Abstract
Background
Resistance to anti-malarials is a major threat to the control and elimination of malaria. Sulfadoxine–pyrimethamine (SP) anti-malarial treatment was used as a national policy for treatment of uncomplicated falciparum malaria in Thailand from 1973 to 1990. In order to determine whether withdrawal of this antifolate drug has led to restoration of SP sensitivity, the prevalence of genetic markers of SP resistance was assessed in historical Thai samples.
Methods
Plasmodium falciparum DNA was collected from the Thailand–Myanmar, Thailand–Malaysia and Thailand–Cambodia borders during 2008–2016 (N = 233). Semi-nested PCR and nucleotide sequencing were used to assess mutations in Plasmodium falciparum dihydrofolate reductase (pfdhfr), P. falciparum dihydropteroate synthase (pfdhps). Gene amplification of Plasmodium falcipaurm GTP cyclohydrolase-1 (pfgch1) was assessed by quantitative real-time PCR. The association between pfdhfr/pfdhps mutations and pfgch1 copy numbers were evaluated.
Results
Mutations in pfdhfr/pfdhsp and pfgch1 copy number fluctuated overtime through the study period. Altogether, 14 unique pfdhfr–pdfhps haplotypes collectively containing quadruple to octuple mutations were identified. High variation in pfdhfr–pfdhps haplotypes and a high proportion of pfgch1 multiple copy number (51% (73/146)) were observed on the Thailand–Myanmar border compared to other parts of Thailand. Overall, the prevalence of septuple mutations was observed for pfdhfr–pfdhps haplotypes. In particular, the prevalence of pfdhfr–pfdhps, septuple mutation was observed in the Thailand–Myanmar (50%, 73/146) and Thailand–Cambodia (65%, 26/40) border. In Thailand–Malaysia border, majority of the pfdhfr–pfdhps haplotypes transaction from quadruple (90%, 9/10) to quintuple (65%, 24/37) during 2008–2016. Within the pfdhfr–pfdhps haplotypes, during 2008–2013 the pfdhps A/S436F mutation was observed only in Thailand–Myanmar border (9%, 10/107), while it was not identified later. In general, significant correlation was observed between the prevalence of pfdhfr I164L (ϕ = 0.213, p-value = 0.001) or pfdhps K540E/N (ϕ = 0.399, p-value ≤ 0.001) mutations and pfgch1 gene amplification.
Conclusions
Despite withdrawal of SP as anti-malarial treatment for 17 years, the border regions of Thailand continue to display high prevalence of antifolate and anti-sulfonamide resistance markers in falciparum malaria. Significant association between pfgch1 amplification and pfdhfr (I164L) or pfdhps (K540E) resistance markers were observed, suggesting a compensatory mutation.
Similar content being viewed by others
Background
Both falciparum and vivax malaria remains an important public health problem in border regions of Thailand. Resistance to anti-malarials presents a major hurdle for eradication of the disease [1]. Drug resistance in both Plasmodium falciparum and Plasmodium vivax has been reported as early as the 1950s [2]. Sulfadoxine–pyrimethamine (SP), a folate pathway inhibitor was deployed in Thailand for the treatment of uncomplicated falciparum malaria from 1973 until 1991 [3]. By 1991, substantial loss of the SP drug efficacy prompted a change in first-line treatment in Thailand [3]. Molecular investigations revealed that mutations in P. falciparum dihydrofolate reductase (pfdhfr) and P. falciparum dihydropteroate synthase (pfdhps) were associated with SP treatment failures and could be used as molecular markers for SP resistance [4, 5]. The mutations in pfdhfr and pfdhps often occurred in a step-wise progressive manner resulting in increased levels of drug resistance [4, 6]. Resistance to antifolates has also been linked to gene amplification of P. falciparum guanosine triphosphate cyclohydrolase 1 (pfgch1)-an enzyme responsible for coding a crucial enzyme in the folate pathway [7]. Parasites with pfgch1 amplification were reportedly less susceptible to antifolates as elevated expression of enzymes assisted antifolate resistance by competing with the drugs [7], and compensating the loss of fitness caused by mutations in pfdhfr and pfdhps by increasing the flux of metabolic products in the folate pathway [8]. An earlier study from Thailand reported a high proportion of parasites carrying multiple copies of pfgch1 and suggested an association between pfgch1 copy number variation (CNV) and the pfdhfr (I164L) mutations [8]. Several studies conducted between 1995 and 2008 have identified varying levels of triple or quadruple mutations in pfdhfr and pfdhps [5, 8, 9]. A more recent survey conducted in Ubonratchathani province close the Thailand–Cambodia borders, which had a lot of reports in many anti-malarial drug resistances [2, 3], showed high levels of pfdhfr (N51I, C59R, and S108N, ≥ 76%) and pfdhps (A437G, K540E, A581G or A437G, K540N, A581G or S436A, A437G, K540E, ≥ 90%) triple mutations [10]. These border areas are malaria endemic regions. Each site is geographically distant from other and often experiences high migration of diverse human population. However, data on the current status of antifolate and anti-sulfonamide resistance markers in P. falciparum in other major border regions of Thailand is scarce. Presumably, the persistence of highly mutations on SP-resistant markers related to the using of other drugs that may also induced pressure on pfdhfr and pfdhps of malaria parasite. The trimethoprim–sulfamethoxazole, which is used to treat acute respiratory infections, presented cross-resistance with pyrimethamine and sulfadoxine [11, 12]. Reemergence of chloroquine-sensitive P. falciparum in Malawi after a decade-long cessation of drug use shows that for some anti-malarials restoration of drug sensitivity is possible after removal of the drug pressure [13]. However, several factors including drug target, nature of genes and host/parasite genetic background may differently affect the persistence of SP resistance after removal of SP use.
The present study is a retrospective molecular surveillance of three antifolate and anti-sulfonamide resistance markers in samples obtained from the malaria endemic border provinces of Thailand between 2008 and 2016. It aimed at describing the current status of resistance markers after long-term cessation of SP as an anti-malarial treatment in Thailand.
Methods
Study sites and samples collection
Between 2008 and 2016, dried blood spots samples were collected using Whatman 3MM CHR filter paper from P. falciparum malaria patients in nine provinces bordering Thailand (Maehongson N = 24; Tak N = 16; Kanchanaburi N = 70; Ranong N = 34; Suratthani N = 2; Ubonratchathani N = 9; Sisaket N = 19; Trat N = 12; and Yala N = 47 (Additional file 1: Table S3). All samples were acquired during anti-malarial drug therapeutic efficacy monitoring projects that were approved by the Research Ethics Committee of the Department of Disease Control, Ministry of Public Health, and Faculty of Tropical Medicine, Mahidol University, Thailand. DNA was extracted using QIAamp DNA Mini Kit (QIAGEN, Germany) and stored at 4 °C before analysis.
Pfdhfr and pfdhps sequencing and quality control
The pfdhfr and pfdhps genes were amplified using semi-nested PCR as described previously [14,15,16,17] (Additional file 1: Table S1). Amplified final products were sequenced to assess pfdhfr (A16V, C50R, N51I, C59R, S108N and I164L) and pfdhfr (A/S436F, A437G, K540E/N, A581G and A613S/T) mutations. The initial semi-nested PCR generated products measured 880 bp for pfdhfr and 715 bp for pfdhps. The nested PCR products were purified by using FavorPrep GEL/PCR Purification Kit (FAVORGEN®BIOTECH CORP., Taiwan) and sequenced by Macrogen Inc. (South Korea). The sequencing results were analysed using BioEdit software version 7.2.6. Resistance marker sequences were compared with reference sequences for pfdhfrs (DHFR-TS PF3D7_0417200) and pfdhsp (PPPK-DHPS PF3D7_0810800).
Plasmodium falciparum gch1 copy number estimation
To assess pfgch1 amplification, quantitative real-time PCR (qPCR) of the pfgch1 gene was compared with a single copy reference gene (seryl tRNA ligase) (Accession No: A0A0L1I323). The specific probes and primers used have been described previously [8] (Additional file 1: Table S1). A laboratory pfgch1 single copy strain and pfgch1 multiple copies strain of P. falciparum were used as reference standard. The copy number of pfgch1 gene was analysed from the threshold cycle (Ct) and ΔΔCt values calculated as follows: ΔΔCt = (Ct of pfgch1 − Ct of pfSerRs) of sample − (Ct of pfgch1 − Ct of pfSerRs) of Calibrator. The copy number of pfgch1 were subsequently calculated using 2−ΔΔCt method [18]. The pfgch1 amplification cut off value was set at 1.5 copies number.
Statistical analysis
The pfdhfr/pfdhps mutations and pfgch1 copy numbers were described as the proportion of each haplotype present at each border region. The changing trend of antifolate resistance markers after drug withdrawal was analysed by comparing the prevalence of markers over time by year. Chi-square (χ2) and the non-parametric phi correlation coefficient statistics (ϕ) was used to evaluate the difference in frequency and associations respectively among the group of each antifolate resistance marker. A p-value ≤ 0.05 was considered statistically significant.
Results
Mutations in pfdhfr and pfdhps genes in isolates from the Thailand–Myanmar border
A total of 146 P. falciparum samples collected from malaria clinics in five provinces between 2008 and 2016 were analysed (Table 1). The six point mutations of pfdhfr gene and five point mutations of pfdhps associated with resistance were assessed in all samples. Septuple mutations at N51I, C59R, S108N and I164L of pfdhfr with A437G, K450E and A581G of pfdhps presented as the main haplotype that was found in 51% (18/35), 50% (36/72) and 46% (18/39) from samples between 2008 and 2010, 2011–2013 and 2014–2016, respectively. The secondary group of the main haplotype was the parasite containing sextuple mutations at N51I, C59R, S108N and I164L for pfdhfr with A437G and K450E mutation in pfdhps that showed 29% (10/35), 21% (15/72) and 41% (16/39), respectively. However, the highly resistance falciparum parasites were the Octuple haplotype which consisted of the mutation at N51I, C59R, S108N and I164L for pfdhfr and A437G, K450E, A581G and A/S436F or A613S/T for pfdhps were detected only in 2008–2010 (6%, 2/35) and 2011–2013 (3%, 2/72) (Fig. 1).
Prevalence of SP resistance markers pfdhfr, pfdhps, and pfgch1 in P. falciparum obtained from malaria patients between 2008 and 2016. (TH Thailand, MY Myanmar, CH Cambodia, ML Malaysia, MH Maehongson Province, TK Tak Province, KN Kanchanaburi Province, RN Ranong Province, ST Suratthani Province, YL Yala Province, TR Trat Province, SK Srisaket Province, UB Ubonratchathani Province)
The proportion of P. falciparum samples with pfgch1 gene amplification from the Thailand–Myanmar border increased from 37% (13/35) in year 2008 to 67% (26/39) in year 2016 (Fig. 2).
The pfgch1 copy number variation in P. falciparum obtained from malaria patients between 2008 and 2016. (TH Thailand, MY Myanmar, CH Cambodia, ML Malaysia, MH Maehongson Province, TK Tak Province, KN Kanchanaburi Province, RN Ranong Province, ST Suratthani Province, YL Yala Province, TR Trat Province, SK Srisaket Province, UB Ubonratchathani Province)
Mutations in pfdhfr and pfdhps genes in isolates from the Thailand–Cambodia border
Plasmodium falciparum isolates (N = 40) from malaria clinics in three provinces along Thailand–Cambodia border were collected between 2008 and 2016 (Table 1). Major haplotype was the septuple mutants that comprised of mutations at N51I, C59R, S108N and I164L in pfdhfr gene and A437G, K450N and A581G in pfdhps gene. The P. falciparum samples with septuple mutation pfdhfr–pfdhps were detected 50% (6/12) in 2008–2010 and 71% (20/28) in 2014–2016. The secondary haplotype in 2008–2010 was sextuple mutations haplotype that was composed of two haplotypes, namely, ACIRNL-A/SGEAA (25% (3/12)) and ACIRNI-A/SGNGA (17% (2/12)) then the prevalence of falciparum parasites with ACIRNL-A/SGEAA decreased to 4% (1/28) in 2014–2016. Whereas, prevalence of falciparum parasite with quintuple mutations (ACIRNI-A/SGEAA) increased from 8% (1/12) in 2008–2010 to 18% (5/28) in 2014–2016 (Fig. 1). The multiple copy number of pfgch1 gene were observed in 7% (2/28)–8% (1/12) in this region (Fig. 2).
Mutations in pfdhfr and pfdhps genes in isolates from the Thailand–Malaysia border
All 47 P. falciparum isolates from Yala province represented the samples from Thailand–Malaysia border (Table 1). All falciparum samples carried quadruple or quintuple mutation of pfdhfr–pfdhps gene. The major haplotype had mutations at N51I, C59R and S108N of pfdhfr and A437G of pfdhps that showed 67% (2/3) in 2008–2010, 100% (7/7) in 2011–2013 and 35% (13/37) in 2014–2016. Quintuple mutations (ACIRNI-A/SGKGA) haplotype became the main haplotype in the last period of this study (65%, 24/37) (Fig. 1). In addition, pfgch1 gene amplification was found in 14% (5/37) of genes in 2014–2016 in contrast to absence of multiple copies in earlier samples (Fig. 2).
Comparing frequencies of mutations from three areas, prevalence of septuple (ACIRNL-A/SGE/NGA) and sextuple mutations (ACIRNL-A/SGEAA, ACNRNL-A/SGEGA and ACIRNI-A/SGE/NGA) were found in the falciparum samples from Thailand–Myanmar and Thailand–Cambodia border as the major group. Whereas, the falciparum samples of Thailand–Malaysia border presented only quintuple and quadruple mutations haplotypes. The octuple mutations which had the mutation at A/S436F or A613S/T were detected only in Thailand–Myanmar border. However, both of octuple haplotype observed below 8% in 2008–2013 were absented in 2014–2016 (Fig. 3).
Associations between pfgch1 copy number and antifolate resistance markers in pfdhfr and pfdhps
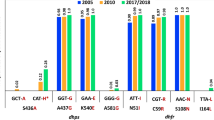
The association between individual six mutation points in pfdhfr and gene amplification of pfgch1 were assessed in all 233 samples. Only the I164L pfdhfr point mutation was significantly associated with pfgch1 amplification (ϕ = 0.213, p-value= 0.001). Plasmodium falciparum carrying both the pfdhfr I164 allele and pfgch1 multiple copies were observed in only 21% (15/73) of samples, while P. falciparum isolates that carried both pfdhfr 164L and pfgch1 amplification were observed in 42% (67/160) of samples (Fig. 3).
Similarly, evaluation of the association between five mutation points in pfdhps and pfgch1 gene amplification showed that the pfdhps K540E/N allele was significantly correlated to pfgch1 amplification (ϕ = 0.399, p-value ≤ 0.001). Plasmodium falciparum carrying the pfdhps 540E allele and pfgch1 multiple copies was observed in 50% (75/151) of samples.
Discussion
This longitudinal study of antifolate resistance markers in Thailand P. falciparum infections showed persisting high prevalence of antifolate resistance haplotypes between 2008 and 2016, despite long-term cessation of SP treatment regimen. Studies from early 1990s to 2014, revealed the pfdhfr point mutations transitioning from double mutations (S108N with N51I/C59R/164L, ≥ 72%) to triple (N51I, C59R, S108N or C59R, S108N, I164L) and eventually quadruple (N51I, C59R, S108N/T, I164L) mutations in provinces of Thailand bordering Myanmar and Cambodia [5, 10, 19,20,21]. Present findings are in agreement with previously observed trends showing that most P. falciparum parasites up to 75% carried triple or quadruple mutations in pfdhfr. However, limited data from 1994 to 1995 on the Thailand–Malaysia border indicated a high prevalence of 88% pfdhfr triple mutations, whereas no quadruple mutations were reported [22].
The pfdhps mutations based on four codons (S436F/A, A437G, A581G, and A613S/T) were first reported in mid 1990s along the Thailand–Myanmar border [5]. Multiple studies conducted between 2001 and 2007 collectively indicated fluctuating dominance of the pfdhps triple (2001–2003, > 95%), quadruple (2002–2003, > 67%) and triple (2004–2007, > 97%) mutations [9, 19, 23]. Taken together, previous reports [9, 10] and present findings indicate emerging trend in shift from pfdhps double to triple mutations as major mutant group along the Thailand–Myanmar and Thailand–Cambodia borders.
Resistance level to antifolate drug in falciparum malaria was increased and conferred by mutations in dhfr and dhps gene. The combination of pfdhfr N51I, C59R and S108N and pfdhps A437G and K540E/N (quintuple mutant) strongly predicts clinical outcome [24]. The mutation in pfdhfr I164L and pfdhps A581G and A613S/T developed later and were associated with increase in antifolate resistance [6].
Effect of mutations in pfdhfr on the inhibition constant (Ki) for pyrimethamine has been well described [25,26,27,28]. This suggests that quadruple mutations including the 164 position were critically important for the development of resistance to pyrimethamine. In addition, it was shown that Ki for sulfadoxine differed significantly between wild type and triple mutant parasites carrying K540E mutation [4, 29]. Strong association between elevated copy number of the pfgch1 gene and the pfdhfr-I164L mutation has been shown previously [8]. In this study, an association between pfgch1 gene multiple copy number and the pfdhfr-I164L mutation was also observed. However, in Ghana with low level of pfgch1 gene amplification (6%; 12/192), no dhfr-I164L was detected in samples with amplified pfgch1 [30]. The positive correlation between pfdhps (540E) and pfgch1 multiple copies observed in this study was not observed before. These correlations in parasites with high-level antifolate and anti-sulfonamide resistance may indicate functional linkage and fitness epistasis between genes on different chromosomes. The observed positive correlations between pfgch1 amplification and pfdhfr (I164L)/pfdhps (K540E/N) mutations may represent adaptation of the parasite dissociated from the anti-malarial drug pressure or arguably the pressure from non-anti-malarial antifolate drugs.
Present study provides indication of co-increase in quintuple mutants and pfgch1 multiple copies in five isolates during 2014–2016. However, relatively smaller sample size (in total 37 isolates) from Thailand–Malaysia border might also have influenced the observed results. Persistence of high prevalence of antifolate resistance haplotypes in Thailand may be explained by several factors. Persistence of antifolate resistance markers might result from continued drug pressure from non-malarial antifolate drugs such as trimethoprim and sulfamethoxazole [11]. Trimethoprim–sulfamethoxazole (a.k.a Bactrim™) is still used in Thailand [31] as a part of standard package to care people with HIV/AIDS [32], urinary tract infection [33], melioidosis [34] and pneumonia [35]. The antifolate gene mutations might have been sustained because of continued presence of this antifolate drug pressure [11].
Human migration contributed to the spread of chloroquine resistant malaria from Southeast Asia to Africa during the 1970s, and SP resistance during the 1980s–1990s [36,37,38]. Studies conducted during 1998–2001 [39] and 2007–2009 [40], identified a high proportion of pfdhfr/pfdhps resistance haplotypes in parts of Myanmar sharing border regions with Thailand. This period coincided with the increased prevalence of pfdhfr/pfdhps mutants in Thailand [9, 19, 20]. The main haplotypes then were quadruple mutants for pfdhfr and triple mutants for pfdhps, respectively (Additional file 1: Table S2). The Thailand–Cambodia border showed a slight difference in the proportion of pfdhfr resistance haplotypes containing quadruple mutations, which was higher than the proportion of triple mutations observed between 2003 and 2007 [21], whereas the prevalence of pfdhps mutations was similar to Cambodia [20] (Additional file 1: Table S2). Present findings are in agreement with the low reported pfdhfr–pfdhps mutations on the Thailand–Malaysia border compared to other regions [9, 22]. It is likely that the parasite population in this border area, which is a conflict zone, is separate from the other border areas, with limited human migration carrying parasites between these areas.
Conclusions
Decades after cessation of SP as anti-malarial treatment in Thailand, there is a persistent high proportion of P. falciparum carrying both sulfadoxine and pyrimethamine resistance markers, with the exception of the Thailand–Malaysia border. Amplification of pfgch1 correlated with pfdhfr (I164L) and pfdhps (K540E) mutations, suggesting pfgch1 amplification might be compensatory to mutations in the pfdhfr and pfdhps genes.
Availability of data and materials
The dataset generated during the current study are available from corresponding author on reasonable request.
Abbreviations
- pfdhfr :
-
Plasmodium falciparum dihydrofolate reductase
- pfdhps :
-
P. falciparum dihydropteroate synthase
- pfgch1 :
-
P. falciparum GTP cyclohydrolase-1
- SNPs:
-
Single nucleotide polymorphisms
- CNV:
-
Copy number variation
References
WHO. Country cooperation strategy, Thailand: 2017–2021. Geneva: World Health Organization; 2017.
Harinasuta T, Suntharasamai P, Viravan C. Chloroquine-resistant falciparum malaria in Thailand. Lancet. 1965;2:657–60.
Wongsrichanalai C, Sirichaisinthop J, Karwacki JJ, Congpuong K, Miller RS, Pang L, et al. Drug resistant malaria on the Thai-Myanmar and Thai–Cambodian borders. Southeast Asian J Trop Med Public Health. 2001;32:41–9.
Wang P, Read M, Sims PF, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997;23:979–86.
Basco LK, Eldin de Pecoulas P, Wilson CM, Le Bras J, Mazabraud A. Point mutations in the dihydrofolate reductase-thymidylate synthase gene and pyrimethamine and cycloguanil resistance in Plasmodium falciparum. Mol Biochem Parasitol. 1995;69:135–8.
Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine–sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–6.
Heinberg A, Siu E, Stern C, Lawrence EA, Ferdig MT, Deitsch KW, et al. Direct evidence for the adaptive role of copy number variation on antifolate susceptibility in Plasmodium falciparum. Mol Microbiol. 2013;88:702–12.
Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, et al. Adaptive copy number evolution in malaria parasites. PLoS Genet. 2008;4:e1000243.
Alam MT, Vinayak S, Congpuong K, Wongsrichanalai C, Satimai W, Slutsker L, et al. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob Agents Chemother. 2011;55:155–64.
Imwong M, Jindakhad T, Kunasol C, Sutawong K, Vejakama P, Dondorp AM. An outbreak of artemisinin resistant falciparum malaria in Eastern Thailand. Sci Rep. 2015;5:17412.
Iyer JK, Milhous WK, Cortese JF, Kublin JG, Plowe CV. Plasmodium falciparum cross-resistance between trimethoprim and pyrimethamine. Lancet. 2001;358:1066–7.
Khim N, Bouchier C, Ekala MT, Incardona S, Lim P, Legrand E, et al. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob Agents Chemother. 2005;49:3147–52.
Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–5.
Duraisingh MT, Curtis J, Warhurst DC. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol. 1998;89:1–8.
Parola P, Pradines B, Simon F, Carlotti MP, Minodier P, Ranjeva MP, et al. Antimalarial drug susceptibility and point mutations associated with drug resistance in 248 Plasmodium falciparum isolates imported from Comoros to Marseille, France in 2004–2006. Am J Trop Med Hyg. 2007;77:431–7.
Tanomsing N, Imwong M, Pukrittayakamee S, Chotivanich K, Looareesuwan S, Mayxay M, et al. Genetic analysis of the dihydrofolate reductase-thymidylate synthase gene from geographically diverse isolates of Plasmodium malariae. Antimicrob Agents Chemother. 2007;51:3523–30.
Vinayak S, Alam MT, Mixson-Hayden T, McCollum AM, Sem R, Shah NK, et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 2010;6:e1000830.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
Krudsood S, Imwong M, Wilairatana P, Pukrittayakamee S, Nonprasert A, Snounou G, et al. Artesunate–dapsone–proguanil treatment of falciparum malaria: genotypic determinants of therapeutic response. Trans R Soc Trop Med Hyg. 2005;99:142–9.
Mita T, Tanabe K, Takahashi N, Tsukahara T, Eto H, Dysoley L, et al. Independent evolution of pyrimethamine resistance in Plasmodium falciparum isolates in Melanesia. Antimicrob Agents Chemother. 2007;51:1071–7.
Cheychom J, Kanchanakhan N, Vijaykadga S, Harnyuttanakorn P. Antifolate resistance mutation and proguanil susceptibility among Plasmodium falciparum isolates in Thai-Cambodia border. J Health Res. 2013;27:301–5.
Cox-Singh J, Zakaria R, Abdullah MS, Rahman HA, Nagappan S, Singh B. Short report: differences in dihydrofolate reductase but not dihydropteroate synthase alleles in Plasmodium falciparum isolates from geographically distinct areas in Malaysia. Am J Trop Med Hyg. 2001;64:28–31.
Mita T, Venkatesan M, Ohashi J, Culleton R, Takahashi N, Tsukahara T, et al. Limited geographical origin and global spread of sulfadoxine-resistant dhps alleles in Plasmodium falciparum populations. J Infect Dis. 2011;204:1980–8.
Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. Molecular markers for failure of sulfadoxine–pyrimethamine and chlorproguanil–dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8.
Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci USA. 1997;94:1124–9.
Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–13.
Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–8.
Thaithong S, Chan S-W, Songsomboon S, Wilairat P, Seesod N, Sueblinwong T, et al. Pyrimethamine resistant mutations in Plasmodium falciparum. Mol Biochem Parasitol. 1992;52:149–57.
Triglia T, Cowman AF. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994;91:7149–53.
Osei M, Ansah F, Matrevi SA, Asante KP, Awandare GA, Quashie NB, et al. Amplification of GTP-cyclohydrolase 1 gene in Plasmodium falciparum isolates with the quadruple mutant of dihydrofolate reductase and dihydropteroate synthase genes in Ghana. PLoS ONE. 2018;13:e0204871.
Sumpradit N, Suthajit S, Poonpolsub S, Chuanchuen R, Prakongsai P. Landscape of antimicrobial resistance situations and actions in Thailand. Bangkok: Ministry of Public Health; 2015.
WHO. Guidelines on post-exposure prophylaxis for HIV and the use of co-trimoxazole prophylaxis for HIV-related infections among adults, adolescents and children. Geneva: World Health Organization; 2014.
Guneysel O, Onur O, Erdede M, Denizbasi A. Trimethoprim/sulfamethoxazole resistance in urinary tract infections. J Emerg Med. 2009;36:338–41.
Cheng AC, McBryde ES, Wuthiekanun V, Chierakul W, Amornchai P, Day NP, et al. Dosing regimens of cotrimoxazole (trimethoprim-sulfamethoxazole) for melioidosis. Antimicrob Agents Chemother. 2009;53:4193–9.
Calderón EJ, Gutiérrez-Rivero S, Durand-Joly I, Dei-Cas E. Pneumocystis infection in humans: diagnosis and treatment. Expert Rev Anti Infect Ther. 2010;8:683–701.
Anderson TJ, Roper C. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 2005;94:269–80.
Naidoo I, Roper C. Following the path of most resistance: dhps K540E dispersal in African Plasmodium falciparum. Trends Parasitol. 2010;26:447–56.
Martens P, Hall L. Malaria on the move: human population movement and malaria transmission. Emerg Infect Dis. 2000;6:103–9.
Nair S, Brockman A, Paiphun L, Nosten F, Anderson TJ. Rapid genotyping of loci involved in antifolate drug resistance in Plasmodium falciparum by primer extension. Int J Parasitol. 2002;32:852–8.
Yang Z, Li C, Miao M, Zhang Z, Sun X, Meng H, et al. Multidrug-resistant genotypes of Plasmodium falciparum, Myanmar. Emerg Infect Dis. 2011;17:498–501.
Acknowledgements
This study was supported by Thailand Science Research and Innovation (TSRI), RTA6280006 and Dean-MORU Scholarship, Faculty of Tropical Medicine, Mahidol University Thailand and the Wellcome Trust Mahidol University Oxford Tropical Medicine Research Programme.
Funding
This study was supported by Thailand Science Research and Innovation (TSRI), RTA6280006 and Dean-MORU Scholarship, Faculty of Tropical Medicine, Mahidol University Thailand and the Wellcome Trust Mahidol University Oxford Tropical Medicine Research Programme.
Author information
Authors and Affiliations
Contributions
MI, NJD, AMD and PS designed the study. RS undertook the laboratory experiment. RS, PP, PS collected the samples. RS, KS, CK, and VM conducted data analysis. RS, VM, AMD and MI wrote the first draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical and Scientific procedure of this study were certificated by the ethics committee of the Faculty of Tropical Medicine, Mahidol University, Thailand. The sample materials used in this study were permitted by responsible person of the Antimalarial Drug Therapeutic Efficacy Monitoring Projects that were approved by the ethical committee from Department of Disease Control, Ministry of Public Health, Thailand.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Primer and Probe sequences for detection of SP resistance markers in P. falciparum. Table S2. The prevalence reports of pfdhfr and pfdhps mutations close the border of Thailand and neighboring countries since 1990 to 2016. Table S3. The Pfdhfr/Pfdhps haplotypes and gch1 copy number variation of P. falciparum in 9 provinces along the border of Thailand-neighboring countries between 2008 and 2016.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sugaram, R., Suwannasin, K., Kunasol, C. et al. Molecular characterization of Plasmodium falciparum antifolate resistance markers in Thailand between 2008 and 2016. Malar J 19, 107 (2020). https://doi.org/10.1186/s12936-020-03176-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-020-03176-x