Abstract
Background
The two main plasmodial species in French Guiana are Plasmodium vivax and Plasmodium falciparum whose respective prevalence influences the frequency of mixed plasmodial infections. The accuracy of their diagnosis is influenced by the sensitivity of the method used, whereas neither microscopy nor rapid diagnostic tests allow a satisfactory evaluation of mixed plasmodial infections.
Methods
In the present study, the frequency of mixed infections in different part of French Guiana was determined using real time PCR, a sensitive and specific technique.
Results
From 400 cases of malaria initially diagnosed by microscopy, real time PCR showed that 10.75 % of the cases were mixed infections. Their prevalence varied considerably between geographical areas. The presence, in equivalent proportions, of the two plasmodial species in eastern French Guiana was associated with a much higher prevalence of mixed plasmodial infections than in western French Guiana, where the majority of the population was Duffy negative and thus resistant to vivax malaria.
Conclusion
Clinicians must be more vigilant regarding mixed infections in co-endemic P. falciparum/P. vivax areas, in order to deliver optimal care for patients suffering from malaria. This may involve the use of rapid diagnostic tests capable of detecting mixed infections or low density single infections. This is important as French Guiana moves towards malaria elimination.
Similar content being viewed by others
Background
In the early 2000s, French Guiana was one of the most malaria-affected territories among the South American regions [1]. Between 2000 and 2009, the average yearly number of cases was 3920, although, the decline was rapid and marked thereafter to reach only 445 cases in 2014 [2]. The dominant plasmodial species are Plasmodium vivax and Plasmodium falciparum. Plasmodium malariae is much rarer, only representing 1 % of cases [3, 4]. For the past 30 years, malaria transmission nearly no longer affects the coastal area of French Guiana [5], but it persists in the interior regions [6]. In these areas, where only 15 % of the 230,000 inhabitants of French Guiana live, infections were mainly observed among populations living along the Maroni River in the western part of French Guiana bordering Suriname, and the Oyapock in the eastern part bordering Brazil. Nowadays, infections are mainly related to mining activities [7].
The presence of P. falciparum was more frequent in western French Guiana, where the Maroon populations live. This population is resistant to P. vivax because, as most Africans, they do not express the Duffy antigen. Eastern French Guiana was mostly populated by Amerindians and the incidence of P. vivax is similar to that of P. falciparum [8]. During the 2000s, there has been an increase in P. vivax malaria in eastern French Guiana and a decrease of P. falciparum malaria in western French Guiana, notably along the Maroni River [3]. Overall, the proportion of P. falciparum in French Guiana has thus decreased from 46 % of all malaria cases in 2005 to 30 % in 2014 [2, 9].
The specific treatment used in French Guiana depends on the infecting species, the severity of disease and the patient’s condition. Chloroquine is administered in uncomplicated vivax malaria, in association with primaquine. For P. falciparum, artemether and lumefantrine (Riamet®) is employed since 2007. Thus, the identification of the infecting plasmodial species is essential to select the appropriate treatment.
Although errors in species identification are rare, it is common to miss mixed species infections by the microscopic examination of blood smears, particularly when one of the species is predominant in the patient’s blood, which is a frequent situation [10, 11]. In addition, most rapid diagnostic tests, and notably the one used in French Guiana (SD Bioline® Pf/Pan), cannot distinguish between single P. falciparum infections and a mixed P. falciparum/P. vivax infections. This may lead to inadequate treatment, since misdiagnosis of a P. falciparum/P. vivax mixed infection as a P. falciparum infection may lead to failure to administer primaquine and hence will lead to vivax relapses, while misdiagnosis of P. falciparum/P. vivax as P. vivax infection may lead to the use of chloroquine for resistant, potentially severe, P. falciparum infections. Therefore, it is important for clinicians to have local data on the frequency of mixed infections. This problem concerns areas with resistant P. falciparum and/or P. vivax with frequent relapses, which was the case in French Guiana [12, 13]. It is thus important to use more sensitive and discriminant techniques such as PCR [14–19].
In the present study, the frequency of mixed P. falciparum/P. vivax infections in eastern and western French Guiana were thus determined using real-time PCR and compared to microscopic results.
Methods
Samples
Between 2000 and 2008, malaria diagnoses in the health centres and at the Hospital were performed using thin and thick blood smears, and were then confirmed by an experienced microscopist at the Department of Parasitology and Mycology at the Hospital. Given the low prevalence of mixed infections using microscopy, PCR diagnosis was implemented in order to estimate the prevalence of mixed infections.
The study included samples from patients with clinical malaria having consulted remote health centres or Cayenne Hospital between 2000 and 2008. Samples were collected for diagnostic purposes on filter paper (Whatman®) or in EDTA vacutainers, through the Cayenne Hospital or the National Reference Centre for Malaria, respectively. When filter paper was available and when the physician on site was informed (fast rotation of professionals in these remote centers, with newcomers not aware of all ongoing protocols) the filter paper was sent to the Hospital where PCR was performed. Thus, whether a patient eventually got PCR, it was not linked to the particulars of the patient or the malaria episode leading to the consultation, but to the health care professional rotation. Thus, this was not likely to be a recruitment bias.
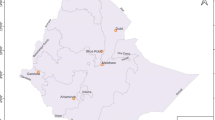
Among the 400 samples collected (representing 10 % of French Guiana cases), 331 came from health centres located in the eastern part of French Guiana: Cacao, Saint-Georges-de-l’Oyapock, Régina, Camopi, Trois-Sauts, and Cayenne Hospital (Fig. 1). In the latter, samples came from patients living in Cayenne or in other towns of central or eastern French Guiana.
The 69 other samples came from health centres located in the western part of French Guiana: Antecume Pata, Maripasoula, Papaïchton, Grand-Santi, and Apatou (Fig. 1). All samples were anonymized before transmission.
DNA extraction
The DNA from the samples conserved on filter paper or at −20 °C were extracted using the DNeasy® Blood and Tissue Kit or the Qiaamp DNA blood kit (Qiagen, Crawley, UK), as recommended by the manufacturer.
Real-time PCR
DNA was amplified using real time PCR as described by Veron et al. [20]. The amplified region corresponds to the small 18S RNA sub-unit. The P. falciparum primer sense sequence was: Pf1 5′-ATTGCTTTTGAGAGGTTTTGTTACTTT-3′, that of the antisense primer was: Pf-2 5′-GCTGTAGTATTCAAACACAATGAACTCAA-3′ and that of the probe was: Pf-probe FAM-CATAACAGACGGGTAGTCAT-MGB. The size of the amplicon was 95 pb. The sequence of the P. vivax sense primer was: Pv-1 5′-CGCTTCTAGCTTAATCCACA TAACTG-3′, that of the antisense primer was: Pv-2 5′-AATTTACTCAAAGTAA CAAGGACTTCCAAG-3′ and that of the probe was: Pv-probe VIC-CGCATTTT GCTATTATGT-MGB. The amplicon size was 142 pb. The amplification and the detection of DNA were performed in duplex, using the PCR Applied Biosystem 7300 analytic system. Real-time PCR was performed in a final volume of 25 µl, in presence of an internal positive control (IPC) (Applied Biosystem, Courtaboeuf, France). Samples were analysed in two tubes, the first one contained P. falciparum and P. vivax primers and probes and the second one the internal control of the absence of inhibitor, the IPC. Each reaction mix contained 12.5 µl of Master Mix Gene Expression (Applied Biosystem), 300 nM of each primer (P. falciparum and P. vivax), 150 nM of each probe (P. falciparum and P. vivax) and 5 µl of matrix DNA. The IPC was used as recommended by the manufacturer. PCR starts with a 10-min phase at 95 °C, followed by 50 cycles of 15 s at 95 °C and 1 min at 60 °C.
Statistical analysis
Proportions of mixed infections were compared between regions using the χ2 test.
Ethical consideration
The retrospective use of anonymous patient files on the site of patient care is authorized by the French National Commission on Informatics and Liberties (CNIL). All the human blood samples and the data collected retrospectively were anonymized in a standardized case report form and in database.
Results
The quantities of the different plasmodia species detected for microscopy and real-time PCR are presented in Table 1. The real-time PCR results (Table 1) showed the presence of 149 P. falciparum and 140 P. vivax infections in the eastern French Guiana and 62 P. falciparum and 6 P. vivax infections in western French Guiana. Forty-three samples, within the 400 collected samples, corresponded to mixed P. falciparum/P. vivax infections, which represented a frequency of 10.75 %, versus 2 % in microscopy. Forty-two originated from Eastern French Guiana and only one came from the western part. The latter was an isolate from Maripasoula, a municipality which marks the end of the Maroon territory and the beginning of Amerindian territory. Thus, in eastern French Guiana the proportion of mixed infections was 12.7 % whereas in western French Guiana the prevalence was only 1.4 %. This difference was statistically significant (P < 0.0001). The municipalities of Saint-Georges-de-l’Oyapock (n = 11), Camopi (n = 9), Trois-Sauts (n = 10), and Cayenne (n = 10) were the sites with the highest number of mixed infections observed (Fig. 1).
Mixed infections misdiagnosed by microscopy were only observed in eastern French Guiana. In 15 cases the association was diagnosed as P. vivax and in 20 cases, as P. falciparum (Table 1).
Discussion
Overall, mixed plasmodial infections were frequent in French Guiana with 10.75 % of malaria cases having mixed P. vivax/P. falciparum malaria. This overall figure however, masks a very heterogeneous situation between eastern, where most mixed infections came from, and western French Guiana, where there was only 1.4 % mixed infection. The incidence of P. vivax malaria is very low among Maroon populations, who are Duffy negative and the main ethnic group living on the Maroni River. This ethnic particularity could explain the low prevalence of mixed infections in western French Guiana [21, 22].
Other authors have observed that over a quarter of P. falciparum infections were in fact mixed infections [23]. Studies conducted in different endemic areas also had different designs often involving cross-sectional studies of exposed populations, and not microscopically confirmed malaria patients as in this study. Molecular studies from Brazil showed that P. falciparum mixed species were detected in 30 % [24], 23.4 % [25] and 10 % [26] in Rondônia for the first two, and Apiacas, respectively. The proportion of mixed infections was lower in Brazil than in some studies from Thailand (24.2–51.6 %) [27] and Papua New Guinea (65.3 %) [17], and similar in Laos (23.1 %) [28].
Mixed infections have been associated with less severe malaria by some [29, 30] and with severe malaria [31] or higher fever [32] by other authors. Apart from the immunologic and pathophysiologic consequences of mixed infections, their misdiagnosis could lead to treatment that is not effective against the hidden species. Thus, missing a hidden P. falciparum infection leads to treatment with chloroquine with potential risks for the patient owing to the 25 % of chloroquine-resistant parasites circulating in the region [33]. Conversely, when P. vivax is hidden, although artemisinin-based combination therapy (ACT) will kill P. vivax, treatment of latent hypnozoites with primaquine will be omitted thus leading to the risk of P. vivax relapses, notably as P. falciparum malaria re-activates latent hypnozoites [13, 34, 35]. As the French and Brazilian Ministers of health announced in July 2015, malaria elimination was a common goal, the capacity to diagnose low density infections is capital [36]. However, when comparing the cases of mixed infections in 2006 and 2007, there was a significant decrease, respectively 18.7 and 9.8 % (p = 0.03), presumably following the overall incidence decrease.
PCR is much more sensitive than microscopy and rapid diagnostic tests, notably to detect mixed infections [14–19]. However, the high costs of this technique are still an obstacle for its use in remote health centres of French Guiana. Nevertheless, depending on the microscopist experience, TDR could be a more efficient technique than microscopy [37] and could be applied to remote areas, provided it is sufficiently sensitive to detect the two Plasmodium species at low parasite densities.
Conclusion
Microscopy often fails to reveal mixed infections or low density single infections. In French Guiana, there is a singular situation with inhabitants of different ethnic origins and therefore malaria susceptibilities, and with differences in local epidemiology, which should be known by clinicians who in routine care do not have access to real-time PCR and thus have a non-negligible risk of overlooking mixed infections. The detection or the anticipation of mixed P. vivax/P. falciparum infections is of clinical importance because interactions between the different species simultaneously infecting the same patient could result in significant changes in the course of the infection and disease, and thus affect therapeutic strategies. The development of rapid diagnostic tests for the detection of mixed infections or the systematic and simultaneous use of two rapid diagnostic tests allowing: (1) the detection of low densities of P. vivax, and, (2) the detection of low densities of P. falciparum, to improve mixed infections diagnosis and treatment, and further decrease malaria as French Guiana moves towards malaria elimination.
References
Carme B, Venturin C. Malaria in the Americas (in French). Med Trop (Mars). 1999;59:298–302.
Ardillon V, Carvalho L, Prince C, Abboud P, Djossou F. Bilans 2013 et 2014 de la situation épidémiologique du paludisme en Guyane (in French). BVS. 2015;1:16–20.
Carme B, Ardillon V, Girod R, Grenier C, Joubert M, Djossou F, et al. Update on the epidemiology of malaria in French Guiana (in French). Med Trop (Mars). 2009;69:19–25.
WHO/PAHO. Malaria in the Americas (in French). Epidemiol Bull. 1997;18:8.
Esterre P, Cordoliani G, Germanetto P, Robin Y. Epidemiology of malaria in French Guiana (in French). Bull Soc Pathol Exot. 1990;83:193–205.
Lepelletier L, Gay F, Nadire-Galliot M, Poman JP, Bellony S, Claustre J, et al. Malaria in Guiana. I. General status of the endemic (in French). Bull Soc Pathol Exot. 1989;82:385–92.
Musset L, Pelleau S, Girod R, Ardillon V, Carvalho L, Dusfour I, et al. Malaria on the Guiana Shield: a review of the situation in French Guiana. Mem Inst Oswaldo Cruz. 2014;109:525–33.
Carme B. Substantial increase of malaria in inland areas of eastern French Guiana. Trop Med Int Health. 2005;10:154–9.
Ardillon V, Eltges F, Chocho A, Chantilly S, Carvalho L, Flamand C, et al. Evolution de la situation épidémiologique du paludisme en Guyane de 2005 à 2011 (in French). BVS. 2012;1–2:5–11.
Boyd MF, Kitchen SF. Simultaneous inoculation with Plasmodium vivax and Plasmodium falciparum. Am J Trop Med. 1937;17:855–61.
Mayne B, Young MD. Antagonism between species of malaria parasites in induced mixed infections. Public Health Reports (1896–1970). 1938;53:1289–91.
Esterre P, Volney B, Meynard JB, Legrand E. Importance of a regional observatory of malarial chemoresistance, an emerging public health problem in the Guyanas region (in French). Bull Soc Pathol Exot. 2009;102:179–84.
Hanf M, Stephani A, Basurko C, Nacher M, Carme B. Determination of the Plasmodium vivax relapse pattern in Camopi. French Guiana. Malar J. 2009;8:278.
Gupta B, Gupta P, Sharma A, Singh V, Dash AP, Das A. High proportion of mixed-species Plasmodium infections in India revealed by PCR diagnostic assay. Trop Med Int Health. 2010;15:819–24.
Ebrahimzadeh A, Fouladi B, Fazaeli A. High rate of detection of mixed infections of Plasmodium vivax and Plasmodium falciparum in South-East of Iran, using nested PCR. Parasitol Int. 2007;56:61–4.
Mayxay M, Pukrittayakamee S, Newton PN, White NJ. Mixed-species malaria infections in humans. Trends Parasitol. 2004;20:233–40.
Mehlotra RK, Lorry K, Kastens W, Miller SM, Alpers MP, Bockarie M, et al. Random distribution of mixed species malaria infections in Papua New Guinea. Am J Trop Med Hyg. 2000;62:225–31.
Zakeri S, Najafabadi ST, Zare A, Djadid ND. Detection of malaria parasites by nested PCR in south-eastern, Iran: evidence of highly mixed infections in Chahbahar district. Malar J. 2002;1:2.
Zakeri S, Kakar Q, Ghasemi F, Raeisi A, Butt W, Safi N, et al. Detection of mixed Plasmodium falciparum and P. vivax infections by nested-PCR in Pakistan, Iran and Afghanistan. Indian J Med Res. 2010;132:31–5.
Veron V, Simon S, Carme B. Multiplex real-time PCR detection of P. falciparum, P. vivax and P. malariae in human blood samples. Exp Parasitol. 2009;121:346–51.
Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–4.
Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–3.
Lorenzetti A, Fornazari PA, Bonini-Domingos AC, de Souza Rodrigues Penhalbel R, Fugikaha É, Bonini-Domingos CR, et al. Mixed Plasmodium falciparum infections and its clinical implications in four areas of the Brazilian Amazon region. Acta Trop. 2008;107:8–12.
Cavasini MT, Ribeiro WL, Kawamoto F, Ferreira MU. How prevalent is Plasmodium malariae in Rondonia, western Brazilian Amazon? Rev Soc Bras Med Trop. 2000;33:489–92.
Alves FP, Durlacher RR, Menezes MJ, Krieger H, Silva LHP, Camargo EP. High prevalence of asymptomatic Plasmodium vivax and Plasmodium falciparum infections in native Amazonian populations. Am J Trop Med Hyg. 2002;66:641–8.
Scopel KK, Fontes CJ, Nunes AC, Horta MF, Braga EM. Low sensitivity of nested PCR using Plasmodium DNA extracted from stained thick blood smears: an epidemiological retrospective study among subjects with low parasitaemia in an endemic area of the Brazilian Amazon region. Malar J. 2004;3:8.
Zhou M, Liu Q, Wongsrichanalai C, Suwonkerd W, Panart K, Prajakwong S, et al. High prevalence of Plasmodium malariae and Plasmodium ovale in malaria patients along the Thai-Myanmar border, as revealed by acridine orange staining and PCR-based diagnoses. Trop Med Int Health. 1998;3:304–12.
Toma H, Kobayashi J, Vannachone B, Arakawa T, Sato Y, Nambanya S, et al. A field study on malaria prevalence in southeastern Laos by polymerase chain reaction assay. Am J Trop Med Hyg. 2001;64:257–61.
Luxemburger C, Ricci F, Nosten F, Raimond D, Bathet S, White NJ. The epidemiology of severe malaria in an area of low transmission in Thailand. Trans R Soc Trop Med Hyg. 1997;91:256–62.
Maitland K, Williams TN, Peto TEA, Day KP, Clegg JB, Weatherall DJ, et al. Absence of malaria-specific mortality in children in an area of hyperendemic malaria. Trans R Soc Trop Med Hyg. 1997;91:562–6.
Genton B, D’Acremont V, Rare L, Baea K, Reeder JC, Alpers MP, et al. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med. 2008;5:e127.
McKenzie FE, Smith DL, O’Meara WP, Forney JR, Magill AJ, Permpanich B, et al. Fever in patients with mixed-species malaria. CID. 2006;42:1713–8.
Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA. 2015;112:11672–7.
Nacher M, Stefani A, Basurko C, Lemonnier D, Djossou F, Demar M, et al. The burden of Plasmodium vivax relapses in an Amerindian village in French Guiana. Malaria J. 2013;12:367.
White NJ. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J. 2011;10:297.
Intervention de Marisol Touraine, Ministre des Affaires sociales, de la Santé et des Droits des femmes aux Assises franco-brésiliennes de la santé (in French). http://www.social-sante.gouv.fr/actualite-presse,42/discours,2333/intervention-de-marisol-touraine,17978.html.
Ehtesham R, Heidari A, Raeisi A, Fazaeli A, Keshavarz H. Detection of mixed-species infections of Plasmodium falciparum and Plasmodium vivax by nested PCR and rapid diagnostic tests in southeastern Iran. Am J Trop Med Hyg. 2015;93:181–5.
Authors’ contributions
MG and VV processed to sample collect, carried out the biologic molecular experiments, participated in the design and coordination of this study, and draft of the manuscript. LM carried out the biologic molecular experiments and participated in the improvement of the manuscript. EL processed to sample collect and the improvement of the manuscript. MD, FD and PB processed to sample preparation. AS and MN participated in the design of the study and performed the statistical analysis. GP participated in the draft of the study. BC conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank all the personnel of health centres and Cayenne hospital (physicians, nurses, logisticians) that have allowed the achievement of the sample collection. We thank Stéphane Simon for his manuscript revision suggestions.
Competing interests
Conflicts that the editors consider relevant to the content of the manuscript have been disclosed. The authors declare that they have no competing interests.
Financial support
This work was supported by the University of the French West Indies and French Guiana, the Ministère Français de l’Enseignement Supérieur et de la Recherche Scientifique, the Institut de Veille Sanitaire. It has benefited from an Investissement d’Avenir grant managed by Agence Nationale de la Recherche (CEBA, reference no. ANR-10-LABX-25-01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ginouves, M., Veron, V., Musset, L. et al. Frequency and distribution of mixed Plasmodium falciparum-vivax infections in French Guiana between 2000 and 2008. Malar J 14, 446 (2015). https://doi.org/10.1186/s12936-015-0971-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-015-0971-1