds significant research significance in advancing automatic segmentation of multiple organs. Future research trends can be considered from the following aspects:
About datasets
Data play a crucial role in enhancing segmentation performance. Even the simplest models can achieve outstanding performance when trained on a high-quality dataset. However, compared to natural images, there is a shortage of publicly available datasets for multi-organ segmentation, and most methods are trained and tested on private datasets [249]. As summarized in the supplementary materials, many methods proposed in the literature are trained and validated on their own private datasets. This poses challenges in validating the model's generalization ability. Therefore, it is necessary to create a multi-center public dataset with a large volume of data, extensive coverage, and strong clinical relevance for multi-organ segmentation. In order to fully utilize abundant unlabeled data, combining weakly supervised and semi-supervised techniques, and leveraging human expertise in iterative labeling loops, federated learning techniques can be employed to jointly train models using data from various sites while ensuring privacy.
About fully supervised based methods
Based on the four types of segmentation methods of fully supervised method introduced earlier, future research directions can be considered from the following aspects: firstly, design a new network architecture or investigate how to better integrate different network architectures. Recently, an efficient variant of attention mechanism, Mamba [250, 251], has been proposed, surpassing CNN and Transformer in many medical image analysis tasks. Secondly, considering the respective issues of 2D and 3D architectures, designing lightweight 3D networks while maintaining image information and reducing computational burden is a research approach. Additionally, current multi-view methods only aggregate three perspectives at the result level, with the intermediate feature extraction processes being independent of each other. In the future, it can be explored to leverage the intermediate feature extraction processes or incorporate more view information. Thirdly, combining the characteristics of multiple organs, designing novel plug-and-play modules to enhance multi-organ segmentation performance. Finally, due to differences in organ size, shape irregularity, and imaging quality, deep neural networks exhibit inconsistent performance in medical image multi-organ segmentation. Designing loss functions based on the characteristics of different organs to make the network pay more attention to difficult-to-segment organs is an important research direction.
About weakly supervised based methods
At present, many pioneering works have been proposed to address the issue of partially supervised based method, but current works mainly consider that each dataset only annotates one organ and only considers CT images. However, in a more general situation, many publicly available datasets have multiple annotated organs, different datasets may have same organs annotated, and there are also datasets with another modality [227]. The future trend is how to design a more general architecture to handle cases with overlapping organs and different modalities.
About semi-supervised based methods
In medical science, there is a vast amount of unlabeled datasets, with only a small portion being labeled. However, there is limited discussion on semi-supervised approaches for multi-organ segmentation. However, there are a large number of unlabeled datasets in medicine, with only a small amount of data labeled. Utilizing the latest semi-supervised methods and combining prior information such as organ size and position, to improve the performance of multi-organ segmentation models is an important research direction [252, 253].
About considering inter-organ correlation
In multi-organ segmentation, a significant challenge is the imbalance in size and categories among different organs. Therefore, designing a model that can simultaneously segment large organs and fine structures is also challenging. To address this issue, researchers have proposed models specifically tailored for small organs, such as those involving localization before segmentation or the fusion of multiscale features for segmentation. In medical image analysis, segmenting structures with similar sizes or possessing prior spatial relationships can help improve segmentation accuracy. For example, Ren et al.[156] focused on segmenting small tissues like the optic chiasm and left/right optic nerves. They employed a convolutional neural network (CNN)-based approach with interleaved and cascaded processing to handle various tissues, allowing preliminary segmentation results of one organ to assist in improving the segmentation of other organs and its own segmentation. Qin et al.[254] considered the correlation between structures when segmenting the trachea, arteries, and veins, including the proximity of arteries to airways and the similarity in strength between airway walls and vessels. Additionally, some researchers [255] took into account that the spatial relationships between internal structures in medical images are often relatively fixed, such as the spleen always being located at the tail of the pancreas. These prior knowledge can serve as latent variables to transfer knowledge shared across multiple domains, thereby enhancing segmentation accuracy and stability.
About combining foundation model
Traditional methods involve training models for specific tasks on specific datasets. However, the current trend is to fine-tune pretrained foundation models for specific tasks. In recent years, there has been a surge in the development of foundation model, including the Generative Pre-trained Transformer (GPT) model [256], CLIP [222], and Segmentation Anything Model (SAM) tailored for segmentation tasks [59]. These models have achieved breakthrough results on natural images. However, due to their training samples being mostly natural images with only a small portion of medical images, the generalization ability of these models in medical images is limited [257, 258]. Recently, there have been many ongoing efforts to fine-tune these models to adapt to medical images [58, 257]. For the problem of multi-organ segmentation, it is possible to train a specialized segmentation model for medical images by integrating more medical datasets, or study better fine-tuning methods, as well as integrate knowledge from multiple foundation models to improve the segmentation performance.
Conclusion
We provide a systematic review of 195 studies on multi-organ segmentation-based on deep learning. It covers two main aspects: datasets and methods, encompassing multiple body regions such as the head, neck, chest, and abdomen. We also propose tailored solutions for some of the current challenges and limitations in this field, highlighting future research directions. Our review indicates that deep learning-based multi-organ segmentation algorithms are rapidly advancing towards a new era of more precise, detailed, and automated analysis.
Availability of data and materials
Not applicable.
References
van Ginneken B, Schaefer-Prokop CM, Prokop M. Computer-aided diagnosis: how to move from the laboratory to the clinic. Radiology. 2011;261:719–32.
Sykes J. Reflections on the current status of commercial automated segmentation systems in clinical practice. J Med Radiat Sci. 2014;61:131–4.
Pfister DG, Spencer S, Adelstein D, Adkins D, Anzai Y, Brizel DM, et al. Head and neck cancers, version 2.2020, NCCN clinical practice guidelines in oncology. J Natl Comprehensive Cancer Netw. 2020;18:873–98.
Molitoris JK, Diwanji T, Snider JW III, Mossahebi S, Samanta S, Badiyan SN, et al. Advances in the use of motion management and image guidance in radiation therapy treatment for lung cancer. J Thorac Dis. 2018;10:S2437–50.
Vyfhuis MAL, Onyeuku N, Diwanji T, Mossahebi S, Amin NP, Badiyan SN, et al. Advances in proton therapy in lung cancer. Ther Adv Respir Dis. 2018;12:175346661878387.
La Macchia M. Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat Oncol. 2012.
Round CE, Williams MV, Mee T, Kirkby NF, Cooper T, Hoskin P, Jena R. Radiotherapy demand and activity in England 2006–2020. Clin Oncol. 2013.
Hurkmans CW, Borger JH, Pieters BR, Russell NS, Jansen EPM, Mijnheer BJ. Variability in target volume delineation on CT scans of the breast. Int J Radn Oncol Biol Phys. 2001;50:1366–72.
Rasch C, Steenbakkers R, van Herk M. Target definition in prostate, head, and neck. Seminar Radn Oncol. 2005;15:136–45.
Van de Steene J, Linthout N, de Mey J, Vinh-Hung V, Claassens C, Noppen M, et al. Definition of gross tumor volume in lung cancer: inter-observer variability. Radiother Oncol. 2002;62:37–49.
Breunig J, Hernandez S, Lin J, Alsager S, Dumstorf C, Price J, et al. A system for continual quality improvement of normal tissue delineation for radiation therapy treatment planning. Int J Radn Oncol Biol Phys. 2012;83:703–8.
Chen X, Pan L. A survey of graph cuts/graph search based medical image segmentation. IEEE Rev Biomed Eng. 2018;11:112–24.
El Naqa I, Yang D, Apte A, Khullar D, Mutic S, Zheng J, et al. Concurrent multimodality image segmentation by active contours for radiotherapy treatment planninga): concurrent multimodality image segmentation for radiotherapy treatment planning. Med Phys. 2007;34:4738–49.
Pratondo A, Chui C-K, Ong S-H. Robust edge-stop functions for edge-based active contour models in medical image segmentation. IEEE Signal Process Lett. 2016;23:222–6.
Tsai A, Yezzi A, Wells W, Tempany C, Tucker D, Fan A, et al. A shape-based approach to the segmentation of medical imagery using level sets. IEEE Trans Med Imaging. 2003;22:137–54.
Saranathan AM, Parente M. Threshold based segmentation method for hyperspectral images. 2013 5th workshop on hyperspectral image and signal processing: evolution in remote sensing (WHISPERS). Gainesville, FL, USA: IEEE; 2013. p. 1–4. http://ieeexplore.ieee.org/document/8080656/. Accessed 8 Oct 2022.
Shi J, Malik J. Normalized cuts and image segmentation. IEEE Trans Pattern Anal Machine Intell. 2000;22:888–905.
Thool RC, Vyavahare AJ. Segmentation using region growing algorithm based on CLAHE for medical images. Fourth International Conference on Advances in Recent Technologies in Communication and Computing (ARTCom2012). Bangalore, India: Institution of Engineering and Technology; 2012. p. 182–5. https://doi.org/10.1049/cp.2012.2522
Isgum I, Staring M, Rutten A, Prokop M, Viergever MA, van Ginneken B. Multi-Atlas-based segmentation with local decision fusion—application to cardiac and aortic segmentation in CT scans. IEEE Trans Med Imaging. 2009;28:1000–10.
Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage. 2009;46:726–38.
Ecabert O, Peters J, Schramm H, Lorenz C, von Berg J, Walker MJ, et al. Automatic model-based segmentation of the heart in CT images. IEEE Trans Med Imaging. 2008;27:1189–201.
Qazi AA, Pekar V, Kim J, Xie J, Breen SL, Jaffray DA. Auto-segmentation of normal and target structures in head and neck CT images: a feature-driven model-based approach: feature-driven model-based segmentation. Med Phys. 2011;38:6160–70.
Heimann T, Meinzer H-P. Statistical shape models for 3D medical image segmentation: a review. Med Image Anal. 2009.
Smirnov EA, Timoshenko DM, Andrianov SN. Comparison of regularization methods for ImageNet classification with deep convolutional neural networks. AASRI Procedia. 2014;6:89–94.
Mobiny A, Van Nguyen H. Fast CapsNet for Lung Cancer Screening. In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G, editors. Medical image computing and computer assisted intervention—MICCAI 2018. Cham: Springer International Publishing; 2018. p. 741–9. https://doi.org/10.1007/978-3-030-00934-2_82
Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imag. 2019;6:1.
Wang R, Lei T, Cui R, Zhang B, Meng H, Nandi AK. Medical image segmentation using deep learning: a survey. IET Image Proc. 2022;16:1243–67.
Huang B, Yang F, Yin M, Mo X, Zhong C. A review of multimodal medical image fusion techniques. Comput Math Methods Med. 2020;2020:8279342.
Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X. Deep learning in medical image registration: a review. Phys Med Biol. 2020;65:20TR01.
Lei Y, Fu Y, Wang T, Qiu RLJ, Curran WJ, Liu T, et al. Deep Learning in multi-organ segmentation. 2020; https://arxiv.org/abs/2001.10619. Accessed 30 Sep 2022.
Vrtovec T, Močnik D, Strojan P, Pernuš F, Ibragimov B. Auto-segmentation of organs at risk for head and neck radiotherapy planning: from atlas-based to deep learning methods. Med Phys. 2020.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. arXiv:170603762 [cs]. 2017; http://arxiv.org/abs/1706.03762.
Rishi Bommasani, Drew A. Hudson,et al. On the opportunities and risks of foundation models. https://doi.org/10.48550/arXiv.2108.07258.
(Author Name Not Available). Segmentation Outside the Cranial Vault Challenge. 2015; https://repo-prod.prod.sagebase.org/repo/v1/doi/locate?id=syn3193805&type=ENTITY. Accessed 3 Oct 2022.
Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ. 2021.
Raudaschl PF, Zaffino P, Sharp GC, Spadea MF, Chen A, Dawant BM, et al. Evaluation of segmentation methods on head and neck CT: auto-segmentation challenge 2015. Med Phys. 2017;44:2020–36.
Yang J, Veeraraghavan H, Armato SG, Farahani K, Kirby JS, Kalpathy-Kramer J, et al. Autosegmentation for thoracic radiation treatment planning: a grand challenge at AAPM 2017. Med Phys. 2018;45:4568–81.
Kavur AE, Gezer NS, Barış M, Aslan S, Conze P-H, Groza V, et al. CHAOS challenge—combined (CT-MR) healthy abdominal organ segmentation. Med Image Anal. 2021;69:101950.
Ma J, Zhang Y, Gu S, Zhu C, Ge C, Zhang Y, et al. AbdomenCT-1K: is abdominal organ segmentation a solved problem. IEEE Trans Pattern Anal Mach Intell. 2021;44:1–1.
Babier A, Zhang B, Mahmood R, Moore KL, Purdie TG, McNiven AL, Chan TC. OpenKBP: The open-access knowledge-based planning grand challenge. Medical Physics. 2021; https://doi.org/10.48550/arXiv.2011.14076.
Antonelli M, Reinke A, Bakas S, et al. The medical segmentation Decathlon. https://doi.org/10.48550/arXiv.2106.05735.
Ji Y, Bai H, Ge C, Yang J, Zhu Y, Zhang R, Li Z, Zhanng L, Ma W, Wan X, Luo P. AMOS: a large-scale abdominal multi-organ benchmark for versatile medical image segmentation. 2022; https://doi.org/10.48550/arXiv.2206.08023.
Luo X, Liao W, Xiao J, Chen J, Song T, Zhang X, Li K, Metaxas DN, Wang G, Zhang S. WORD: revisiting organs segmentation in the whole abdominal region. 2023; https://doi.org/10.48550/arXiv.2111.02403.
Rister B, Yi D, Shivakumar K, Nobashi T, Rubin DL. CT-ORG, a new dataset for multiple organ segmentation in computed tomography. 2020; https://doi.org/10.1038/s41597-020-00715-8
Lambert Z, Petitjean C, Dubray B, Ruan S. SegTHOR: segmentation of thoracic organs at risk in CT images. 2019; https://arxiv.org/abs/1912.05950. Accessed 3 Oct 2022.
Podobnik G, Strojan P, Peterlin P, Ibragimov B, Vrtovec T. HaN-Seg: the head and neck organ-at-risk CT and MR segmentation dataset. Med Phys. 2023;50:1917–27.
Luo X et al. SegRap2023: a benchmark of organs-at-risk and gross tumor volume segmentation for radiotherapy planning of nasopharyngeal carcinoma. arXiv: 231209576. 2023;
Ma J, Wang B. Fast, low-resource, and accurate organ and pan-cancer segmentation in abdomen ct. international conference on medical image computing and computer assisted intervention (MICCAI) 2023 (MICCAI 2023). 2023.
Wasserthal J et al. TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. Radiol Artif Intell. 2023.
Hofmanninger J, Prayer F, Pan J, Röhrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp. 2020.
Xingjiao Wu, Xiao L, Sun Y, Zhang J, Ma T, He L. A survey of human-in-the-loop for machine learning. Futur Gener Comput Syst. 2022;135:364–81.
Rauniyar A, Hagos DH, Jha D, Håkegård JE, Bagci U, Rawat DB, Vlassov V. Federated learning for medical applications: a taxonomy, current trends, challenges, and future research directions. IEEE Int Things J. 2023. https://doi.org/10.1109/JIOT.2023.3329061.
Budd S, Robinson EC, Kainz B. A survey on active learning and human-in-the-loop deep learning for medical image analysis. Med Image Anal. 2021. https://doi.org/10.1016/j.media.2021.102062.
Zhang Y, Chen J, Ma X, Wang G, Bhatti UA, Huang M. Interactive medical image annotation using improved attention U-net with compound geodesic distance. Expert systems with applications. 2024.
Wang H, Jin Q, Li S, Liu S, Wang M, Song Z. A comprehensive survey on deep active learning and its applications in medical image analysis. arXiv:231014230. 2023;
AbdomenAtlas-8K: Annotating 8,000 CT Volumes for Multi-Organ Segmentation in Three Weeks. Conference on Neural Information Processing Systems (NeurIPS 2023). https://doi.org/10.48550/arXiv.2305.09666
Zhou T, Li L, Bredell G, Li J, Unkelbach J, Konukoglu E. Volumetric memory network for interactive medical image segmentation. Medical Image Analysis. 2023.
Wu J, Fu R, Fang H, Liu Y, Wang Z, Xu Y, Jin Y, Arbel T. The Segment Anything Model (SAM) has recently gained popularity in the field of image segmentation. Thanks to its. Medical SAM adapter: adapting segment anything model for medical image segmentation. 2023; https://doi.org/10.48550/arXiv.2304.12620.
Kirillov A, Mintun E, Ravi N, Mao H, Rolland C, Gustafson L, et al. Segment Anything. 2023; https://arxiv.org/abs/2304.02643. Accessed 21 May 2023.
McMahan B, Moore E, Ramage D, Hampson S, y Arcas BA. Communication-efficient learning of deep networks from decentralized data. Proceedings of the 20th international conference on artificial intelligence and statistics, PMLR. 2017; 54:1273–82.
Xu A, Li W, Guo P, Yang D, Roth HR, Hatamizadeh A, Zhao C, Xu D, Huang H, Xu Z. Closing the generalization gap of cross-silo federated medical image segmentation. CVPR. 2022.
Qu L, Zhou Y, Liang PP, Xia Y, Wang F, Adeli E, Fei-Fei L, Rubin D. Rethinking architecture design for tackling data heterogeneity in federated learning. CVPR. 2022.
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, et al. Backpropagation applied to handwritten Zip code recognition. Neural Comput. 1989;1:541–51.
Karthik R, Menaka R, Johnson A, Anand S. Neuroimaging and deep learning for brain stroke detection—a review of recent advancements and future prospects. Comput Methods Programs Biomed. 2020;197:105728.
Zhao X, Chen K, Wu G, Zhang G, Zhou X, Lv C, et al. Deep learning shows good reliability for automatic segmentation and volume measurement of brain hemorrhage, intraventricular extension, and peripheral edema. Eur Radiol. 2021;31:5012–20.
Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, et al. Towards image-guided pancreas and biliary endoscopy: automatic multi-organ segmentation on abdominal CT with dense dilated networks. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, editors., et al., Medical image computing and computer assisted intervention—MICCAI 2017. Cham: Springer International Publishing; 2017. p. 728–36. https://doi.org/10.1007/978-3-319-66182-7_83.
Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44:547–57.
Fritscher K, Raudaschl P, Zaffino P, Spadea MF, Sharp GC, Schubert R. Deep neural networks for fast segmentation of 3D medical images. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. Medical image computing and computer-assisted intervention—MICCAI 2016. Cham: Springer International Publishing; 2016. p. 158–65. https://doi.org/10.1007/978-3-319-46723-8_19.
Moeskops P, Wolterink JM, van der Velden BHM, Gilhuijs KGA, Leiner T, Viergever MA, et al. Deep learning for multi-task medical image segmentation in multiple modalities. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors., et al., Medical image computing and computer-assisted intervention—MICCAI 2016. Cham: Springer International Publishing; 2016. p. 478–86. https://doi.org/10.1007/978-3-319-46723-8_55.
Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:640–51.
Wang Y, Zhou Y, Tang P, Shen W, Fishman EK, Yuille AL. Training multi-organ segmentation networks with sample selection by relaxed upper confident bound. 2018; https://arxiv.org/abs/1804.02595. Accessed 14 Nov 2022.
Trullo R, Petitjean C, Ruan S, Dubray B, Nie D, Shen D. Segmentation of Organs at Risk in thoracic CT images using a SharpMask architecture and conditional random fields. 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017). Melbourne, Australia: IEEE; 2017. p. 1003–6. http://ieeexplore.ieee.org/document/7950685/. Accessed 3 Oct 2022.
Pinheiro PO, Lin T-Y, Collobert R, Dollàr P. Learning to refine object segments. 2016; https://arxiv.org/abs/1603.08695. Accessed 20 Nov 2022.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Medical image computing and computer-assisted intervention—MICCAI 2015. Cham: Springer International Publishing; 2015. p. 234–41. https://doi.org/10.1007/978-3-319-24574-4_28.
Zhu W, Huang Y, Zeng L, Chen X, Liu Y, Qian Z, et al. AnatomyNet: deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys. 2019;46:576–89.
van Rooij W, Dahele M, Ribeiro Brandao H, Delaney AR, Slotman BJ, Verbakel WF. Deep learning-based delineation of head and neck organs at risk: geometric and dosimetric evaluation. Int J Radiation Oncol Biol Phys. 2019;104:677–84.
Gou S, Tong N, Qi S, Yang S, Chin R, Sheng K. Self-channel-and-spatial-attention neural network for automated multi-organ segmentation on head and neck CT images. Phys Med Biol. 2020;65:245034.
Zhang Z, Zhao T, Gay H, Zhang W, Sun B. Weaving attention U-net: a novel hybrid CNN and attention-based method for organs-at-risk segmentation in head and neck CT images. Med Phys. 2021;48:7052–62.
Roth HR, Shen C, Oda H, Oda M, Hayashi Y, Misawa K, et al. Deep learning and its application to medical image segmentation. 2018; https://doi.org/10.11409/mit.36.63
Jia C, Wei J. AMO-Net: abdominal multi-organ segmentation in MRI with a extend Unet. 2021 IEEE 4th advanced information management, communicates, electronic and automation control conference (IMCEC) [Internet]. Chongqing, China: IEEE; 2021. p. 1770–5. https://ieeexplore.ieee.org/document/9482216/. Accessed 14 Nov 2022.
Berzoini R, Colombo AA, Bardini S, Conelli A, D’Arnese E, Santambrogio MD. An optimized U-Net for unbalanced multi-organ segmentation. 2022 44th annual international conference of the IEEE engineering in medicine and biology society (EMBC). Glasgow, Scotland, United Kingdom: IEEE; 2022. p. 3764–7. https://ieeexplore.ieee.org/document/9871288/. Accessed 8 Nov 2022.
Liu Y, Lei Y, Fu Y, Wang T, Zhou J, Jiang X, et al. Head and neck multi-organ auto-segmentation on CT images aided by synthetic MRI. Med Phys. 2020;47:4294–302.
Cros S, Vorontsov E, Kadoury S. Managing class imbalance in multi-organ CT segmentation in head and neck cancer patients. 2021 IEEE 18th international symposium on biomedical imaging (ISBI). Nice, France: IEEE; 2021. p. 1360–4. https://ieeexplore.ieee.org/document/9433991/. Accessed 14 Nov 2022.
Jiang J, Elguindi S, Berry SL, Onochie I, Cervino L, Deasy JO, et al. Nested block self-attention multiple resolution residual network for multiorgan segmentation from CT. Med Phys. 2022;49:5244–57.
Francis S, Pooloth G, Singam SBS, Puzhakkal N, Pulinthanathu Narayanan P, Pottekkattuvalappil BJ. SABOS-Net : self-supervised attention based network for automatic organ segmentation of head and neck CT images. Int J Imaging Syst Tech. 2022. https://doi.org/10.1002/ima.22802.
Vu CC, Siddiqui ZA, Zamdborg L, Thompson AB, Quinn TJ, Castillo E, et al. Deep convolutional neural networks for automatic segmentation of thoracic organs-at-risk in radiation oncology—use of non-domain transfer learning. J Appl Clin Med Phys. 2020;21:108–13.
Mahmood H, Shamsul Islam SM, Hill J, Tay G. Rapid segmentation of thoracic organs using U-net architecture. 2021 digital image computing: techniques and applications (DICTA). Gold Coast, Australia: IEEE; 2021. p. 01–6. https://ieeexplore.ieee.org/document/9647312/. Accessed 14 Nov 2022.
Hong J, Zhang Y-D, Chen W. Source-free unsupervised domain adaptation for cross-modality abdominal multi-organ segmentation. Knowl-Based Syst. 2022;250:109155.
Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44:6377–89.
Milletari F, Navab N, Ahmadi S-A. V-Net: fully convolutional neural networks for volumetric medical image segmentation. 2016; https://arxiv.org/abs/1606.04797. Accessed 20 Nov 2022.
Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, et al. Automatic multi-organ segmentation on abdominal CT With dense V-networks. IEEE Trans Med Imaging. 2018;37:1822–34.
Xu M, Guo H, Zhang J, Yan K, Lu L. A New Probabilistic V-Net model with hierarchical spatial feature transform for efficient abdominal multi-organ segmentation. 2022; https://arxiv.org/abs/2208.01382. Accessed 3 Oct 2022.
Fu S, Lu Y, Wang Y, Zhou Y, Shen W, Fishman E, et al. Domain adaptive relational reasoning for 3D multi-organ segmentation. 2020; https://arxiv.org/abs/2005.09120. Accessed 8 Oct 2022.
Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18:203–11.
Podobnik G, Ibragimov B, Strojan P, Peterlin P, Vrtovec T. Segmentation of organs-at-risk from Ct and Mr images of the head and neck: baseline results. 2022 IEEE 19th international symposium on biomedical imaging (ISBI). Kolkata, India: IEEE; 2022. p. 1–4. https://ieeexplore.ieee.org/document/9761433/. Accessed 14 Nov 2022.
Isensee F, Jäger PF, Full PM, Vollmuth P, Maier-Hein KH. nnU-Net for Brain Tumor Segmentation. In: Crimi A, Bakas S, editors. Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. Cham: Springer International Publishing; 2021.
Zhang G, Yang Z, Huo B, Chai S, Jiang S. Multiorgan segmentation from partially labeled datasets with conditional nnU-Net. Comput Biol Med. 2021;136:104658.
Altini N, Brunetti A, Napoletano VP, Girardi F, Allegretti E, Hussain SM, et al. A fusion biopsy framework for prostate cancer based on deformable superellipses and nnU-Net. Bioengineering. 2022;9:343.
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial networks. arXiv; 2014. http://arxiv.org/abs/1406.2661. Accessed 3 Oct 2022.
Tong N, Gou S, Yang S, Cao M, Sheng K. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low-field MR images. Med Phys. 2019;46:2669–82.
Cai J, Xia Y, Yang D, Xu D, Yang L, Roth H. End-to-end adversarial shape learning for abdomen organ deep segmentation. In: Suk H-I, Liu M, Yan P, Lian C, editors. Machine learning in medical imaging. Cham: Springer International Publishing; 2019. p. 124–32. https://doi.org/10.1007/978-3-030-32692-0_15.
Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys. 2019;46:2157–68.
Trullo R, Petitjean C, Dubray B, Ruan S. Multiorgan segmentation using distance-aware adversarial networks. J Med Imag. 2019;6:1.
Mahmood F, Borders D, Chen RJ, Mckay GN, Salimian KJ, Baras A, et al. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans Med Imaging. 2020;39:3257–67.
Gao Y, Huang R, Yang Y, Zhang J, Shao K, Tao C, et al. FocusNetv2: imbalanced large and small organ segmentation with adversarial shape constraint for head and neck CT images. Med Image Anal. 2021;67:101831.
Fang H, Fang Y, Yang X. Multi-organ segmentation network with adversarial performance validator. 2022; https://arxiv.org/abs/2204.07850. Accessed 17 Nov 2022.
Kumar V, Sharma MK, Jehadeesan R, Venkatraman B, Sheet D. Adversarial training of deep convolutional neural network for multi-organ segmentation from multi-sequence MRI of the abdomen. 2021 international conference on intelligent technologies (CONIT). Hubli, India: IEEE; 2021. p. 1–6. https://ieeexplore.ieee.org/document/9498339/. Accessed 14 Nov 2022.
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. 2017 IEEE Conference on computer vision and pattern recognition (CVPR). Honolulu, HI: IEEE; 2017. p. 2261–9. https://ieeexplore.ieee.org/document/8099726/. Accessed 14 Nov 2022.
Jeong JJ, Tariq A, Adejumo T, Trivedi H, Gichoya JW, Banerjee I. Systematic review of generative adversarial networks (GANs) for medical image classification and segmentation. J Digit Imaging. 2022;35:137–52.
Gao Y, Zhou M, Metaxas D. UTNet: a hybrid transformer architecture for medical image segmentation. 2021; https://arxiv.org/abs/2107.00781. Accessed 3 Oct 2022.
Chang Y, Menghan H, Guangtao Z, Xiao-Ping Z. TransClaw U-Net: Claw U-Net with transformers for medical image segmentation. 2021; https://arxiv.org/abs/2107.05188. Accessed 3 Oct 2022.
Valanarasu JMJ, Oza P, Hacihaliloglu I, Patel VM. Medical transformer: gated axial-attention for medical image segmentation. arXiv; 2021. http://arxiv.org/abs/2102.10662. Accessed 3 Oct 2022.
Pan S, Lei Y, Wang T, Wynne J, Chang C-W, Roper J, et al. Male pelvic multi-organ segmentation using token-based transformer Vnet. Phys Med Biol. 2022;67:205012.
Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, et al. Swin-Unet: Unet-like pure transformer for medical image segmentation. arXiv; 2021. http://arxiv.org/abs/2105.05537. Accessed 3 Oct 2022.
Huang X, Deng Z, Li D, Yuan X. MISSFormer: an effective medical image segmentation transformer. arXiv; 2021. http://arxiv.org/abs/2109.07162. Accessed 3 Oct 2022.
Tang Y, Yang D, Li W, Roth HR, Landman B, Xu D, Nath V, Hatamizadeh A.. Self-Supervised Pre-Training of Swin Transformers for 3D Medical Image Analysis. CVPR. 2022;
Zheng S, Lu J, Zhao H, Zhu X, Luo Z, Wang Y, Fu Y, Feng J, Xiang T, Torr PH, Zhang L. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. CVPR. 2021;
Shamshad F, Khan S, Zamir SW, Khan MH, Hayat M, Khan FS, Fu H. Transformers in medical imaging: a survey. Med Image Anal. 2023;88:102802.
Xie Y, Zhang J, Shen C, Xia Y. CoTr: Efficiently bridging CNN and transformer for 3D medical image segmentation. arXiv; 2021. http://arxiv.org/abs/2103.03024. Accessed 3 Oct 2022.
Wang H, Cao P, Wang J, Zaiane OR. UCTransNet: Rethinking the skip connections in U-Net from a channel-wise perspective with transformer. arXiv; 2022. http://arxiv.org/abs/2109.04335. Accessed 3 Oct 2022.
Wang H, Xie S, Lin L, Iwamoto Y, Han X-H, Chen Y-W, et al. Mixed transformer U-Net For medical image segmentation. 2021; https://arxiv.org/abs/2111.04734 Accessed 3 Oct 2022.
Xu G, Wu X, Zhang X, He X. LeViT-UNet: Make faster encoders with transformer for medical image segmentation. arXiv; 2021. http://arxiv.org/abs/2107.08623 Accessed 3 Oct 2022.
Zhang Y, Liu H, Hu Q. TransFuse: fusing transformers and CNNs for Medical image segmentation. 2021; https://arxiv.org/abs/2102.08005 Accessed 23 Oct 2022.
Suo C, Li X, Tan D, Zhang Y, Gao X. I2-Net: intra- and inter-scale collaborative learning network for abdominal multi-organ segmentation. proceedings of the 2022 international conference on multimedia retrieval. Newark NJ USA: ACM; 2022. p. 654–60. https://doi.org/10.1145/3512527.3531420. Accessed 3 Oct 2022.
Kan H, Shi J, Zhao M, Wang Z, Han W, An H, et al. ITUnet: integration of transformers and unet for organs-at-risk segmentation. 2022 44th annual international conference of the IEEE engineering in medicine & biology society (EMBC). Glasgow, Scotland, United Kingdom: IEEE; 2022. p. 2123–7. https://ieeexplore.ieee.org/document/9871945/ Accessed 3 Oct 2022.
Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. TransUNet: transformers make strong encoders for medical image segmentation. 2021; https://arxiv.org/abs/2102.04306 Accessed 3 Oct 2022.
Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, et al. UNETR: Transformers for 3D medical image segmentation. 2022 IEEE/CVF winter conference on applications of computer vision (WACV). Waikoloa, HI, USA: IEEE; 2022. p. 1748–58. https://ieeexplore.ieee.org/document/9706678/ Accessed 3 Oct 2022.
Ali Hatamizadeh, Vishwesh Nath, Yucheng Tang, Dong Yang, Holger Roth, Daguang Xu. Swin UNETR: Swin transformers for semantic segmentation of brain tumors in MRI images. arXiv:220101266. 2022.
Chen P-H, Huang C-H, Hung S-K, Chen L-C, Hsieh H-L, Chiou W-Y, et al. Attention-LSTM Fused U-Net architecture for organ segmentation in CT images. 2020 international symposium on computer, consumer and control (IS3C). Taichung city, Taiwan: IEEE; 2020. p. 304–7. https://ieeexplore.ieee.org/document/9394166/ Accessed 20 Nov 2022.
Chakravarty A, Sivaswamy J. RACE-Net: a recurrent neural network for biomedical image segmentation. IEEE J Biomed Health Inform. 2019;23:1151–62.
Tappeiner E, Pröll S, Hönig M, Raudaschl PF, Zaffino P, Spadea MF, et al. Multi-organ segmentation of the head and neck area: an efficient hierarchical neural networks approach. Int J CARS. 2019;14:745–54.
Hu P, Wu F, Peng J, Bao Y, Chen F, Kong D. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int J CARS. 2017;12:399–411.
Zhang L, Zhang J, Shen P, Zhu G, Li P, Lu X, et al. Block level skip connections across cascaded V-Net for multi-organ segmentation. IEEE Trans Med Imaging. 2020;39:2782–93.
Xie L, Yu Q, Zhou Y, Wang Y, Fishman EK, Yuille AL. Recurrent saliency transformation network for tiny target segmentation in abdominal CT scans. IEEE Trans Med Imaging. 2020;39:514–25.
Lee HH, Tang Y, Bao S, Abramson RG, Huo Y, Landman BA. Rap-Net: Coarse-To-Fine multi-organ segmentation with single random anatomical prior. 2021 IEEE 18th international symposium on biomedical imaging (ISBI). Nice, France: IEEE; 2021. p. 1491–4. https://ieeexplore.ieee.org/document/9433975/. Accessed 3 Oct 2022.
Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, et al. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. 2016. p. 415–23. http://arxiv.org/abs/1610.02177. Accessed 3 Oct 2022.
Lachinov D, Vasiliev E, Turlapov V. Glioma Segmentation with Cascaded UNet. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T, editors. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Cham: Springer International Publishing; 2019. p. 189–98. https://doi.org/10.1007/978-3-030-11726-9_17
Li S, Chen Y, Yang S, Luo W. Cascade Dense-Unet for Prostate Segmentation in MR Images. In: Huang D-S, Bevilacqua V, Premaratne P, editors. Intelligent Computing Theories and Application. Cham: Springer International Publishing; 2019. p. 481–90. https://doi.org/10.1007/978-3-030-26763-6_46.
Kakeya H, Okada T, Oshiro Y. 3D U-JAPA-Net: Mixture of Convolutional Networks for Abdominal Multi-organ CT Segmentation. In: Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C, Fichtinger G, editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Cham: Springer International Publishing; 2018 [cited 2022 Oct 8]. p. 426–33. https://doi.org/10.1007/978-3-030-00937-3_49
Trullo R, Petitjean C, Nie D, Shen D, Ruan S. Joint segmentation of multiple thoracic organs in CT images with two collaborative deep architectures. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support. 2017;2017(10553):21–9.
Cao Z, Yu B, Lei B, Ying H, Zhang X, Chen DZ, et al. Cascaded SE-ResUnet for segmentation of thoracic organs at risk. Neurocomputing. 2021;453:357–68.
Wang Y, Zhao L, Wang M, Song Z. Organ at risk segmentation in head and neck CT images using a two-stage segmentation framework based on 3D U-Net. IEEE Access. 2019;7:144591–602.
Men K, Geng H, Cheng C, Zhong H, Huang M, Fan Y, et al. Technical note: more accurate and efficient segmentation of organs-at-risk in radiotherapy with convolutional neural networks cascades. Med Phys. 2018. https://doi.org/10.1002/mp.13296.
Tang H, Chen X, Liu Y, Lu Z, You J, Yang M, et al. Clinically applicable deep learning framework for organs at risk delineation in CT images. Nat Mach Intell. 2019;1:480–91.
Yang Q, Zhang S, Sun X, Sun J, Yuan K. Automatic segmentation of head-neck organs by Multi-MODE CNNs for radiation therapy. 2019 international conference on medical imaging physics and engineering (ICMIPE). Shenzhen, China: IEEE; 2019. p. 1–5. https://ieeexplore.ieee.org/document/9098166/. Accessed 8 Oct 2022.
Liang S, Tang F, Huang X, Yang K, Zhong T, Hu R, et al. Deep-learning-based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. Eur Radiol. 2019;29:1961–7.
Gao Y, Huang R, Chen M, Wang Z, Deng J, Chen Y, et al. FocusNet: imbalanced large and small organ segmentation with an end-to-end deep neural network for head and neck CT images. arXiv; 2019. http://arxiv.org/abs/1907.12056. Accessed 3 Oct 2022.
Liang S, Thung K-H, Nie D, Zhang Y, Shen D. Multi-view spatial aggregation framework for joint localization and segmentation of organs at risk in head and neck CT images. IEEE Trans Med Imaging. 2020;39:2794–805.
Lei Y, Zhou J, Dong X, Wang T, Mao H, McDonald M, et al. Multi-organ segmentation in head and neck MRI using U-Faster-RCNN. In: Landman BA, Išgum I, editors. Medical Imaging 2020: Image Processing. Houston, United States: SPIE; 2020 [cited 2022 Oct 3]. p. 117. Available from: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11313/2549596/Multi-organ-segmentation-in-head-and-neck-MRI-using-U/https://doi.org/10.1117/12.2549596.full
Huang B, Ye Y, Xu Z, Cai Z, He Y, Zhong Z, et al. 3D Lightweight Network for Simultaneous Registration and Segmentation of Organs-at-Risk in CT Images of Head and Neck Cancer. IEEE Trans Med Imaging. 2022;41:951–64.
Korte JC, Hardcastle N, Ng SP, Clark B, Kron T, Jackson P. Cascaded deep learning-based auto-segmentation for head and neck cancer patients: Organs at risk on T2-weighted magnetic resonance imaging. Med Phys. 2021;48:7757–72.
Larsson M, Zhang Y, Kahl F. Robust abdominal organ segmentation using regional convolutional neural networks. Appl Soft Comput. 2018;70:465–71.
Zhao Y, Li H, Wan S, Sekuboyina A, Hu X, Tetteh G, et al. Knowledge-aided convolutional neural network for small organ segmentation. IEEE J Biomed Health Inform. 2019;23:1363–73.
Ma Q, Zu C, Wu X, Zhou J, Wang Y, et al. Coarse-to-fine segmentation of organs at risk in nasopharyngeal carcinoma radiotherapy. In: de Bruijne M, Cattin PC, Cotin S, Padoy N, Speidel S, Zheng Y, et al., editors. Medical image computing and computer assisted intervention—MICCAI 2021. Cham: Springer International Publishing; 2021. p. 358–68. https://doi.org/10.1007/978-3-030-87193-2_34.
Francis S, Jayaraj PB, Pournami PN, Thomas M, Jose AT, Binu AJ, et al. ThoraxNet: a 3D U-Net based two-stage framework for OAR segmentation on thoracic CT images. Phys Eng Sci Med. 2022;45:189–203.
Ren X, Xiang L, Nie D, Shao Y, Zhang H, Shen D, et al. Interleaved 3D-CNNs for joint segmentation of small-volume structures in head and neck CT images. Med Phys. 2018;45:2063–75.
Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys. 2018;45:4558–67.
Roth HR, Oda H, Hayashi Y, Oda M, Shimizu N, Fujiwara M, et al. Hierarchical 3D fully convolutional networks for multi-organ segmentation. arXiv; 2017. http://arxiv.org/abs/1704.06382. Accessed 3 Oct 2022.
Kéchichian, R., Valette, S., Sdika, M., Desvignes, M. Automatic 3D Multiorgan Segmentation via Clustering and Graph Cut Using Spatial Relations and Hierarchically-Registered Atlases. Medical Computer Vision: Algorithms for Big Data MCV. Lecture Notes in Computer Science. 2014; https://doi.org/10.1007/978-3-319-13972-2_19
Hammon M, Cavallaro A, Erdt M, Dankerl P, Kirschner M, Drechsler K, Wesarg S, Uder M, Janka R. Model-based pancreas segmentation in portal venous phase contrast-enhanced CT images. J Digit Imaging. 2013;26:1082–90.
Zhao Z, Chen H, Wang L. A coarse-to-fine framework for the 2021 kidney and kidney tumor segmentation challenge. Kidney and kidney. Tumor Seg. 2021;2021:13168.
Zhou L, Meng X, Huang Y, et al. An interpretable deep learning workflow for discovering subvisual abnormalities in CT scans of COVID-19 inpatients and survivors. Nat Mach Intell. 2022;4:494–503.
Guo D, Jin D, Zhu Z, Ho T-Y, Harrison AP, Chao C-H, et al. Organ at risk segmentation for head and neck cancer using stratified learning and neural architecture search. 2020 IEEE/CVF conference on computer vision and pattern recognition (CVPR). Seattle, WA, USA: IEEE; 2020. p. 4222–31. https://ieeexplore.ieee.org/document/9156960/. Accessed 23 Nov 2022.
Chen Z, Li C, He J, Ye J, Song D, Wang S, et al. A Novel hybrid convolutional neural network for accurate organ segmentation in 3D head and neck CT images. arXiv; 2021. http://arxiv.org/abs/2109.12634. Accessed 3 Oct 2022.
Chen Y, Ruan D, Xiao J, Wang L, Sun B, Saouaf R, et al. Fully automated multi-organ segmentation in abdominal magnetic resonance imaging with deep neural networks. Med Phys. 2020;47:4971–82.
Jain R, Sutradhar A, Dash AK, Das S. Automatic Multi-organ Segmentation on Abdominal CT scans using Deep U-Net Model. 2021 19th OITS international conference on information Technology (OCIT). Bhubaneswar, India: IEEE; 2021. p. 48–53. https://ieeexplore.ieee.org/document/9719516/. Accessed 3 Oct 2022.
Ahn Y, Yoon JS, Lee SS, Suk H-I, Son JH, Sung YS, et al. Deep learning algorithm for automated segmentation and volume measurement of the liver and spleen using portal venous phase computed tomography images. Korean J Radiol. 2020;21:987.
Shi J, Wen K, Hao X, Xue X, An H, Zhang H. A Novel U-Like Network For The segmentation of thoracic organs. 2020 IEEE 17th international symposium on biomedical imaging workshops (ISBI Workshops). Iowa City, IA, USA: IEEE; 2020 [cited 2022 Oct 8]. p. 1–4. https://ieeexplore.ieee.org/document/9153358/.
Pu Y, Kamata S-I, Wang Y. A Coarse to Fine Framework for Multi-organ Segmentation in Head and Neck Images. 2020 Joint 9th International Conference on Informatics, Electronics & Vision (ICIEV) and 2020 4th International Conference on Imaging, Vision & Pattern Recognition (icIVPR). Kitakyushu, Japan: IEEE; 2020 [cited 2022 Oct 3]. p. 1–6. https://ieeexplore.ieee.org/document/9306647/.
Zhou X, Takayama R, Wang S, Hara T, Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys. 2017;44:5221–33.
Wang Y, Zhou Y, Shen W, Park S, Fishman EK, Yuille AL. Abdominal multi-organ segmentation with organ-attention networks and statistical fusion. Med Image Anal. 2019;55:88–102.
Tang H, Liu X, Han K, Sun S, Bai N, Chen X, et al. Spatial context-aware self-attention model for multi-organ segmentation. arXiv; 2020. http://arxiv.org/abs/2012.09279. Accessed 3 Oct 2022.
Zhao Q, et al. Efficient multi-organ segmentation from 3D abdominal CT images with lightweight network and knowledge distillation. IEEE Trans Med Imaging. 2023;42:2513.
Cheng ZS, Zeng TY, Huang SJ, Yang X. A Novel Hybrid Network for H&N Organs at Risk Segmentation. Proceedings of the 2020 5th International Conference on Biomedical Signal and Image Processing. Suzhou China: ACM; 2020 [cited 2022 Oct 3]. p. 7–13. https://doi.org/10.1145/3417519.3417522
Zhou Y, Li Z, Bai S, Chen X, Han M, Wang C, et al. Prior-aware neural network for partially-supervised multi-organ segmentation. 2019 IEEE/CVF International conference on computer vision (ICCV). Seoul, Korea (South): IEEE; 2019. p. 10671–80. https://ieeexplore.ieee.org/document/9009566/. Accessed 8 Oct 2022.
Lian S, Li L, Luo Z, Zhong Z, Wang B, Li S. Learning multi-organ segmentation via partial—and mutual-prior from single-organ datasets. Biomed Signal Process Control. 2023;80:104339.
Tappeiner E, Pröll S, Fritscher K, et al. Training of head and neck segmentation networks with shape prior on small datasets. Int J CARS. 2020;15:1417–25.
Ho J, Jain A, Abbeel P. Denoising diffusion probabilistic models. 2020; https://arxiv.org/abs/2006.11239. Accessed 23 Nov 2022.
Song J, Meng C, Ermon S. Denoising diffusion implicit models. 2020; https://arxiv.org/abs/2010.02502. Accessed 23 Nov 2022.
Oktay O, Ferrante E, Kamnitsas K, Heinrich M, Bai W, Caballero J, et al. Anatomically constrained neural networks (ACNNs): application to cardiac image enhancement and segmentation. IEEE Trans Med Imaging. 2018;37:384–95.
Isler I, Lisle C, Rineer J, Kelly P, Turgut D, Ricci J, et al. Enhancing organ at risk segmentation with improved deep neural networks. arXiv; 2022. http://arxiv.org/abs/2202.01866. Accessed 3 Oct 2022.
Vesal S, Ravikumar N, Maier A. A 2D dilated residual U-Net for multi-organ segmentation in thoracic CT. 2019; https://arxiv.org/abs/1905.07710. Accessed 8 Oct 2022.
Li, W., Wang, G., Fidon, L., Ourselin, S., Cardoso, M.J., Vercauteren, T. On the compactness, efficiency, and representation of 3D convolutional networks: brain parcellation as a pretext task. Information processing in medical imaging IPMI. 2017; 10265.
Lin T-Y, Dollar P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. 2017 IEEE conference on computer vision and pattern recognition (CVPR). Honolulu, HI: IEEE; 2017. p. 936–44. http://ieeexplore.ieee.org/document/8099589/. Accessed 20 Mar 2022.
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40:834–48.
Srivastava A, Jha D, Keles E, Aydogan B, Abazeed M, Bagci U. An efficient multi-scale fusion network for 3D organ at Risk (OAR) Segmentation. 2022; https://arxiv.org/abs/2208.07417. Accessed 3 Oct 2022.
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, et al. Attention U-net: learning where to look for the pancreas. arXiv; 2018. http://arxiv.org/abs/1804.03999. Accessed 20 Jun 2022.
Hu J, Shen L, Albanie S, Sun G, Wu E. Squeeze-and-excitation networks. IEEE Trans Pattern Anal Mach Intell. 2020;42:2011–23.
Liu Z, Wang H, Lei W, Wang G. CSAF-CNN: Cross-Layer Spatial Attention Map Fusion Network for Organ-at-Risk Segmentation in Head and Neck CT Images. 2020 IEEE 17th international symposium on biomedical imaging (ISBI). Iowa City, IA, USA: IEEE; 2020. p. 1522–5. https://ieeexplore.ieee.org/document/9098711/. Accessed 3 Oct 2022.
Lin H, Li Z, Yang Z, Wang Y. Variance-aware attention U-Net for multi-organ segmentation. Med Phys. 2021;48:7864–76.
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J, et al. UNet++: a nested U-net architecture for medical image segmentation. In: Stoyanov D, Taylor Z, Carneiro G, Syeda-Mahmood T, Martel A, Maier-Hein L, et al., editors. Deep learning in medical image analysis and multimodal learning for clinical decision support. Cham: Springer International Publishing; 2018. p. 3–11. https://doi.org/10.1007/978-3-030-00889-5_1.
He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. arXiv:151203385 [cs]. 2015; http://arxiv.org/abs/1512.03385. Accessed 14 Nov 2022.
Dai J, Qi H, Xiong Y, Li Y, Zhang G, Hu H, et al. Deformable convolutional networks. 2017 IEEE international conference on computer vision (ICCV). Venice: IEEE; 2017. p. 764–73. http://ieeexplore.ieee.org/document/8237351/. Accessed 14 Nov 2022.
Heinrich MP, Oktay O, Bouteldja N. OBELISK-Net: Fewer layers to solve 3D multi-organ segmentation with sparse deformable convolutions. Med Image Anal. 2019;54:1–9.
Shen N, Wang Z, Li J, Gao H, Lu W, Hu P, et al. Multi-organ segmentation network for abdominal CT images based on spatial attention and deformable convolution. Expert Syst Appl. 2023;211:118625.
Hou Q, Zhang L, Cheng M-M, Feng J. Strip pooling: rethinking spatial pooling for scene parsing. 2020; https://arxiv.org/abs/2003.13328. Accessed 17 Nov 2022.
Zhang F, Wang Y, Yang H. Efficient context-aware network for abdominal multi-organ segmentation. 2021; https://arxiv.org/abs/2109.10601. Accessed 14 Nov 2022.
Jadon S. A survey of loss functions for semantic segmentation. 2020 IEEE conference on computational intelligence in bioinformatics and computational biology (CIBCB). 2020. p. 1–7. http://arxiv.org/abs/2006.14822. Accessed 3 Oct 2022.
Ma J, Chen J, Ng M, Huang R, Li Y, Li C, Yang X, Martel AL. Loss odyssey in medical image segmentation. Med Image Anal. 2021;71:102035.
Yi-de Ma, Qing Liu, Zhi-bai Quan. Automated image segmentation using improved PCNN model based on cross-entropy. Proceedings of 2004 international symposium on intelligent multimedia, video and speech processing, 2004. Hong Kong, China: IEEE; 2004. p. 743–6. http://ieeexplore.ieee.org/document/1434171/. Accessed 3 Oct 2022.
Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. 2017. p. 240–8. http://arxiv.org/abs/1707.03237. Accessed 3 Oct 2022.
Salehi SSM, Erdogmus D, Gholipour A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In: Wang Q, Shi Y, Suk H-I, Suzuki K, editors. Machine learning in medical imaging. Cham: Springer International Publishing; 2017. p. 379–87. https://doi.org/10.1007/978-3-319-67389-9_44.
Lin T-Y, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. 2017; https://arxiv.org/abs/1708.02002. Accessed 3 Oct 2022.
Pihur V, Datta S, Datta S. Weighted rank aggregation of cluster validation measures: a Monte Carlo cross-entropy approach. Bioinformatics. 2007;23:1607–15.
Shen C, Roth HR, Oda H, Oda M, Hayashi Y, Misawa K, et al. On the influence of dice loss function in multi-class organ segmentation of abdominal CT using 3D fully convolutional networks. 2018; https://arxiv.org/abs/1801.05912. Accessed 3 Oct 2022.
Tappeiner E, Welk M, Schubert R. Tackling the class imbalance problem of deep learning-based head and neck organ segmentation. Int J CARS. 2022;17:2103–11.
Kodym O, Španěl M, Herout A. Segmentation of head and neck organs at risk using CNN with batch dice loss. Pattern Recogn GCPR. 2018;2018:11269.
Abraham N, Khan NM. A novel focal Tversky loss function with improved attention U-Net for lesion segmentation. arXiv; 2018. http://arxiv.org/abs/1810.07842. Accessed 3 Oct 2022.
Wong KCL, Moradi M, Tang H, Syeda-Mahmood T. 3D segmentation with exponential logarithmic loss for highly unbalanced object sizes. 2018. p. 612–9. http://arxiv.org/abs/1809.00076. Accessed 3 Oct 2022.
Song J, Chen X, Zhu Q, Shi F, Xiang D, Chen Z, et al. Global and local feature reconstruction for medical image segmentation. IEEE Trans Med Imaging. 2022;41:2273–84.
Lei W, Mei H, Sun Z, Ye S, Gu R, Wang H, et al. Automatic segmentation of organs-at-risk from head-and-neck CT using separable convolutional neural network with hard-region-weighted loss. arXiv; 2021. http://arxiv.org/abs/2102.01897. Accessed 3 Oct 2022.
Y. Song, J. Y. -C. Teoh, K. -S. Choi and J. Qin. Dynamic loss weighting for multiorgan segmentation in medical images. IEEE transactions on neural networks and learning systems.
Bilic P, Christ PF, Vorontsov E, Chlebus G, Chen H, Dou Q, et al. The liver tumor segmentation benchmark (LiTS). 2019; https://arxiv.org/abs/1901.04056. Accessed 19 Oct 2022.
Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, et al. The KiTS19 challenge data: 300 kidney tumor cases with clinical context, ct semantic segmentations, and surgical outcomes. 2019; https://arxiv.org/abs/1904.00445. Accessed 19 Oct 2022.
Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, van Ginneken B, et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. 2019; https://arxiv.org/abs/1902.09063. Accessed 3 Oct 2022.
Chen S, Ma K, Zheng Y. Med3D: Transfer Learning for 3D Medical Image Analysis. arXiv:190400625 [cs]. 2019; http://arxiv.org/abs/1904.00625. Accessed 20 Mar 2022.
Zhang J, Xie Y, Xia Y, Shen C. DoDNet: learning to segment multi-organ and tumors from multiple partially labeled datasets. 2020; https://arxiv.org/abs/2011.10217. Accessed 8 Oct 2022.
Xie Y, Zhang J, Xia Y, Shen C. Learning from partially labeled data for multi-organ and tumor segmentation. 2022; https://arxiv.org/abs/2211.06894. Accessed 20 Nov 2022.
Dmitriev K, Kaufman AE. Learning multi-class segmentations from single-class datasets. 2019 IEEE/CVF conference on computer vision and pattern recognition (CVPR). Long Beach, CA, USA: IEEE; 2019. p. 9493–503. https://ieeexplore.ieee.org/document/8953428/. Accessed 19 Oct 2022.
Wu H, Pang S, Sowmya A. Tgnet: a task-guided network architecture for multi-organ and tumour segmentation from partially labelled datasets. 2022 IEEE 19th international symposium on biomedical imaging (ISBI). Kolkata, India: IEEE; 2022. p. 1–5. https://ieeexplore.ieee.org/document/9761582/. Accessed 7 Dec 2022.
Liu X, Wen B, Yang S. CCQ: cross-class query network for partially labeled organ segmentation. AAAI. 2023;37:1755–63.
Radford A, Kim JW, Hallacy C, Ramesh A, Goh G, Agarwal S, et al. Learning transferable visual models from natural language supervision. 2021: https://arxiv.org/abs/2103.00020. Accessed 21 May 2023.
Liu J, Zhang Y, Chen J-N, Xiao J, Lu Y, Landman BA, et al. CLIP-driven universal model for organ segmentation and tumor detection. arXiv; 2023. http://arxiv.org/abs/2301.00785. Accessed 21 May 2023.
Ye Y, Xie Y, Zhang J, Chen Z, Xia Y. UniSeg: a prompt-driven universal segmentation model as well as a strong representation learner. arXiv; 2023. http://arxiv.org/abs/2304.03493. Accessed 21 May 2023.
Huang R, Zheng Y, Hu Z, Zhang S, Li H. Multi-organ segmentation via co-training weight-averaged models from few-organ datasets. 2020; https://arxiv.org/abs/2008.07149. Accessed 8 Oct 2022.
Zhang L, Feng S, Wang Y, Wang Y, Zhang Y, Chen X, et al. Unsupervised ensemble distillation for multi-organ segmentation. 2022 IEEE 19th international symposium on biomedical imaging (ISBI). Kolkata, India: IEEE; 2022. p. 1–5. https://ieeexplore.ieee.org/document/9761568/. Accessed 19 Oct 2022.
Liu H, Xu Z, Gao R, Li H, Wang J, Chabin G, et al. COSST: Multi-organ segmentation with partially labeled datasets using comprehensive supervisions and self-training. arXiv; 2023. http://arxiv.org/abs/2304.14030. Accessed 18 Jul 2023.
Cermelli F, Mancini M, Bulò SR, Ricci E, Caputo B. Modeling the Background for incremental learning in semantic segmentation. 2020; https://arxiv.org/abs/2002.00718. Accessed 21 May 2023.
Liu P, Xiao L, Zhou SK. Incremental learning for multi-organ segmentation with partially labeled datasets. 2021; https://arxiv.org/abs/2103.04526. Accessed 15 Nov 2022.
Xu X, Yan P. Federated multi-organ segmentation with partially labeled data. 2022; https://arxiv.org/abs/2206.07156. Accessed 15 Nov 2022.
Zhang Y, Li X, Chen H, Yuille A, Liu Y, Zhou Z. Continual learning for abdominal multi-organ and tumor segmentation. 2023; https://arxiv.org/abs/2306.00988. Accessed 3 Aug 2023.
Ji Z, et al. Continual Segment: Towards a Single, Unified and Non-forgetting Continual Segmentation Model of 143 Whole-body Organs in CT Scans. IEEE/CVF Int Conf Comp Vis. 2023;2023:21083–94.
Fang X, Yan P. Multi-organ segmentation over partially labeled datasets with multi-scale feature abstraction. IEEE Trans Med Imaging. 2020;39:3619–29.
Shi G, Xiao L, Chen Y, Zhou SK. Marginal loss and exclusion loss for partially supervised multi-organ segmentation. Med Image Anal. 2021;70:101979.
Wu Q, Chen Y, Huang N, Yue X. Weakly-supervised Cerebrovascular Segmentation Network with Shape Prior and Model Indicator. Proceedings of the 2022 International Conference on Multimedia Retrieval. Newark NJ USA: ACM; 2022 [cited 2022 Nov 15]. p. 668–76. https://doi.org/10.1145/3512527.3531377.
Kanavati F, Misawa K, Fujiwara M, Mori K, Rueckert D, Glocker B. Joint supervoxel classification forest for weakly-supervised organ segmentation. In: Wang Q, Shi Y, Suk H-I, Suzuki K, editors. Machine learning in medical imaging. Cham: Springer International Publishing; 2017. p. 79–87. https://doi.org/10.1007/978-3-319-67389-9_10.
Bai W, Oktay O, Sinclair M, Suzuki H, Rajchl M, Tarroni G, et al. Semi-supervised learning for network-based cardiac MR image segmentation. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, editors., et al., Medical image computing and computer-assisted intervention—MICCAI 2017. Cham: Springer International Publishing; 2017. p. 253–60. https://doi.org/10.1007/978-3-319-66185-8_29.
Luo X, Hu M, Song T, Wang G, Zhang S. Semi-supervised medical image segmentation via cross teaching between CNN and transformer. 2021; https://arxiv.org/abs/2112.04894. Accessed 7 Dec 2022.
Chen J, Zhang J, Debattista K, Han J. Semi-supervised unpaired medical image segmentation through task-affinity consistency. IEEE Trans Med Imaging. 2022. https://doi.org/10.1109/TMI.2022.3213372.
Wu Y, Ge Z, Zhang D, Xu M, Zhang L, Xia Y, et al. Mutual consistency learning for semi-supervised medical image segmentation. Med Image Anal. 2022;81:102530.
Luo X, Liao W, Chen J, Song T, Chen Y, Zhang S, et al. Efficient semi-supervised gross target volume of nasopharyngeal carcinoma segmentation via uncertainty rectified pyramid consistency. In: de Bruijne M, Cattin PC, Cotin S, Padoy N, Speidel S, Zheng Y, et al., editors. Medical image computing and computer assisted intervention—MICCAI 2021. Cham: Springer International Publishing; 2021. p. 318–29. https://doi.org/10.1007/978-3-030-87196-3_30.
Jiao R, Zhang Y, Ding L, Cai R, Zhang J. Learning with limited annotations: a survey on deep semi-supervised learning for medical image segmentation. 2022; https://arxiv.org/abs/2207.14191. Accessed 6 Dec 2022.
Zhou Y, Wang Y, Tang P, Bai S, Shen W, Fishman E, et al. Semi-Supervised 3D Abdominal multi-organ segmentation Via deep multi-planar co-training. 2019 IEEE winter conference on applications of computer vision (WACV). Waikoloa Village, HI, USA: IEEE; 2019. p. 121–40. https://ieeexplore.ieee.org/document/8658899/. Accessed 8 Dec 2022.
Xia Y, Yang D, Yu Z, Liu F, Cai J, Yu L, et al. Uncertainty-aware multi-view co-training for semi-supervised medical image segmentation and domain adaptation. Med Image Anal. 2020;65:101766.
Lai H, Wang T, Zhou S. DLUNet: Semi-supervised learning based dual-light UNet for multi-organ segmentation. 2022. https://arxiv.org/abs/2209.10984. Accessed 7 Dec 2022.
Chen D, Bai Y, Shen W, Li Q, Yu L, Wang Y. MagicNet: semi-supervised multi-organ segmentation via magic-cube partition and recovery. 2023; https://doi.org/10.48550/arXiv.2212.14310.
Lee HH, Tang Y, Tang O, Xu Y, Chen Y, Gao D, et al. Semi-supervised multi-organ segmentation through quality assurance supervision. In: Landman BA, Išgum I. (Eds). Medical Imaging 2020: Image Processing [Internet]. Houston, United States: SPIE; 2020 [cited 2022 Nov 15]. p. 53. Available from: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11313/2549033/Semi-supervised-multi-organ-segmentation-through-quality-assurance-supervision/https://doi.org/10.1117/12.2549033.full
Raju A, Cheng C-T, Huo Y, Cai J, Huang J, Xiao J, et al. Co-heterogeneous and adaptive segmentation from multi-source and multi-phase ct imaging data: a study on pathological liver and lesion segmentation. In: Vedaldi A, Bischof H, Brox T, Frahm J-M, editors., et al., Computer vision—ECCV 2020. Cham: Springer International Publishing; 2020. p. 448–65. https://doi.org/10.1007/978-3-030-58592-1_27.
Litjens G, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Gu A, Dao T. Mamba: linear-time sequence modeling with selective state spaces. arXiv:231200752.
Wang Z, Zheng J-Q, Zhang Y, Cui G, Li L. Mamba-UNet: UNet-Like pure visual mamba for medical image segmentation. arXiv:240205079. 2024.
Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z, Ding X. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med Image Anal. 2020;63:101693.
Qu L, Liu S, Liu X, Wang M, Song Z. Towards Label-efficient Automatic Diagnosis and Analysis: A Comprehensive Survey of Advanced Deep Learning-based Weakly-supervised, Semi-supervised and Self-supervised Techniques in Histopathological Image Analysis. arXiv; 2022. http://arxiv.org/abs/2208.08789. Accessed 19 Aug 2022.
Qin Y, et al. Learning tubule-sensitive CNNs for pulmonary airway and artery-vein segmentation in CT. IEEE Trans Med Imaging. 2021;40:1603–17.
Cerrolaza JJ, Picazo ML, Humbert L, Sato Y, Rueckert D, Ballester MA, Linguraru MG. Computational anatomy for multi-organ analysis in medical imaging: a review. Med Image Anal. 2019;56:44–67.
Brown TB, Mann B, Ryder N, Subbiah M, Kaplan J, Dhariwal P, et al. Language models are few-shot learners. arXiv; 2020. http://arxiv.org/abs/2005.14165. Accessed 21 May 2023.
Ma J, He Y, Li F, Han L, You C, Wang B. Segment anything in medical images. Nat Commun. 2023;15:654. https://doi.org/10.48550/arXiv.2304.12306.
Zhao Z et al. CLIP in medical imaging: a comprehensive survey. arXiv:231207353. 2023.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China under grant 82072021. This work was also supported by the medical–industrial integration project of Fudan University under grant XM03211181.
Author information
Authors and Affiliations
Contributions
Xiaoyu Liu and Linhao Qu conceived and wrote the manuscript. Ziyue Xie, Jiayue Zhao, Yonghong Shi, Zhijian Song revising manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, X., Qu, L., Xie, Z. et al. Towards more precise automatic analysis: a systematic review of deep learning-based multi-organ segmentation. BioMed Eng OnLine 23, 52 (2024). https://doi.org/10.1186/s12938-024-01238-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12938-024-01238-8