Abstract
Background
Over a third of the world’s population is at risk of Plasmodium vivax-induced malaria. The unique aspect of the parasite’s biology and interactions with the human host make it harder to control and eliminate the disease. Glucose-6-phosphate dehydrogenase (G6PD) deficiency and Duffy-negative blood groups are two red blood cell (RBC) variations that can confer protection against malaria.
Methods
Molecular genotyping of G6PD and Duffy variants was performed in 225 unrelated patients (97 with uncomplicated and 128 with severe vivax malaria) recruited at a Reference Centre for Infectious Diseases in Manaus. G6PD and Duffy variants characterizations were performed using Real Time PCR (qPCR) and PCR–RFLP, respectively.
Results
The Duffy blood group system showed a phenotypic distribution Fy(a + b−) of 70 (31.1%), Fy(a + b +) 96 (42.7%), Fy(a−b +) 56 (24.9%) and Fy(a−b−) 1 (0.44%.) The genotype FY*A/FY*B was predominant in both uncomplicated (45.3%) and severe malaria (39.2%). Only one Duffy phenotype Fy(a-b) was found and this involved uncomplicated vivax malaria. The G6PD c.202G > A variant was found in 11 (4.88%) females and 18 (8.0%) males, while c.376A > G was found in 20 females (8.88%) and 23 (10.22%) male patients. When combined GATA mutated and c.202G > A and c.376A > G mutated, was observed at a lower frequency in uncomplicated (3.7%) in comparison to severe malaria (37.9%). The phenotype Fy(a−b +) (p = 0.022) with FY*B/FY*B (p = 0.015) genotype correlated with higher parasitaemia.
Conclusions
A high prevalence of G6PD c202G > A and c.376A > G and Duffy variants is observed in Manaus, an endemic area for vivax malaria. In addition, this study reports for the first time the Duffy null phenotype Fy(a-b-) in the population of the Amazonas state. Moreover, it is understood that the relationship between G6PD and Duffy variants can modify clinical symptoms in malaria caused by P. vivax and this deserves to be further investigated and explored among this population.
Similar content being viewed by others
Background
Malaria is one of the most serious public health problems worldwide, and in the tropical and subtropical regions, this parasitic disease is the leading cause of social and economic problems [1, 2]. In 2020, 140,974 cases of malaria were recorded in Brazil, with a total number of Plasmodium vivax cases of 118,651 (84.2% of the total cases) and Plasmodium falciparum of 22,182 (15.8%) [3].
Susceptibility or resistance to malarial infection depends on the host-parasite interactions, the Plasmodium sp., the parasite load, the host genetics factor, the immunological status of the host [4, 5]. The clinical manifestation of malaria may include severe anaemia, coagulation disorders, prominent thrombocytopenia, and numerical or functional alterations in leukocytes with spleen involvement [6]. In regions where P. vivax infections are endemic, clinical complications and mortality are reported and this has led to the characterization of P. vivax malaria as a serious or even fatal disease [7, 8].
Although little is known of the pathophysiology, the progression and aggravation of P. vivax malaria is mainly associated with anaemia, which is occasionally due to severe haemolysis [9,10,11]. Other pathophysiological events, such as oxidative stress, may influence the development of clinical conditions [12, 13].
The Duffy Antigen Receptor for Chemokines (DARC), recently renamed Atypical Chemokine Receptor 1 (ACKR1), acts as a possible facilitator in the process of erythrocyte invasion by P. vivax, and blood system antigens act as receptors for P. vivax merozoite ligands that contain Duffy-Binding-Like (DBL) domains through alleles. These Duffy blood group system antigens (Fya, Fyb, Fy3, Fy5, and Fy6) are encoded by two co-dominant allelic forms FY*A and FY*B that differ by the single nucleotide polymorphism (SNP) in position c.125G > A of the exon 2 [14]. The SNP located at position c.-67 T > C in the GATA promoter region is characterized by the allele FY*BN.01 that silences Fyb expression in erythroid cells, and homozygous individuals (FY*BN.01 / FY*BN.01) bear the Fy(a−b−) phenotype that is known to be a protective factor against vivax malaria infections [15].
Another phenotype associated with weak Fyb antigen expression is determined by SNP c.265C > T and c.298G > A. These polymorphisms occur within the first intracellular loop of the Duffy protein, which result in reduced expression of the Fyb antigen. The frequency of Fyb weak, characterized as FY*BW.01 is approximately 2% in Caucasians [16]. Some polymorphisms resulting in the Fya weak (FY*AW.01) allele are yet to be observed in inhabitants of the Amazonas state, Brazil [17, 18].
Glucose 6-phosphate dehydrogenase (G6PD) is an enzyme involved in the pentose monophosphate pathway. Deficiency of this enzyme leads to free radical-mediated oxidative damage to erythrocytes, thus causing haemolysis. G6PD deficiency, linked to the inheritance of X chromosome(s) with disease causing variant(s), is mostly prevalent in people of African, Asian, and Mediterranean descent [19, 20]. In females, there are selective advantages with G6PD A-, which is characterized by the combination of variants A376G (c.376A > G) with variants G202A (c.202G > A), A542T, G680T or T968C [21, 22]. It is suggested that the heterozygous state offers women a selective advantage against severe malaria [23,24,25], and it is known that G6PD deficiency has a prevalence of 8% in malaria-endemic regions [26].
The clinical severity between malaria endemic areas can be complex with multifactorial influence and genetic factors [27]. In endemic areas of P. vivax malaria, comorbidities such as inherited haemoglobinopathies and G6PD deficiency require investigation since they may also be factors that affect heterogeneity in clinical manifestations, and may be characterized by subtypes or endophenotypes [28].
Vivax malaria is a challenge for malaria control strategies and its elimination [29]. The unique parasite biology that involves the formation and subsequent reactivation of latent forms in the liver, and the ability of P. vivax to infect the vector before symptoms occur, favours the perpetuation of the parasitic life cycle and, due to subpatent infections, this causes difficulty in tracking infected individuals [30]. Many studies show the G6PD c.202G > A and c.376A > G variants and Duffy-negative blood group are two RBC variants that confer protection against malaria [19, 31, 32]. Many authors postulate that this is a result of the increased sensitivity of P. vivax to oxidative stress in G6PD deficiency, with negative influence on the parasite, as well as the association of different phenotypes of the Duffy blood group in the invasion of RBCs [33,34,35]. Furthermore, the mechanism of action between G6PD and Duffy group is not fully known. Based on these facts, this study aimed to determine the prevalence of Duffy alleles and G6PD c.202G > A and c.376A > G variants in uncomplicated and severe malaria patients, in order to answer if these erythrocyte variants deserve to be better investigated with clinical signs, susceptibility, protection and parasitaemia against of P. vivax infections.
Methods
This is a retrospective study that is based on a convenience sample for the period between March 2013 and April 2016, which was obtained from the Tropical Medicine Foundation Dr. Heitor Vieira Dourado (FMT-HDV), a reference center for infectious diseases in Manaus, capital of the Amazonas state, Brazil. Blood samples were collected from all the participants. Inclusion criteria were patients over 18 years of age, of either sex and of any skin colour with severe (hospitalized) or uncomplicated (outpatient) malaria, and without any associated diseases. All patients were treated at the clinical research ward at the hospital. All the patients included in the study are unrelated individuals, non-smokers and non-diabetic. Patients with comorbidities, haemoglobinopathies, mixed Plasmodium infections, viral infections and pregnant women were excluded. Patients were classified as either uncomplicated malaria or severe malaria following the World Health Organization (WHO) recommendations (Additional file 1: Table S1) [36, 37].
Approximately 2.5 mL of peripheral blood were collected in tube with EDTA (ethylenediaminetetraacetic acid disodium salt) at a concentration of 1.5 mg/mL for blood counts. Aliquots of 0.3 mL blood in 1.5 mL tubes were kept for extraction of nuclear DNA. An additional 2.5 mL were collected in a tube without anticoagulants for biochemical analyses. Immediately after blood collection, haematological determinations were performed using an automated counter (ABX Pentra 80, Horiba Diagnostics, Montpellier, France) and biochemistry was performed on a Beckman Coulter (Inc, CA, USA).
The diagnosis of the malaria infection followed the standardized protocol established by FMT-HVD, which is a nationally recognized centre for the diagnosis of malaria. In summary, the malaria diagnosis was performed under light microscopy with multiple readings for the parasitaemia. Identification of Plasmodium species (Plasmodium vivax, Plasmodium falciparum and Plasmodium ovale) was done by polymerase chain reaction (PCR) and Real Time PCR (qPCR) methods [38]. The determination of parasitaemia was based on the count of asexual parasites per 200 or maximum 500 white blood cells. The total number of leukocytes of each patient was used for the determination of the parasite density using the following formula [18]:
DNA analysis
DNA was extracted from 200 µL of whole blood according to the QIAamp DNA Mini Kit (Qiagen, Hilden, DE) manufacturer's protocol (Cat No./ID 51,304). After extraction, the DNA was quantified using a spectrophotometer (NanoDrop™ 2000, Thermo Fisher Scientific, Massachusetts, USA), and then stored at − 20 ºC.
Duffy genotyping by PCR–RFLP
Duffy genotyping was performed using conventional PCR for amplification of the sequence of interest, followed by restriction enzyme digestion; one for Duffy blood group (Duffy PCR) and one for the GATA box variant (GATA PCR). The Fy(a − b −) phenotype is known as “erythrocyte silent” and arises from homozygosity FY*BN.01 allele from c.1-67 T > C mutation [15]. This mutation impairs promoter activity in erythroid cells by disrupting a binding site for the GATA-1 erythroid transcription factor, stopping Fyb antigen expression in red blood cells [39, 40].
Genotyping for Duffy blood system groups was performed with synthetic oligonucleotides FYAB1 (5 'TCC CCC TCA ACT GAG AAC TC 3') and FAB2 (5 'AAG GCT GAG CCA TAC CAG AC 3'). Amplified products were visualized on a 1.5% agarose gel stained with ethidium bromide. After PCR amplification was confirmed, the products were processed using BanI restriction enzyme digestion and incubated for at least 4 h at 37 ºC. The enzyme digestion product was electrophoresed on a 1.5% agarose gel stained with ethidium bromide for discrimination of the alleles.
Duffy genotypes, as well as phenotype characterizations, were named according to the FY (ISBT 008) Blood Group Alleles (http://www.isbtweb.org/fileadmin/userupload/Workingparties/WPonRedCellImmunogeneticsand/008FYAllelesv4.1.pdf) [41].
For verification of c.-67 T > C SNP in the GATA box promoter region, FY*A/FY*B and FY*B/FY*B genotype samples were used. Synthetic oligonucleotides FYN1 (5 'CAA GGC TGA CCC TA 3') and FYN2 (5 'CAT GGC ACC GTT TGG TTC AG 3') were used for the GATA PCR.
The GATA PCR product was treated with an StyI restriction enzyme and incubated for at least 4 h at 37 °C. The enzyme digestion product was observed on a 2.5% agarose gel. GATA normal genotypes appeared as 108 and 81 bp bands, while GATA mutated showed an additional 61 bp band. Samples with genotypes FY*A/FY*A and FY*A/FY*B were used to verify the SNPs c.265C > T and c.298G > A in the FY*AW.02 coding region and for Fyx.
The PCR product was treated with the restriction enzyme MspAI in order to verify the SNP c.265C > T, and was then incubated for 4 h at 60 °C. For the SNP c.298G > A, the Mwol restriction enzyme was used and then incubated for 4 h at 37 °C. Both were discriminated on an 8% polyacrylamide gel. The mutated genotypes for the SNP c.265C > T had an additional band of 161 bp, and c.298G > A had an additional band of 343 bp.
G6PD genotyping
For characterization of the variants, Real Time PCR (qPCR) was performed (QuantStudio™ 3 Real-Time PCR System, Applied Biosystems, Thermo Fisher Scientific®) using TaqMan® probes that were specific for each polymorphism. The amplification reaction was performed for a final volume of 12 uL/reaction, which contained 5 μL of 2 × TaqMan Universal Master Mix, 0.3 μL of 20 × SNP genotyping assay, 4.8 μL of sterile water and 2.0 μL of DNA (~ 100 ng) of the sample. The G6PD variants analysed in this project were chosen based on globally observed frequencies and their clinical importance according to WHO classification. The inclusion of the G6PD variants c.202G > A and c.376A > G was based on several studies that showed that both account for more than 95% of all mutations demonstrated in the Brazilian population, as well as in the population of the Amazonas state. For the qPCR technique, the (A-) V68M (202G > A) (rs1050828) and (A +) N126D (376A > G) (rs1050829) probes were used. To confirm the mutations found, amplification by PCR of the relevant DNA segments was performed and was followed by DNA sequencing (ABI 3100, Applied Biosystems, Foster City, CA, USA) [42,43,44].
Statistical analysis
Data were entered into a database using Graphpad-Prism 5.0 software (Graphpad Software, San Diego, CA, USA) and IBM SPSS Statistics, version 19 (IBM Corp., Armonk, NY, USA), and organized by variable type. The analysis of qualitative or categorical variables of three or more groups was performed using the non-parametric Chi-square test (χ2) test using Yates' continuity correction. For the analysis categorical or dichotomous data was used Fisher's exact test when the number of subjects per cell was less than 5. All the statistical tests were performed using a significance level of 5%. The Hardy–Weinberg equilibrium (HWE) was evaluated by comparing expected genotypic frequencies with the observed ones using a Chi-square test.
Results
Clinical and laboratory data
A total of 225 patients diagnosed with P. vivax malaria were included in the study. Ninety-seven patients (43.1%) had uncomplicated malaria and 128 (56,9%) had severe malaria. Table 1 showed the epidemiological and physical parameters for uncomplicated and severe P. vivax malaria stratified by gender. Most of the severe malaria male cases had pallor (82.5%), choluria (77.8), headache (76.2%), loss of appetite (74.6%) and hepatosplenomegaly (71.4%). Among the female patients, headache (92.9%), loss of appetite (85.7%), pallor (83.3%), vomiting (83.3%) and choluria (73.8%) were the common symptoms.
Patients with severe malaria presented a significant decrease in RBCs (female 3.51 and 3.48 male × 106 uL), haemoglobin (female 9.72 and male 10.06 g/dL) and haematocrit (female 30.45 and male 30.22%). High bilirubin levels were also observed (12.5, 10.36, female 12.5 and male 10.36) (ANOVA—p < 0.001). Hypoglycaemia (< 70 mg/dL), a clinical and laboratory features of severe malaria, was detected in 16% of patients with severe malaria (Table 2).
Duffy genotyping
Analysis of the Duffy system (Table 3) showed a phenotypic distribution of Fy(a + b-) of 31.1% (70/225), Fy(a + b +) 42.70% (96/225), Fy(a-b +) 24. 89% (56/225) and Fy(a-b-) 0.44% (1/225). The genotype FY*A/FY*B was predominant in both uncomplicated (45.3%) and severe malaria (39.2%) (Fig. 1). The phenotypic frequency distribution of Duffy genotypes among uncomplicated and severe vivax malaria by gender is shown in Additional file 2: Table S2.
G6PD genotyping
Twenty-nine patients were carriers of the G6PD c.202G > A variant; 18 (8.0%) males were hemizygous, 10 (4.44%) and 1 (0.44%) female was heterozygous and homozygous, respectively. The c.376A > G was identified in 43 (19.11%) patients. Eighteen women (8.0%) were heterozygous and 2 (0.88%) were homozygous. Twenty-three (10.22%) males were hemizygous. Both SNPs were in HWE as calculated from the frequencies of the alleles among the females, since the G6PD gene is located on the X chromosome.
Duffy and G6PD genotypes with analysis of clinical characteristics
The FY*BN.01/FY*BN.01, responsible for the Fy(a−b−) phenotype, was observed in one patient (female) with uncomplicated malaria who was not a carrier of the G6PD variants. Among patients with uncomplicated malaria, the frequency of c.202G > A and c.376A > G variants was low, particularly when these polymorphisms were concomitant. Severe malaria patients showed no variations in frequency for the c.202G > A variant. However, in the presence of A376G, there was a slight decrease in frequency.
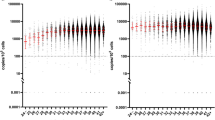
When combined with the GATA variant, the c.202G > A and c.376A > G variants were observed at a lower frequency in uncomplicated (3.7%) malaria in comparison to severe malaria (37.9%) (Table 4). The levels of gametocytes and mean parasitaemia (± standard deviation), considering the number of parasites found per mm3, is shown in Figs. 2 and 3. The minimum and the maximum parasitaemia were 18/µL–101,904/µL among uncomplicated malaria and 29/µL–102,358/µL in severe malaria. Statistical significance (p = 0.022) was observed for parasitaemia. Higher parasitaemia was observed to phenotype Fy(a−b +) (p = 0.022) and genotypes FY*A/ FY*A and FY*B/ FY*B, with lower parasitaemia in the presence of the FY*A/FY*BN.01 (p = 0.015).
Discussion
Death due to malaria, regardless of the Plasmodium species, is not related to gender, though children and pregnant women are more susceptible to malaria. However, socio-economic and cultural factors play a central factor in determining differences in gender vulnerability to malaria [45,46,47].
Six (19.35%) female patients and sixteen (39.02%) male patients had both G6PD c.202G > A/c.376A > G mutations concomitantly c.202G > A and c.376A > G are responsible for the majority of the observed prevalence of G6PD deficiency in Brazil. The occurrence of these concomitant mutations, which can be in heterozygous or homozygous states in females and hemizygous in males, deserves attention in genetic association studies in order to investigate possible clinical phenotypes of this deficiency among patients receiving treatment with malaria medication [48,49,50].
The higher the parasitaemia in the host, the higher the consumption of glucose that may lead to hypoglycaemia, mainly in severe malaria. Hypoglycaemia may provoke dizziness, palpitations, tremors and loss of consciousness [51]. Careful glucose monitoring should be targeted in these complications, especially in those patients with G6PD deficiency. However, none of the G6PD alleles correlated with low levels of glucose among the severe malaria patients [52].
In this study, FY*A/FY*B was the most frequent genotype in both malaria patients, uncomplicated (45.3%) and severe (39.2%). These findings corroborate the study reported by Cavasini et al. [53] which correlated the high frequency of the FY*A and FY*B alleles among P. vivax malaria patients. The FY*A/FY*B and FY*A/FY*A genotypes were associated with a high frequency of P. vivax infection, thus suggesting that these individuals have a higher risk of developing malaria disease. The FY*A/FY*A and FY*A/FY*B genotypes are associated with increased frequency of P. vivax infection, while FY*A/FY*BW.01 and FY*B/FY*BW.01 were associated with low parasitic density levels [18].
The presence of a single case with null Duffy phenotype Fy(a−b−) is first described in the population of the Amazonas state. In endemic areas, the Duffy negative blood group is reported as a protective factor against P. vivax malaria infection [54]. In a study conducted in São Paulo, the phenotypic frequencies found in blood donors for Duffy blood system antigens were 19.8% for the phenotype Fy(a + b−) in Caucasians and 14.0% in African-Brazilians, Fy(a + b +) in 41.4% of Caucasians and 1.6% of African-Brazilians, Fy(a−b +) in 37.8% of Caucasians and 17.5% of African-Brazilians and Fy(a−b−) in 1.1% of Caucasians and 66.9% of African-Brazilians [55]. These results indicate that the Manaus/Amazonas region has individuals who express three Duffy phenotypes Fy(a + b +), Fy(a + b−) and Fy(a−b +) more frequently, with expression of FY*A and FY*B antigens.
For the process of parasite invasion into RBCs, Duffy protein is functionally important. In regions of high malaria transmission rates, as in the inhabitants of the Amazon, Duffy protein is naturally immunogenic. Woolley et al. [56] demonstrated (in vitro) that the expression level of the Fy6 epitopes was significantly lower in reticulocytes and RBCs from individual carriers of the FY*B/FY*B genotypes compared with individuals of the FY*A/FY*A or FY*A/FY*B genotypes. Another similar study showed that the presence of the FY*BN.01 allele resulted in a 50% reduction of that protein when invaded by P. vivax [57].
In a study with P. vivax malaria patients, the FY*A and FY*B alleles were found to have low, medium, and high parasitic density [18]. However, in the presence of the GATA variant, genotypes with FY*BN.01 and FY*BW.01 alleles were found only in patients with low parasitic density and low symptomatology.
A study performed in the state of Pará-Brazil, with a population of African descent demonstrated that the presence of the c.202G > A variant was 0.060, the Duffy blood group included 24.3% Duffy negative and 41.3% of the individuals were heterozygous for FY*BES (ES: erythrocyte silent). The frequency of the FY*BES allele was 41.0% [44]. These findings support the monitoring of individuals with G6PD deficiency for use of primaquine during the routine care of afro-descendant communities of the Trombetas, Erepecuru, and Cumná Rivers, in order to assess the risk of haemolytic crisis in recurrent cases of malaria in the region.
Several studies have revealed that the complexity of phenotypic and genotypic variation of the Duffy system and the variants of G6PD (A-) is geographically variable across human populations in areas in which malaria is endemic [26, 58,59,60,61]. The Duffy system and G6PD are polymorphic systems that offer great challenges to researchers not only due to their academic importance, but also due to their potential applications to treatment of vivax malaria [62]. Whenever natural selection occurs within a population in an area of endemicity for malaria, natural adaptations may result from genetic variation that provides a partial defense mechanism against P. vivax infections [56].
One of the major confounders and a limitation in this study is the inherent disadvantage of G6PD deficiency due to the associated haemolysis, which may be one of the factors that account for the absence of consensus on the G6PD-malaria protection hypothesis. In some cases, this can potentially protect against uncomplicated malaria, but not severe malaria [63, 64]. Despite most G6PD-deficient persons being asymptomatic, haemolytic anaemia, a main clinical sign, can occur 1–3 days after exposure, after eating fava beans, or can be triggered by infections or by certain drugs, such as those used to treat malaria [65, 66]. In many cases, this acute haemolytic anaemia is usually self-limiting, thus, resolving on its own [67]. In addition, the authors acknowledge that an important limitation of this study is the small sample size and they understand that additional studies of larger samples will be required to confirm the results.
Conclusion
A high prevalence of G6PD c202G > A and c.376A > G and Duffy variants is observed in Manaus, Amazonas state, Brazil, principally in severe vivax malaria patients. This presents a new viewpoint in the protection or not in vivax malaria. In addition, this study reports for the first time the Duffy null phenotype Fy(a−b−) in the population of the Amazonas state. Moreover, it is understood that the relationship between G6PD and Duffy variants can modify clinical symptoms in malaria caused by P. vivax and this deserves to be further investigated and explored among this population.
Availability of data and materials
The full data used to support the findings of this study, including, consent terms, electronic files, as well as the lab techniques and materials used may be released upon reasonable request to my Institutional email address: jpmn@ufam.edu.br.
References
Greenwood BM, Fidock DA, Kyle DE, Kappe SH, Alonso PL, Collins FH, Duffy PE. Malaria: progress, perils, and prospects for eradication. J Clin Invest. 2008;118:1266–76.
WHO. World Malaria Report. Geneva, World Health Organization, 2015.
Ministério da Saúde. Secretaria de Vigilância em Saúde. (Internet) www.saude.gov.br/svs Versão 1, 23 de abril de 2021.
Akinosoglou KS, Solomou EE, Gogos CA. Malaria: a haematological disease. Hematology. 2012;17:106–14.
Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, et al. Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am J Trop Med Hyg. 2009;80:194–8.
Oliveira-Ferreira J, Lacerda MV, Brasil P, Ladislau JL, Tauil PL, Daniel-Ribeiro CT. Malaria in Brazil: an overview. Malar J. 2010;9:115.
Lacerda MV, Mourão MP, Alexandre MA, Siqueira AM, Magalhães BM, Martinez-Espinosa FE, et al. Understanding the clinical spectrum of complicated Plasmodium vivax malaria: a systematic review on the contributions of the Brazilian literature. Malar J. 2012;11:12.
Baird JK. Evidence and implications of mortality associated with acute Plasmodium vivax malaria. Clin Microbiol Rev. 2013;26:36–57.
Alexandre MA, Ferreira CO, Siqueira AM, Magalhães BL, Mourão MP, Lacerda MV, et al. Severe Plasmodium vivax malaria, Brazilian Amazon. Emerg Infect Dis. 2010;16:1611–4.
Quintero JP, Siqueira AM, Tobón A, Blair S, Moreno A, Arévalo-Herrera M, et al. Malaria-related anaemia: a Latin American perspective. Mem Inst Oswaldo Cruz. 2011;106:91–104.
Douglas NM, Anstey NM, Buffet PA, Poespoprodjo JR, Yeo TW, White NJ, et al. The anaemia of Plasmodium vivax malaria. Malar J. 2012;11:135.
Carvalho BO, Lopes SC, Nogueira PA, Orlandi PP, Bargieri DY, Blanco YC, et al. On the cytoadhesion of Plasmodium vivax-infected erythrocytes. J Infect Dis. 2010;15(202):638–47.
Yeo TW, Lampah DA, Tjitra E, Piera K, Gitawati R, Kenangalem E, et al. Greater endothelial activation, Weibel-Palade body release and host inflammatory response to Plasmodium vivax, compared with Plasmodium falciparum: a prospective study in Papua. Indonesia J Infect Dis. 2010;202:109–12.
Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–4.
Tournamille C, Colin Y, Cartron JP, Le Van KC. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8.
Cruz BR, Chiba AK, Moritz E, Bordin JO. RHD alleles in Brazilian blood donors with weak D or D-negative phenotypes. Transfus Med. 2012;22:84–9.
Albuquerque SR, Cavalcante Fde O, Sanguino EC, Tezza L, Chacon F, Castilho L, et al. FY polymorphisms and vivax malaria in inhabitants of Amazonas State. Brazil Parasitol Res. 2010;106:1049–53.
Abou-Ali RK, Dhyani A, Terço AL, Toro DM, Gomes KS, Tezza LC, et al. Impact of Duffy polymorphisms on parasite density in Brazilian Amazonian patients infected by Plasmodium vivax. Malar J. 2019;18:289.
Nkhoma ET, Poole C, Vannappagari V, Hall SA, Beutler E. The global prevalence of glucose-6-phosphate dehydrogenase deficiency: a systematic review and meta-analysis. Blood Cells Mol Dis. 2009;42:267–78.
Luzzatto L. G6PD deficiency and malaria selection. Heredity. 2012;108:456.
Beutler E, Kuhl W, Vives-Corrons JL, Prchal JT. Molecular heterogeneity of glucose-6-phosphate dehydrogenase A-. Blood. 1989;74:2550–5.
De Araujo C, Migot-Nabias F, Guitard J, Pelleau S, Vulliamy T, Ducrocq R. The role of the G6PD AEth376G/968C allele in glucose-6-phosphate dehydrogenase deficiency in the seerer population of Senegal. Haematologica. 2006;91:262–3.
Sirugo G, Predazzi IM, Bartlett J, Tacconelli A, Walther M, Williams SM. G6PD A- deficiency and severe malaria in The Gambia: heterozygote advantage and possible homozygote disadvantage. Am J Trop Med Hyg. 2014;90:856–9.
Manjurano A, Sepulveda N, Nadjm B, Mtove G, Wangai H, Maxwell C, et al. African glucose-6-phosphate dehydrogenase alleles associated with protection from severe malaria in heterozygous females in Tanzania. PLoS Genet. 2015;11: e1004960.
Nafa K, Reghis A, Osmani N, Baghli L, Aït-Abbes H, Benabadji M, et al. At least five polymorphic mutants account for the prevalence of glucose-6-phosphate dehydrogenase deficiency in Algeria. Hum Genet. 1994;94:513–7.
Howes RE, Piel FB, Patil AP, Nyangiri OA, Gething PW, Dewi M, et al. G6PD deficiency prevalence and estimates of affected populations in malaria endemic countries: a geostatistical model-based map. PLoS Med. 2012;9: e1001339.
Driss A, Hibbert JM, Wilson NO, Iqbal SA, Adamkiewicz TV, Stiles JK. Genetic polymorphisms linked to susceptibility to malaria. Malar J. 2011;10:271.
Ramos Júnior WM, Sardinha JF, Costa MR, Santana MS, Alecrim MG, Lacerda MV. Clinical aspects of hemolysis in patients with P. vivax malaria treated with primaquine, in the Brazilian Amazon. Braz J Infect Dis. 2010;14:410–2.
Hedrick PW. Population genetics of malaria resistance in humans. Heredity (Edinb). 2011;107:283–304.
Adams JH, Mueller I. The biology of Plasmodium vivax. Cold Spring Harb Perspect Med. 2017;7: a025585.
Ruwende C, Khoo SC, Snow RW, Yates SN, Kwiatkowski D, Gupta S, et al. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature. 1995;376:246–9.
Leslie T, Briceño M, Mayan I, Mohammed N, Klinkenberg E, Sibley CH, et al. The impact of phenotypic and genotypic G6PD deficiency on risk of Plasmodium vivax infection: a case-control study amongst Afghan refugees in Pakistan. PLoS Med. 2010;7: e1000283.
Wajcman H, Galactéros F. Le déficit en glucose-6 phosphate déshydrogénase: protection contre le paludisme et risque d’accidents hémolytiques [Glucose 6-phosphate dehydrogenase deficiency: a protection against malaria and a risk for hemolytic accidents]. C R Biol. 2004;327:711–20.
Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74.
Adapa SR, Taylor RA, Wang C, Thomson-Luque R, Johnson LR, Jiang RHY. Plasmodium vivax readiness to transmit: implication for malaria eradication. BMC Syst Biol. 2019;13:5.
WHO. Severe malaria. Trop Med Int Health. 2014;19;7–131.
Malaria – drug therapy. 2.Malaria – diagnosis. 3.Antimalarials – administration and dosage. 4. Drug Therapy, Combination. 5.Guideline. Third Edition: WHO, 2015.
Almeida ACG, Kuehn A, Castro AJM, Vitor-Silva S, Figueiredo EFG, Brasil LW, et al. High proportions of asymptomatic and submicroscopic Plasmodium vivax infections in a peri-urban area of low transmission in the Brazilian Amazon. Parasit Vectors. 2018;11:194.
Chaudhuri A, Polyakova J, Zbrzezna V, Pogo AO. The coding sequence of Duffy blood group gene in humans and simians: restriction fragment length polymorphism, antibody and malarial parasite specificities, and expression in non erythroid tissues in Duffy-negative individuals. Blood. 1995;85:615–21.
Peiper SC, Wang ZX, Neote K, Martin AW, Showell HJ, Conklyn MJ, et al. The Duffy antigen/receptor for chemokines (DARC) is expressed in endothelial cells of Duffy negative individuals who lack the erythrocyte receptor. J Exp Med. 1995;181:1311–7.
ISBT http://www.isbtweb.org/fileadmin/user_upload/Working_parties/WP_on_Red_Cell_Immunogenetics_and/008_FY_Alleles_v4.1.pdf. Accessed 2021.
Moura Neto, JP, Dourado MV, Reis MG, Gonçalves MS. A novel c.197T -> A variant among Brazilian neonates with glucose-6-phosphate dehydrogenase deficiency. Genet Mol Biol. 2088;31:33–35.
Ferreira NS, Anselmo FC, Albuquerque SRL, Sanguino ECB, Fraiji NA, Marinho GB, et al. G6PD deficiency in blood donors of Manaus, Amazon Region, northern Brazil. Int J Lab Hematol. 2021;43:e290–3.
Oliveira HS, Silva A, Andrade GB, Gaia KC, Costa GL, Santos A, Guerreiro JF. Molecular genotyping of G6PD mutations and Duffy blood group in Afro-descendant communities from Brazilian Amazon. Genet Mol Biol. 2018;41:758–65.
Domingo GJ, Advani N, Satyagraha AW, Sibley CH, Rowley E, Kalnoky M, et al. Addressing the gender-knowledge gap in glucose-6-phosphate dehydrogenase deficiency: challenges and opportunities. Int Health. 2019;11:7–14.
WHO. Gender, Health and Malaria. Geneva, World Health Organization, 2007.
King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, et al. Fy(a)/Fy(b) antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci USA. 2011;108:20113–8.
Hirono A, Kawate K, Honda A, Fujii H, Miwa S. A single mutation 202G>A in the human glucose-6-phosphate dehydrogenase gene (G6PD) can cause acute hemolysis by itself. Blood. 2002;15(99):1498.
Fanello CI, Karema C, Avellino P, Bancone G, Uwimana A, Lee SJ, et al. High risk of severe anaemia after chlorproguanil-dapsone+artesunate antimalarial treatment in patients with G6PD (A-) deficiency. PLoS ONE. 2008;3: e4031.
Premji Z, Umeh RE, Owusu-Agyei S, Esamai F, Ezedinachi EU, Oguche S, et al. Chlorproguanil-dapsone-artesunate versus artemether-lumefantrine: a randomized, double-blind phase III trial in African children and adolescents with uncomplicated Plasmodium falciparum malaria. PLoS ONE. 2009;4: e6682.
Tovar-Acero C, Velasco MC, Avilés-Vergara PA, Ricardo-Caldera DM, Alvis EM, Ramirez-Montoya J, et al. Liver and kidney dysfunction, hypoglycemia, and thrombocytopenia in Plasmodium vivax malaria patients at a Colombian Northwest region. Parasite Epidemiol Control. 2021;13: e00203.
Siqueira AM, Lacerda MV, Magalhães BM, Mourão MP, Melo GC, Alexandre MA, et al. Characterization of Plasmodium vivax-associated admissions to reference hospitals in Brazil and India. BMC Med. 2015;13:57.
Cavasini CE, Mattos LC, Couto AA, Bonini-Domingos CR, Valencia SH, Neiras WC, et al. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception? Trans R Soc Trop Med Hyg. 2007;101:1042–4.
Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266.
Novaretti MC, Dorlhiac-Llacer PE, Chamone DA. Estudo de grupos sanguíneos em doadores de sangue caucasóides e negróides na cidade de São Paulo. Rev Bras Hematol Hemoter. 2000;22:1.
Woolley IJ, Hotmire KA, Sramkoski RM, Zimmerman PA, Kazura JW. Differential expression of the duffy antigen receptor for chemokines according to RBC age and FY genotype. Transfusion. 2000;40:949–53.
Ceravolo IP, Sanchez BA, Sousa TN, Guerra BM, Soares IS, Braga EM, et al. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin Exp Immunol. 2009;156:502–10.
Chamchod F, Beier JC. Modeling Plasmodium vivax: relapses, treatment, seasonality, and G6PD deficiency. J Theor Biol. 2013;316:25–34.
Kano FS, de Souza AM, Torres LM, Costa MA, Souza-Silva FA, Sanchez BAM, et al. Susceptibility to Plasmodium vivax malaria associated with DARC (Duffy antigen) polymorphisms is influenced by the time of exposure to malaria. Sci Rep. 2018;8:13851.
He WQ, Shakri AR, Bhardwaj R, França CT, Stanisic DI, Healer J, et al. Antibody responses to Plasmodium vivax Duffy binding and Erythrocyte binding proteins predict risk of infection and are associated with protection from clinical malaria. PLoS Negl Trop Dis. 2019;13: e0006987.
DePina AJ, Pires CM, Andrade AJB, Dia AK, Moreira AL, Ferreira MCM, et al. The prevalence of glucose-6-phosphate dehydrogenase deficiency in the Cape Verdean population in the context of malaria elimination. PLoS ONE. 2020;15: e0229574.
Tran TM, Oliveira-Ferreira J, Moreno A, Santos F, Yazdani SS, Chitnis CE, et al. Comparison of IgG reactivities to Plasmodium vivax merozoite invasion antigens in a Brazilian Amazon population. Am J Trop Med Hyg. 2005;73:244–55.
Uyoga S, Ndila CM, Macharia AW, Nyutu G, Shah S, Peshu N, et al. Glucose-6-phosphate dehydrogenase deficiency and the risk of malaria and other diseases in children in Kenya: a case-control and a cohort study. Lancet Haematol. 2015;2:e437–44.
Mbanefo EC, Ahmed AM, Titouna A, Elmaraezy A, Trang NT, Phuoc Long N, et al. Association of glucose-6-phosphate dehydrogenase deficiency and malaria: a systematic review and meta-analysis. Sci Rep. 2017;7:45963.
Luzzatto L, Ally M, Notaro R. Glucose-6-phosphate dehydrogenase deficiency. Blood. 2020;136:1225–40.
Howes RE, Battle KE, Satyagraha AW, Baird JK, Hay SI. G6PD deficiency: global distribution, genetic variants and primaquine therapy. Adv Parasitol. 2013;81:133–201.
Youngster I, Arcavi L, Schechmaster R, Akayzen Y, Popliski H, Shimonov J, et al. Medications and glucose-6-phosphate dehydrogenase deficiency: an evidence-based review. Drug Saf. 2010;33:713–26.
Acknowledgements
We especially thank to all patients involved in this study and staff of the Molecular Biology Laboratory of the Federal University of Amazonas (UFAM) and the Tropical Medicine Foundation (FMT-HD) for the medical and technical support. Finally, the authors thank the Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support.
Funding
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM). Protocol Number: 1094/2013-FAPEAM; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
NSF: Data curation (lead); Formal analysis (lead); Methodology (lead); Software (equal). JLSM: Data curation (lead); Formal analysis (lead); Methodology (lead); Software (equal). SRLA: Validation (equal); Visualization (equal). ACGA: Resources (equal); Visualization (equal). ACD: Resources (equal); Visualization (equal). FCA: Resources (equal); Visualization (equal). ESL: Validation (equal); Visualization (equal). MVGL: Data curation (lead); Visualization (equal). PAN: Formal analysis (lead); Methodology (equal); Writing (equal). RR: Supervision (equal); Writing (equal). MSG: Supervision (equal); Writing (equal). JPMN: Conceptualization (lead); Supervision (lead); Investigation (lead); Project administration (equal); Funding acquisition (equal); Writing- review & editing (lead).
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This research was initiated after approval by the Research Ethics Committee (CEP) at FMT-HVD. To ensure patient welfare, all the patients included in the study signed the informed consent form (ICF), in compliance with CNS Resolutions 196/96.
The samples were from a larger study entitled “Clinical characterization of malaria complicated by Plasmodium vivax”, approved by the National Commission for Ethics in Research (CONEP), in June 2009, opinion No. 343/2009, protocol No. 25.000.011.792/2009–15. This new project was approved by the Research Ethics Committee at FMT-HVD (study number 343/2009) entitled “Study of DUFFY polymorphisms in patients infected with Plasmodium vivax” under approval No. CAAE-0004.0.112.000–11 on 12/29/2011.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
. World Health Organization diagnostic criteria for severe falciparum malaria. Adapted from: Guidelines for the Treatment of Malaria, World Health Organization, 2015.
Additional file 2: Table S2
. Phenotypic frequency distribution of Duffy blood group among uncomplicated and severe vivax malaria according to gender. No significant correlations were demonstrated in the frequency analysis of the Duffy blood group phenotypes between the gender and uncomplicated and severe P. vivax malaria patients.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ferreira, N.S., Mathias, J.L.S., Albuquerque, S.R.L. et al. Duffy blood system and G6PD genetic variants in vivax malaria patients from Manaus, Amazonas, Brazil. Malar J 21, 144 (2022). https://doi.org/10.1186/s12936-022-04165-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-022-04165-y