Abstract
Background
Although a number of Plasmodium vivax proteins have been identified, few have been investigated as potential vaccine candidates. This study characterized the Plasmodium vivax merozoite surface antigen 180 (PvMSA180, PVX_094920), a novel P. vivax antigenic protein.
Methods
The target gene was amplified as four overlapping domains (D1, D2, D3 and D4) to enable expression of the recombinant protein using cell-free and bacterial expression systems. The recombinant PvMSA180 proteins were used in protein microarrays to evaluate the humoral immune response of 72 vivax-infected patients and 24 vivax-naïve individuals. Antibodies produced in mice against the PvMSA180-D1 and -D4 domains were used to assess the subcellular localization of schizont-stage parasites with immunofluorescence assays. A total of 51 pvmsa180 sequences from 12 countries (41 sequences from PlasmoDB and 6 generated in this study) were used to determine the genetic diversity and genealogical relationships with DNAsp and NETWORK software packages, respectively.
Results
PvMSA180 consists of 1603 amino acids with a predicted molecular mass of 182 kDa, and has a signal peptide at the amino-terminus. A total of 70.8% of patients (51/72) showed a specific antibody response to at least one of the PvMSA180 domains, and 20.8% (15/72) exhibited a robust antibody response to at least three of the domains. These findings suggest that PvMSA180 is targeted by the humoral immune response during natural infection with P. vivax. Immunofluorescence analysis demonstrated that PvMSA180 is localized on the merozoite surface of schizont-stage parasites, and pvmsa180 sequences originating from various geographic regions worldwide showed low genetic diversity. Twenty-two haplotypes were found, and haplotype 6 (Hap_6, 77%) of pvmsa180 was detected in isolates from six countries.
Conclusions
A novel P. vivax surface protein, PvMSA180, was characterized in this study. Most of P. vivax-infected patients had specific antibodies against particular antigenic domains, indicating that this protein is immunogenic in naturally exposed populations. Genetic analysis of worldwide isolates showed that pvmsa180 is less polymorphic than other well-known candidates and that some haplotypes are common to several countries. However, additional studies with a larger sample size are necessary to evaluate the antibody responses in geographically separated populations, and to identify the function of PvMSA180 during parasite invasion.
Similar content being viewed by others
Background
Malaria is a public health problem in countries in which it is endemic [1]. In 2015, the number of cases and deaths was 214 million and 438,000, respectively [2]. Plasmodium vivax causes 50% of all malaria cases globally [3], and is prevalent in the tropics and subtropics [4]. A malaria vaccine shows promise for controlling malaria [5]; however, the antigenic diversity and immune-evasion ability of Plasmodium has hampered vaccine development [6]. Molecules expressed on the merozoite surface, such as apical membrane antigen-1 (AMA1), merozoite surface protein-1 (MSP1), and Duffy binding protein, have been the focus of vaccine development efforts [7].
Bioinformatic and genome analysis of P. vivax have led to the identification of malaria antigens, few of which have been investigated as vaccine candidates [8,9,10]. MSPs, such as MSP-1, MSP-9, MSP-4 and MSP-5, have been identified as vaccine candidates [11]. Some hypothetical proteins have been identified as vaccine candidates based on coiled coil structure [10]. Moreover, several proteins of P. vivax that are expressed on the surface or in apical organelles, including MSPs, rhoptry-associated membrane antigen, glycosylphosphatidylinositol (GPI)-anchored micronemal antigen and AMA1, have been proposed as vaccine candidates due to their involvement in merozoite invasion or the longevity of the antibody response [12,13,14,15,16]. Due to the limitations of P. vivax in vitro culture systems, fewer surface proteins have been identified in this pathogen than in P. falciparum. In fact, a number of P. vivax surface proteins have been identified based on their orthologues in P. falciparum [9, 10, 15, 17], and the antibody responses to them have been investigated [18,19,20].
One of hypothetical proteins, named P. vivax merozoite surface antigen 180 (PvMSA180) was previously identified [21]. Of the 96 P. vivax blood-stage proteins, 18 (including PvMSA180) elicited robust antibody responses [21]. Thus, this study has characterized PvMSA180, which is immunogenic in naturally exposed populations, and determined its subcellular localization in P. vivax.
Methods
Sample and serum information
Blood samples were collected from 72 patients in 2012 in the Shwe Kyin area of Myanmar (mean age, 24.5 years; range, 18–42 years). The patients were confirmed to have P. vivax malaria using the malaria rapid diagnostic test (SDFK80; Standard Diagnostics, Gyeonggi, Korea) and microscopy. Samples were centrifuged and the serum was separated. Serum samples from 24 healthy malaria-naïve individuals residing in non-endemic areas in the Republic of Korea (ROK) were also collected and used as controls.
Amplification of full-length pvmsa180
Parasite genomic DNA was extracted using a Qiagen DNA Extraction Kit (Hilden, Germany). The pvmsa180 (PVX_094092) sequence was obtained from PlasmoDB (http://plasmodb.org/). Full-length pvmsa180 was amplified from five Myanmar and one South Korean isolate using the forward primer 5′-GATGACGACACAAACAAAAGGG-3′ and reverse primer 3′-CGCGGCGTAGTTGATGTG-5′. Full-length pvmsa180 was amplified by PCR using high-fidelity Thermococcus kodakaraensis (KOD) DNA polymerase (Toyobo, Osaka, Japan) under the following conditions: 2.0 μL DNA template, 0.4 U KOD DNA polymerase, 0.25 mM of each primer and 500 μM of each dNTP, in a final volume of 20 mL. The cycling conditions were 94 °C for 2 min, followed by 35 cycles at 94 °C for 15 s, at 58 °C for 30 s, at 68 °C for 4.5 min, and a final extension at 68 °C for 10 min.
Recombinant PvMSA180 expression
PvMSA180 was divided into four fragments and expressed using a cell-free system. The four fragments of pvmsa180 were amplified under the aforementioned conditions, with the exception of a final extension for 1.5 min, using the following In-fusion primers:
D1-F: 5′-GGGCGGATAT CTCGAGGATGACGACACAAACAAAAGGG-3′ and D1-R: 5′-GCGGTACCCGG GATCCTTACCCGACATAGTACATTTGCTCA-3′; D2-F: 5′-GGGCGGATAT CTCGAGAAGCTGCACCCAAAGAAGC-3′ and D2-R: 5′-GCGGTACCCGG GATCCTTACATCTTCTCGTACAACAGCATATCA-3′; D3-F: 5′- GGGCGGATAT CTCGAGGTGCTGGAGCTAGTAAATAATGATATG-3′ and D3-R: 5′- GCGGTACCCGG GATCCTCAAAAGGCGCACTTCAAACTCA-3′; D4-F: 5′-GGGCGGATAT CTCGAGGTGCGAAAAGAAAATGGAC-3′ and D4-R: 5′-GCGGTACCCGG GATCCTCACGCGGCGTAGTTGATGTG-3′. Underlining indicates the 15 base pair (bp) homologous vector site; italicized, bolded, and underlined text indicates the XhoI and BamHI sites. The PCR products were cloned into the pEU-E01-His-Tev-N2 vector (Cell-Free Sciences, Matsuyama, Japan) using the In-fusion cloning kit (Clontech, Palo, Alto, CA, USA), and expressed using a wheat germ cell-free system (Cell-Free Sciences) [15, 22,23,24]. The crude recombinant proteins were used for immunoscreening of serum samples. Recombinant PvMSA180-D1 and PvMSA180-D4 proteins were expressed in Escherichia coli. PvMSA180–D1 was expressed using the pGEX-4T-2 expression vector (GE Healthcare, Upsala, Sweden) with a glutathione S transferase (GST) tag, and PvMSA180-D4 was expressed using the pET-28a (+) expression vector (Invitrogen, Carlsbad, CA, USA) with a His-tag. The target domains were amplified using In-fusion primers and High-Fidelity DNA Polymerase (Toyobo), and cloned into the appropriate expression vectors. Positive clones confirmed by DNA sequencing analysis were transformed into E. coli BL21(DE3) cells (Invitrogen). When the cultures reached an optical density of 0.6, expression of the recombinant D1 and D4 fragments was induced by addition of 0.1 and 0.3 mM isopropyl-β-d-thiogalactopyranoside, respectively. The GST-tagged proteins were purified using glutathione Sepharose 4B (GE Healthcare) and 6 His-tagged proteins using nickel-nitrilotriacetic acid (Ni–NTA) (Qiagen), according to the manufacturer’s instructions. The purity of the recombinant proteins was evaluated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
SDS-PAGE and Western blotting
The recombinant PvMSA180 proteins were resolved on 13% SDS-PAGE gels under reducing conditions, and then electrotransferred to 0.45 µm PVDF membranes (Millipore, Billerica, MA, USA) in semi-dry transfer buffer (50 mM Tris, 190 mM glycine, 3.5 mM SDS, 20% methanol) at a constant current of 360 mA for 40 min using a semi-dry blotting system (ATTO Corp., Tokyo, Japan). Recombinant PvMSA180-D1 and PvMSA180-D4 (1 µg each) and PvDBP-RII (0.5 µg) were used to assay antibody responses. The membranes were blocked in 5% skim milk and then incubated with a primary anti-GST antibody (1:10,000), anti-pentahistidine antibody (1:2000), mouse immune serum, or pooled patient serum (1:100), followed by incubation with a secondary IRDye® goat anti-mouse (1:10,000 dilution) or IRDye® goat anti-human (1:20,000) (LI-COR® Bioscience, Lincoln, NE, USA) antibody. An Odyssey infrared imaging system and the accompanying software (LI-COR® Bioscience) were used for data analysis.
Protein microarray
Slides were coated with amine solution [21] and used to screen the serum of 72 vivax-infected patients and 24 healthy individuals by protein microarray. The crude recombinant proteins expressed using the wheat germ cell-free system were applied in duplicate and incubated for 2 h at 37 °C, followed by blocking in 5% BSA in phosphate-buffered saline (PBS) with 0.1% Tween 20 (PBS-T) and incubation for 1 h at 37 °C. Diluted serum (1:25) was added to a slide harbouring recombinant proteins and incubated for 1 h, followed by addition of an Alexa 647-conjugated goat-anti-human IgG (10 µg/mL, Invitrogen). Then slides were imaged using a fluorescence scanner (ScanArray Express, PerkinElmer, Boston, MA, USA). The cut-off value was equal to the mean fluorescence intensity (MFI) plus two standard deviations (SDs) of the negative samples. Normalized MFI values were calculated from the MFI/cut-off values.
Production of mice antibody
Female BALB/c mice at 6–8 weeks of age (DBL, Seoul, ROK) were injected intraperitoneally with 30 µg recombinant PvMSA180-D1 and -D4 in PBS with complete Freund’s adjuvant (Sigma-Aldrich, St. Louis, MO) in a final volume of 100 µL, and subsequently boosted with incomplete Freund’s adjuvant. Three mice were injected three times with each of the D1 and D4 fragments of PvMSA180 at 2-week intervals. Sera were collected 2 weeks after the final boost. All the animal experimental protocols were approved by the Institutional Ethics Committee and were conducted in accordance with the Ethical Guidelines for Animal Experiments of Kangwon National University (ROK).
Immunofluorescence assays
Plasmodium vivax parasites were collected from patients with malaria in Thailand and spotted onto eight-well glass slides. The slides were fixed in ice-cold acetone for 10 min, dried, and blocked in 5% BSA in PBS-T at 37 °C for 30 min. Then the slides were incubated (dual-labelled) with rabbit anti-PvMSP1-19 (1:50 dilution), rabbit anti-PvDBP (1:50 dilution), rabbit anti-PvRAMA (1:50 dilution), and mouse anti-PvMSA180 (1:100) as primary antibodies at 37 °C for 1 h. Next, the slides were stained with Alexa Fluor 568-conjugated anti-rabbit IgG or Alexa Fluor 488-conjugated goat anti-mouse IgG as secondary antibodies (Invitrogen), and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen) at 37 °C for 30 min. The slides were mounted in ProLong Gold antifade reagent (Invitrogen) and visualized under oil immersion using a confocal laser scanning microscope (FV200Olympus, Tokyo, Japan) equipped with 20× dry and 60× oil objectives. Images were captured using the FV10-ASW 3.0 viewer software.
Sequence diversity and haplotype analysis of pvmsa180 in isolates obtained worldwide
The pvmsa180 sequences obtained in this study were compared to those of 46 isolated worldwide in the PlasmoDB database. Pvmsa180 sequences from 12 countries (South Korea, Myanmar, Salvador, Peru, Colombia, Mexico, Thailand, Brazil, India, Papua New Guinea, North Korea, and China) were included in the analysis (Additional file 1). All the raw sequences were analysed and trimmed using the SeqMan software, Lasergene ver. 7.0 (DNASTAR). Sequences were aligned using the CLUSTAL-W program in MegAlign Lasergene ver. 7.0 (DNASTAR) and exported in FASTA format. Sequence diversity (π), defined as the average number of nucleotide differences per site between two sequences, number of polymorphic sites was determined using the DNAsp ver. 5.0 software. The relationships among the haplotypes of pvmsa180 were evaluated with the me-dian-joining method using the NETWORK software ver. 4.6.1.2. (Fluxus Technology Ltd., Suffolk, UK). The C-terminus amino acid sequences of PvMSA180 were also separately aligned using Clustal W2 to assess the sequence homology among two human and simian malaria parasites.
Statistical analysis
Simple scatter regression was applied to construct a standard curve using SigmaPlot (Systat Software Inc., San Jose, CA, USA). Data were analysed using GraphPad Prism (GraphPad, Software, San Diego, CA, USA). The Student’s t test was used to assess differences between means. Values of p < 0.05 were considered statistically significant. The MeV software (MultiExperiment Viewer, http://www.tm4.org/mev.html) was used to visualize the antibody responses of individual patients to the PvMSA180 domains.
Results
Schematic structure of PvMSA180
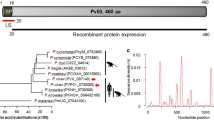
The protein consists of 1603 amino acids with a predicted molecular mass of 182 kDa, and lacks a transmembrane domain and GPI anchor, as determined using the deduced amino acid sequence of P. vivax (Sal-1). The cysteine residues are adjacent to the C-terminus domain. Proteins were expressed without a signal peptide. The D1 domain of PvMSA180 (207–307 amino acids) from worldwide isolates contains a polymorphic region (Fig. 1a). In addition, the C-terminus sequence of PvMSA180 has high sequence homology with the human (P. falciparum) and two simian (Plasmodium knowlesi and Plasmodium cynomolgi) malaria parasites. The amino acid alignment revealed 70% amino acid identity among human and simian malaria parasites (Fig. 1b).
Schematic of recombinant PvMSA180 expression. a Schematic of PvMSA180. The D1 (amino acids [aa] 28–401), D2 (aa 385–800), D3 (aa 785–1180), and D4 (aa 1163–1603) domains were expressed using cell-free and bacterial expression systems. SP signal peptide, P polymorphic region (aa 290–307). b Clustal alignment of the C-terminus sequence of MSA180 of Plasmodium vivax (Pv), Plasmodium falciparum (Pf), Plasmodium knowlesi (Pk), and Plasmodium cynomolgi (Pc). Red bars indicate the conserved aa in the four Plasmodium species, green bars in three species, sky-blue bars in two species, and dark-blue bars in one species. Asterisks denote cysteine residues conserved in the four Plasmodium species
Expression of recombinant PvMSA180 proteins
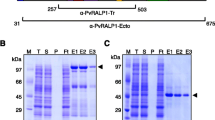
The four domains of PvMSA180 were expressed using a wheat germ cell-free system, resulting in a single band of ~50 to 75 kDa; all the domains were predicted to be <50 kDa (Fig. 2a). PvMSA180-D1 was successfully expressed as a 70.3 kDa GST fusion protein, and PvMSA180-D4 was expressed in E. coli as a 6× His-tagged, 52.7 kDa recombinant protein (Fig. 2b, c). SDS-PAGE and Western blotting showed that recombinant PvMSA180-D1 and PvMSA180-D4 have molecular weights of ~80 and ~60 kDa, respectively (Fig. 2b, c). Purified PvMSA180-D1 and PvMSA180-D4 were used to immunize Balb/c mice with the aim of generating polyclonal antibodies.
Expression of recombinant PvMSA180. a Expression of recombinant domains of PvMSA180 in a wheat germ cell-free expression system. Arrowheads indicate the four domains of PvMSA180. D1 domain 1, D2 domain 2, D3 domain 3, and D4 domain 4. b Expression and purification of PvMSA180-D1 (GST-fused) and D4 (His-tagged) in E. coli. c Recombinant PvMSA180-D1 and D4 were probed with anti-GST and anti-His antibodies under reducing conditions. Arrowheads indicate the PvMSA180-D1 (~80 kDa) and D4 (~60 kDa) bands. d Antibody recognition of PvMSA180-D1 and D4 compare to PvDBP-RII recombinant proteins using pooled vivax-patient sera (lane P), malaria naïve healthy sera (lane H), mouse immune serum (lane M), and anti-GST or anti-His antibody (lane R)
Recognition of recombinant PvMSA180 proteins
To confirm the immunoreactivity of PvMSA180, purified recombinant PvMSA180-D1 and PvMSA180-D4 were incubated with pooled sera of vivax-infected patients. Bands at ~80 and ~60 kDa for PvMSA180-D1 and PvMSA180-D4, respectively, were detected by Western blotting (Fig. 2d). In addition, to confirm antibody production in mice against the D1 and D4 domains of PvMSA180, recombinant proteins were subjected to Western blotting using polyclonal anti-PvMSA180-D1 and PvMSA180-D4 antibodies (Fig. 2d).
Humoral immune response to PvMSA180
The humoral immune response of 72 vivax-infected patients and 24 healthy individuals to the four domains of PvMSA180 was evaluated by protein microarray. Sera from vivax-infected patients showed significantly higher reactivity than that from healthy individuals (Fig. 3a, p < 0.0001). The number of healthy individuals and vivax-infected patients showing a serum immune response to PvMSA180 was determined (Table 1). The antibody response to the four domains did not differ markedly. However, antibodies to PvMSA180 showed positive reactivity against PvMSA180, and there was no correlation between the humoral immune response and patient age or the presence of parasitaemia.
Human IgG response to the four domains of PvMSA180 by protein microarray. a Crude recombinant PvMSA180 was probed with sera from vivax-infected patients (P) and naïve healthy individuals (H). The total prevalence of anti-PvMSA180 IgG significantly differed between patients and naïve healthy individuals. *p < 0.01; ***p < 0.0001 by Student’s t-test. Bars indicate means ± standard deviation. b IgG response to the four domains of PvMSA180 in individual patients. Red and green bars indicate positive and negative reactivity, respectively, in each domain. For total reactivity, red bar indicates 3–4 positive domains; green indicates 1–2 positive domains, and blue indicates the negative domains. The serial numbers of the serum samples are shown on the vertical line
PvMSA180 localized on the merozoite surface
The subcellular localization of PvMSA180 was evaluated by immunofluorescence analysis using an anti-PvMSA180-D1 antibody. Dual-labelled immunofluorescence assays were performed using an anti-PvMSA180-D1 antibody together with an anti-PvMSP1-19 (merozoite surface marker) (Fig. 4a, b), anti-PvDBP (microneme marker) (Fig. 4c), or anti-PvRAMA (rhoptry marker) (Fig. 4d) antibody as controls. The anti-PvMSA180-D1 (N-terminus) and PvMSA180-D4 (C-terminus) antibodies specifically reacted with anti-PvMSP1-19 antibodies in the merozoites of mature schizonts. This suggests that PvMSA180 is localized on the merozoite surface, but not in the rhoptry or microneme.
Localization of PvMSA180 in mature schizont-stage parasites. Schizont-stage parasites were dual-labelled with antisera against PvMSP1-19 (merozoite surface marker) (a and b), PvDBP (microneme marker) (c), or PvRAMA (rhoptry marker) (d). Nuclei are stained with DAPI in the merged images. Scale bar represents 5 µm. DIC differential interference contrast
Nucleotide diversity and haplotype analysis
The 51 pvmsa180 sequences exhibited a low level of diversity (π = 0.00080 ± 0.00012). Alignment of the pvmsa180 sequences revealed 34 (0.84%) polymorphic and 3985 (99.15%) invariant sites. The polymorphic region (i.e., 879–927 nucleotides) contained the highest nucleotide sequence diversity (π = 0.02196 ± 0.0099). The haplotype network analysis of 51 pvmsa180 sequences from worldwide isolates identified 22 unique haplotypes with moderate haplotype diversity (Hd = 0.8698). One haplotype (Hap_6) was shared (Fig. 5) by 17 isolates from Colombia (n = 5), Mexico (n = 2), Peru (n = 4), Myanmar (n = 2), Salvador (n = 1), and Thailand (n = 3). None of the other haplotypes were shared (Fig. 5). The haplotypes obtained in this study are listed in Additional file 2.
Median-joining networks of pvmsa180 haplotypes from isolates worldwide. Genealogical haplotype network showing the relationships among 22 pvmsa180 haplotypes in 51 sequences obtained from P. vivax isolates from 12 countries. H-number, haplotype number. The size of the circles represents the haplotype frequencies and unnumbered circles indicate a single haplotype. Geographical haplotypes are indicated by the colour key. Small red nodes are hypothetical median vectors created by the program to connect sampled haplotypes into a parsimonious network. The distances between nodes are arbitrary
Discussion
Genomics, proteomics, and transcriptomics have been used to identify target antigens in Plasmodium spp. [10, 21,22,23,24]. This has resulted in the identification of proteins expressed by infected red blood cells on the surface or in apical organelles; however, few such P. vivax proteins have been reported [13]. A small number of promising merozoite antigens has been investigated as vaccine candidates, some of which have shown partial efficacy in clinical trials [13]. MSPs are of interest for the development of vaccines as well as drugs that inhibit blood-stage replication of P. falciparum [25,26,27]. In this study, PvMSA180, a novel surface protein of P. vivax, was characterized.
Most of the cysteine residues are in the C-terminus domain of PvMSA180, which has no transmembrane domain or GPI-anchor. The results of the immunofluorescence assay showed that the PvMSA180 protein is localized on the merozoite surface; thus, it might be categorized as being peripherally associated with the surface of P. vivax merozoites as it lacks transmembrane domains and a GPI-anchor [13]. PvMSA180 orthologs in other species have a conserved domain at the C-terminus that shows ~70% sequence similarity with simian malaria parasites (P. knowlesi and Plasmodium chabaudi), but a lower level of identity to Plasmodium falciparum. Nevertheless, P. vivax is genetically, phenotypically, and biologically more similar to P. chabaudi and P. knowlesi than to P. falciparum [28,29,30,31]. Unlike erythrocyte-binding ligand (ebl) and reticulocyte-binding protein (rbp), which are crucial for the invasion of erythrocytes by P. vivax, P. knowlesi, and P. chabaudi, the function of msa180 in other species is unclear.
The amplification and sequence analysis of clinical isolates from diverse geographic regions showed the presence of a short polymorphic domain at the N-terminus region of pvmsa180. The polymorphic region of invasion genes is considered under immune selection pressure [32] and plays a role in protective immunity against clinical isolates [32,33,34]. However, in this study, the polymorphic and conserved domains of PvMSA180 showed similar immunogenicity. Furthermore, 20.8% of the patients (15/72) showed a robust antibody response to more than three domains of PvMSA180, and only 30% did not respond to any of the antigens (Fig. 3b). The development of a malaria vaccine based on merozoite surface antigens has been hindered by their high sequence diversity [35, 36]. In this study, haplotype network analysis resulted in the detection of 22 haplotypes, 1 of which (Hap_6, 17/22) was shared by six countries. However, a greater number of samples are required to confirm this finding, as serum samples from only Myanmar and the ROK were used in this study. These data will facilitate the development of a subunit vaccine based on the domains conserved among strains isolated worldwide. The functional activity of this antibody could not be performed due to the limitation of P. vivax culture system. It remains to be determined as further study in the future.
Conclusions
In this study, a novel P. vivax surface protein, PvMSA180, was identified. Its immunogenicity in a naturally exposed population suggests that it elicits a humoral immune response, and its localization suggests that it may be involved in merozoite attachment to, and invasion of red blood cells. However, further functional and biological investigations of PvMSA180 protein are required prior to its development as a subunit vaccine. Genetic analysis indicated that the gene is less polymorphic than other well-known candidates and a haplotype common to six countries was identified.
References
Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002;415:680–5.
WHO. World malaria report 2015. Geneva: World Health Organization; 2016.
Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009;9:555–66.
Gething PW, Elyazar IRF, Moyes CL, Smith DL, Battle KE, Guerra CA, et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis. 2012;6:e1814.
Arevalo-Herrera M, Herrera S. Plasmodium vivax malaria vaccine development. Mol Immunol. 2001;38:443–55.
Genton B. Malaria vaccines: a toy for travelers or a tool for eradication? Expert Rev Vaccines. 2008;7:597–611.
Arevalo-Herrera M, Chitnis C, Herrera S. Current status of Plasmodium vivax vaccine. Hum Vaccin. 2010;6:124–32.
Versiani FG, Almeida ME, Mariuba LA, Orlandi PP, Nogueira PA. N-terminal Plasmodium vivax merozoite surface protein-1, a potential subunit for malaria vivax vaccine. Clin Dev Immunol. 2013;2013:965841.
Cheng Y, Lu F, Tsuboi T, Han ET. Characterization of a novel merozoite surface protein of Plasmodium vivax, Pv41. Acta Trop. 2013;126:222–8.
Céspedes N, Habel C, Lopez-Perez M, Castellanos A, Kajava AV, Servis C, et al. Plasmodium vivax antigen discovery based on alpha-helical coiled coil protein motif. PLoS ONE. 2014;9:e100440.
Herrera S, Corradin G, Arevalo-Herrera M. An update on the search for a Plasmodium vivax vaccine. Trends Parasitol. 2007;23:122–8.
Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–66.
Beeson JG, Drew DR, Boyle MJ, Feng G, Fowkes FJ, Richards JS. Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol Rev. 2016;40:343–72.
Changrob S, Wang B, Han JH, Lee SK, Nyunt MH, Lim CS, et al. Correction. Naturally acquired immune response against Plasmodium vivax rhoptry-associated membrane antigen. PLoS ONE. 2016;11:e0153110.
Cheng Y, Wang Y, Ito D, Kong D-H, Ha K-S, Chen J-H, et al. The Plasmodium vivax merozoite surface protein 1 paralog is a novel erythrocyte-binding ligand of P. vivax. Infect Immun. 2013;81:1585–95.
Lu F, Li J, Wang B, Cheng Y, Kong DH, Cui L, et al. Profiling the humoral immune responses to Plasmodium vivax infection and identification of candidate immunogenic rhoptry-associated membrane antigen (RAMA). J Proteom. 2014;102:66–82.
Li J, Ito D, Chen JH, Lu F, Cheng Y, Wang B, et al. Pv12, a 6-Cys antigen of Plasmodium vivax, is localized to the merozoite rhoptry. Parasitol Int. 2012;61:443–9.
Tran TM, Oliveira-Ferreira J, Moreno A, Santos F, Yazdani SS, Chitnis CE, et al. Comparison of IgG reactivities to Plasmodium vivax merozoite invasion antigens in a Brazilian amazon population. Am J Trop Med Hyg. 2005;73:244–55.
Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, Mueller I, et al. Strain-specific Duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect Immun. 2009;77:4009–17.
Souza-Silva FA, da Silva-Nunes M, Sanchez BAM, Ceravolo IP, Malafronte RS, Brito CFA, et al. Naturally acquired antibodies to Plasmodium vivax Duffy binding protein (DBP) in rural Brazilian Amazon. Am J Trop Med Hyg. 2010;82:185–93.
Chen JH, Jung JW, Wang Y, Ha KS, Lu F, Lim CS, Takeo S, et al. Immunoproteomics profiling of blood stage Plasmodium vivax infection by high-throughput screening assays. J Proteome Res. 2010;9:6479–89.
Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acd Sci USA. 2008;105:305–10.
Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol Biol Cell. 2001;12:3114–25.
Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci USA. 2008;105:16290–5.
Chandramohanadas R, Basappa Russell B, Liew K, Yau YH, Chong A, et al. Small molecule targeting malaria merozoite surface protein-1 (MSP-1) prevents host invasion of divergent plasmodial species. J Infect Dis. 2014;210:1616–26.
Wilson DW, Goodman CD, Sleebs BE, Weiss GE, de Jong NW, Angrisano F, et al. Macrolides rapidly inhibit red blood cell invasion by the human malaria parasite. Plasmodium falciparum. BMC Biol. 2015;13:52.
Boyle MJ, Wilson DW, Beeson JG. New approaches to studying Plasmodium falciparum merozoite invasion and insights into invasion biology. Int J Parasitol. 2013;43:1–10.
Cornejo OE, Escalante AA. The origin and age of Plasmodium vivax. Trends Parasitol. 2006;22:558–63.
Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, et al. A monkey’s tale. The origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci USA. 2005;102:1980–5.
Tachibana S-I, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 2012;44:1051–5.
Cornejo OE, Fisher D, Escalante AA. Genome-wide patterns of genetic polymorphism and signatures of selection in Plasmodium vivax. Genome Biol Evol. 2015;7:106–19.
Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP. Polymorphism at the merozoite surface protein-3alpha locus of Plasmodium vivax: global and local diversity. Am J Trop Med Hyg. 1999;61:518–25.
Nogueira PA, Piovesan Alves F, Fernandez-Becerra C, Pein O, Rodrigues Santos N, Pereira da Silva LH, et al. A reduced risk of infection with Plasmodium vivax and clinical protection against malaria are associated with antibodies against the N terminus but not the C terminus of merozoite surface protein 1. Infect Immun. 2006;74:2726–33.
Fernandez-Becerra C, Sanz S, Brucet M, Stanisic DI, Alves FP, Camargo EP, et al. Naturally acquired humoral immune responses against the N- and C-termini of the Plasmodium vivax MSP1 protein in endemic regions of Brazil and Papua New Guinea using a multiplex assay. Malar J. 2010;9:29.
Epstein JE, Giersing B, Mullen G, Moorthy V, Richie TL. Malaria vaccines: are we getting closer? Curr Opin Mol Ther. 2007;9:12–24.
Dzikowski R, Deitsch KW. Genetics of antigenic variation in Plasmodium falciparum. Curr Genet. 2009;55:103–10.
Authors’ contributions
FM and ETH conceived and designed the study. MHN collected the samples. FM, HJH, LSK, HYJ, KSH, WSP, SHH, SHN, and TT conducted the laboratory work, data handling and analysis and reviewed the manuscript. FM and MAM performed the genetic analysis. All the authors contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to the staff of the Department of Medical Research for serum collection and the members of the Proteomics Laboratory at the Department of Cellular and Molecular Biology, Kangwon National University, Chunchon, ROK. The authors would also like to express their gratitude to the Indonesia Government Fund for Education for their material and non-material support.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets analysed in this study are available from the research team, but were used under license in this study, and as such, are not publicly available; therefore, restrictions may apply. However, data are available from the corresponding authors upon reasonable request and are subject to obtaining permission from the original research team.
Ethics approval and consent to participate
This study was approved by the Ethics Committee, Kangwon National University Hospital, ROK (IRB No. 2014-08-008-002) and the Department of Medical Research, Myanmar (Approval No. 52/ethic/2012). Informed consent was obtained from all the participants.
Funding
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIP) (NRF-2014R1A2A1A11052079), by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2015R1A4A1038666), and the Indonesia Government Fund for Education (LPDP/20140812021475).
Author information
Authors and Affiliations
Corresponding author
Additional information
Fauzi Muh and Jin-Hee Han are joint first authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Muh, F., Han, JH., Nyunt, M.H. et al. Identification of a novel merozoite surface antigen of Plasmodium vivax, PvMSA180. Malar J 16, 133 (2017). https://doi.org/10.1186/s12936-017-1760-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-017-1760-9